Abstract
Detecting early plant diseases autonomously poses a significant challenge for self-navigating robots and automated systems utilizing Artificial Intelligence (AI) imaging. For instance, Botrytis cinerea, also known as gray mold disease, is a major threat to agriculture, particularly impacting significant crops in the Cucurbitaceae and Solanaceae families, making early and accurate detection essential for effective disease management. This study focuses on the improvement of deep learning (DL) segmentation models capable of early detecting B. cinerea on Cucurbitaceae crops utilizing Vision Transformer (ViT) encoders, which have shown promising segmentation performance, in systemic use with the Cut-and-Paste method that further improves accuracy and efficiency addressing dataset imbalance. Furthermore, to enhance the robustness of AI models for early detection in real-world settings, an advanced imagery dataset was employed. The dataset consists of healthy and artificially inoculated cucumber plants with B. cinerea and captures the disease progression through multi-spectral imaging over the course of days, depicting the full spectrum of symptoms of the infection, ranging from early, non-visible stages to advanced disease manifestations. Research findings, based on a three-class system, identify the combination of U-Net++ with MobileViTV2-125 as the best-performing model. This model achieved a mean Dice Similarity Coefficient (mDSC) of 0.792, a mean Intersection over Union (mIoU) of 0.816, and a recall rate of 0.885, with a high accuracy of 92%. Analyzing the detection capabilities during the initial days post-inoculation demonstrates the ability to identify invisible B. cinerea infections as early as day 2 and increasing up to day 6, reaching an IoU of 67.1%. This study assesses various infection stages, distinguishing them from abiotic stress responses or physiological deterioration, which is crucial for accurate disease management as it separates pathogenic from non-pathogenic stress factors. The findings of this study indicate a significant advancement in agricultural disease monitoring and control, with the potential for adoption in on-site digital systems (robots, mobile apps, etc.) operating in real settings, showcasing the effectiveness of ViT-based DL segmentation models for prompt and precise botrytis detection.
1. Introduction
Botrytis cinerea, commonly known as gray mold, is a fungal pathogen that significantly impacts a wide range of agricultural crops. It is widely recognized that this pathogen can infect a variety of plant tissues, leading to substantial yield losses [1]. This pathogen affects nearly all vegetable and fruit crops, resulting in annual losses estimated at between USD 10 billion and 100 billion worldwide [2]. The disease is particularly challenging due to its rapid spread under favorable conditions, such as high humidity and moderate temperatures [3,4]. Botrytis cinerea can infect crops both in the field and during post-harvest storage [5], making it a persistent threat throughout the agricultural supply chain. Moreover, the pathogen’s capacity to develop resistance to commonly used fungicides has aggravated the challenge [6], necessitating the exploration of alternative detection and management strategies. Early and accurate detection of B. cinerea is essential to implement robust management strategies, reduce crop damage, and decrease dependence on chemical interventions [6,7]. The traditional methods for detecting plant pathogens like B. cinerea have primarily relied on visual inspection and laboratory-based diagnostic tests [8]. However, these approaches are often slow, require significant manual effort, and are prone to human error. The advent of image-based detection techniques, leveraging the power of multi-spectral imaging and Deep Learning (DL), has revolutionized the approach to plant disease management [9].
DL has significantly advanced the domain of plant pathology, facilitating more sophisticated and accurate disease detection methods [10,11]. Utilizing complex algorithms, DL systems can process and analyze vast amounts of image data, identifying patterns and anomalies indicative of plant diseases [12]. This technology has proven particularly effective in diagnosing diseases from digital images [13], a method that offers several advantages over traditional visual inspection. DL models have shown significant potential in improving diagnostic accuracy and adapting to various tasks, such as object detection and semantic segmentation, through their ability to automatically learn features from given datasets and handle transformations [14]. This adaptability is crucial when dealing with the diverse and evolving nature of plant pathogens. Furthermore, DL applications extend to predicting disease spread and severity, providing valuable insights for crop management and decision-making processes [15]. The integration of DL in plant pathology not only enhances disease detection but also improves the effectiveness and longevity of agricultural practices [16].
Vision Transformers (ViTs) have introduced a novel approach to image analysis, marking a significant shift from traditional image processing methods [17]. Unlike previous techniques that rely heavily on local feature extraction, ViTs approach image analysis by dividing images into a series of patches and applying self-attention mechanisms [18]. This allows them to capture and process the complex interrelationships between different parts of an image [19]. In the field of plant pathology, ViTs can offer a more comprehensive analysis of plant health, identifying disease symptoms that might be missed by other conventional methods [20]. Their application in agricultural image analysis is still relatively new, but it holds great promise for improving accuracy and efficiency in identifying diseases, especially within complex and varied datasets [21].
In segmentation, ViTs provide the architecture models, allowing models to not only detect but also comprehend the context of each segment in an image [22]. This ability is crucial to accurately identify different stages of plant diseases, differentiating between healthy and diseased tissue, and offering detailed views of how disease symptoms are distributed across a plant [21]. The incorporation of ViTs into segmentation models represents a significant advancement, providing an insightful and context-sensitive method for image analysis.
Data augmentation is a key aspect of many vision systems and imbalanced datasets. With this aim in mind, a technique called Cut-and-Paste [23] is gaining attention as an effective method to artificially increase datasets by generating synthetic images [24]. The usual application of this technique involves cropping objects from the original images and pasting them to another image, thus creating a whole new image with more objects of the researchers’ interest. The initial implementations of this approach focused on making the models able to distinguish between what is real and what is fake in the image.
The use of multi-spectral imaging in modern agriculture is crucial, offering a non-invasive and efficient means to oversee crop vitality and identify initial indicators of disease [25]. This technology captures images at various wavelengths, including beyond the visible spectrum, allowing for the detection of physiological changes in plants that precede visible symptoms [26]. Multi-spectral imaging can identify subtle variations in plant reflectance, which are often indicative of stress or disease [27]. This capability is valuable in early disease detection, where timely intervention can prevent widespread crop damage. Multi-spectral imaging provides multi-dimensional data that are suited for DL model training and optimization, enhancing their accuracy and effectiveness in disease detection. As such, multi-spectral imaging represents a critical tool in the transition towards precision agriculture, enabling more targeted and sustainable crop management strategies [28].
As proven by recent research, fungal plant diseases may now be detected using Machine Learning (ML) and DL techniques [29,30]. Giakoumoglou et al. (2023) [31] artificially inoculated leaves of cucumber plants with B. cinerea and employed multi-spectral imaging along with DL to detect the fungal response, achieving a mean Average Precision (mAP) of 0.88. Bhujel et al. (2022) [32] employed a DL-based semantic segmentation model to detect the artificially inoculated gray mold disease in strawberries. The U-Net model outperformed traditional image processing techniques in the detection and quantification of gray mold, demonstrating an Intersection over Union (IoU) of 0.821. Sanchez et al. (2020) [33] applied ML techniques, including several image processing techniques, segmentation, feature extraction, and classification, in order to detect and classify B. cinerea infection in pomegranates. The findings indicated that pomegranate selection may be carried out with an accuracy of 96%, which can aid in the automated identification and classification of B. cinerea. Ilyas et al. (2021) [34] proposed a DL-based framework incorporating modules to manage receptive field dimensions, salient feature transmission, and computational complexity to identify and classify different classes of strawberry fruits, including diseased ones. Their approach led to a 3% improvement in mean intersection over union relative to other state-of-the-art semantic segmentation models and identified infected fruits with a precision of 92.45%. Sun et al. (2018) [35] utilized multi-spectral imaging to assess early indicators of B. cinerea in strawberries to predict early gray mold infection. Wang et al. (2021) [36] introduced a two-stage approach that employs DeepLabV3+ for segmenting cucumber leaves from intricate backgrounds, followed by U-Net for segmenting the infected leaves. This model achieved a leaf segmentation accuracy of 93.27% and an average disease severity classification accuracy of 92.85%, demonstrating robustness in complex environments. Qasrawi et al. (2021) [37] applied ML models to cluster, identify, and classify diseases of tomato plants, including, among others, B. cinerea, using smartphone images of five diseases, achieving a clustering accuracy of 70% and prediction accuracies of 70.3% and 68.9% with neural network and logistic regression models, respectively.
In our previous work, Giakoumoglou et al. (2024) [38], we demonstrated initial efforts in developing segmentation models for early detection of B. cinerea using ResNet encoders, which resulted in IoU values of 67.1%, 39.3%, and 53.4% for healthy, B. cinerea-invisible and B. cinerea-visible classes, respectively. However, the current study goes beyond these initial efforts by leveraging ViT encoders, which have shown superior performance over the ResNet family encoders, while also advancing AI modeling perspectives. Our current research involves a comprehensive re-annotation from scratch, incorporating both a 6-class and a 3-class labeling system. This re-annotation refines our earlier approach, where model predictions extended to unannotated leaves, leading to misinterpretations and reduced performance. Moreover, we have introduced and evaluated the performance of two new architectures in combination with ViTs. The primary objective is to detect fungal infection, prior to symptom development, enabling timely intervention to reduce pesticide usage, along with the effort to address dataset imbalance by utilizing the Cut-and-Paste augmentation technique. This customized augmentation technique, detailed in Section 2.6, was specifically designed to augment numerically weaker classes and enhance overall results, leading to significant improvements in the IoU values for all classes. More specifically, this augmentation method led to a 14.29% increase for healthy class, a 56.45% increase for the identification of leaves with no visible botrytis infection, and a 23.27% increase for the detection of leaves with obvious fungal symptoms. The key motivations of this study are: (i) early diagnosis of B. cinerea disease; (ii) address dataset imbalance; (iii) early treatment of affected plants; (iv) reducing pesticide use as a consequence of early diagnosis and treatment; (v) employing optimized AI models to identify symptoms in plants under real conditions.
To achieve these goals, an approach was developed for the effective detection of B. cinerea, focusing on identifying the fungus at various stages of its development, especially during the initial phases. The experiment involved growing cucumber plants under controlled environmental conditions and introducing the pathogen using two distinct inoculation methods. The study captured the evolution of the disease through multi-spectral imaging, which efficiently recorded both the early, invisible symptoms and the later, more advanced stages of the infection. The study utilized linear disease progression models to analyze the disease in individual leaves. The segmentation models were evaluated using a three-class labeling system. Four advanced DL segmentation architectures, namely U-Net++, PAN, MA-Net, and DeepLabV3+, were evaluated in combination with two Vision Transformer (ViT) encoders. To address dataset imbalance and improve the identification and segmentation of all classes, the Cut-and-Paste augmentation technique was employed. This method enhances image content density by increasing the presence of underrepresented classes. By copying objects from one image and pasting them onto another, new images are created, effectively multiplying the training dataset and augmenting the overall augmentation process for better model performance. The results of this study underscore the models’ strength in detecting B. cinerea at various infection stages, showcasing their potential as effective tools for early and accurate disease management in farming practices. This approach has the potential to be implemented in automated systems and self-navigating robots, advancing agricultural disease monitoring and control. The potential integration into on-site digital systems (such as robots and mobile apps) working in real-world environments further showcases the value of ViT-based DL segmentation models for real-time, accurate detection of botrytis disease.
The rest of the paper is structured as follows. Section 2 delineates the materials and methods employed in this study, including plant and fungal material, the DL segmentation models, the Cut-and-Paste augmentation technique, and the assessment metrics used to detect B. cinerea in cucumber plants. Section 3 exhibits the results. The study concludes in Section 4.
2. Materials and Methods
This section describes the methodology of this study, encompassing plant preparation and artificial inoculation, dataset creation, and the development of DL models. Figure 1 provides a comprehensive overview of the proposed methodology employed to develop effective semantic segmentation models for early detection of B. cinerea. Initially, the multi-spectral images along with their masks undergo the Cut-and-Paste augmentation technique to enhance the representation of key classes. These augmented images are then used as input for the DL models, which generate the final segmentation outputs.
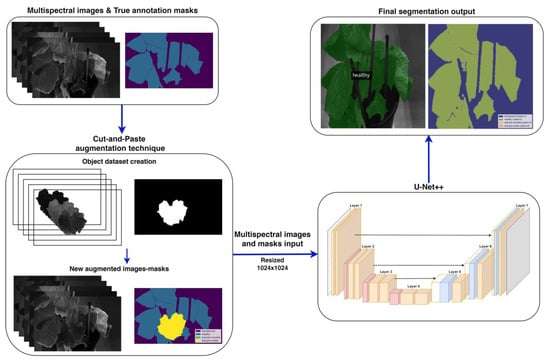
Figure 1.
Flowchart illustrating the proposed methodology. It begins with multi-spectral images paired with true annotation masks sourced from the advanced imagery dataset. The images then undergo Cut-and-Paste augmentation, along with other augmentation techniques as described in Section 2.4. These augmented images and masks are subsequently inputted into an adjusted U-Net++ segmentation model (image source: [39]) with a ViT backbone (Section 2.3), customized to process multi-spectral input, and resized to 1024 × 1024 pixels. The network produces a segmented image as output.
2.1. Experimental Setup and Inoculation Processes
This study utilized the cucumber cultivar Green Baboo (Cucumis sativus L. cv. Hasan). Seedlings were grown in controlled environment chambers with specific conditions (21 ± 1 °C, 16 h photoperiod, 352.81 μmol/sec photosynthetic photon flux, 85–90% humidity) until the first true leaf fully developed. They were then transplanted into sterile compost in plastic pots and maintained under greenhouse-like conditions for about two months. Botrytis cinerea was extracted from cucumber plants naturally infected and displaying characteristic typical gray mold symptoms. The conidial suspension of the fungal isolate was stored at −20 °C in aqueous glycerol and grown on potato dextrose agar (PDA) (Merck, Darmstadt, Germany) at 21 °C before use [40].
Bioassays were performed in two identical controlled environment chambers, one for plants inoculated with B. cinerea and another for mock-inoculated plants. Plants were inoculated with the B. cinerea when they had four fully expanded leaves. For inoculum preparation, B. cinerea was grown on PDA in petri dishes at 21 °C for 10–15 days in darkness. Spore suspensions were made using sterile distilled water (SDW) with 2% sucrose and 0.01% Tween® 20 (Sigma-Aldrich, St. Louis, MO, USA) [41]. The conidial concentration was set to 105 conidia mL−1. Two inoculation techniques were employed: (i) mycelial plugs (5 mm in diameter) were cut from the edges of 10-day-old colonies of the B. cinerea isolate and placed on the adaxial side of the first true leaf near to the base, termed inoculation method “A”, and (ii) the adaxial surfaces of the first and second true leaves were sprayed with the conidial suspension until wet using a low-pressure hand sprayer (about 500 μL per leaf) [42], termed inoculation method “B”. Mock inoculation followed the same process but without the pathogen. The study included 20 plants for B. cinerea inoculation (ten each for both methods) and nine for mock inoculation. After the artificial inoculation, the plants were placed in darkness for a day, while maintaining other environmental conditions.
Disease severity was assessed over 37 days post-inoculation (dpi), quantifying the percentage of leaf area showing visible disease symptoms. The data were analyzed using a logit transformation to linearize disease progression in relation to time [43]. Linear regression was employed to calculate the rate of disease progression and to estimate the infection onset time on each leaf [44].
2.2. Multi-Spectral Imaging and Annotation
For multi-spectral imaging, a customized Qcell (https://qcell.tech (accessed on 15 April 2024)) Phenocheck camera was employed, capturing images at wavelengths of 460, 540, 640, 775, and 875 nm. The camera’s resolution was 3096 × 2080 pixels. Imaging began on the day of B. cinerea inoculation and continued daily for the next five days, followed by weekly captures until 37 dpi. This approach allowed for a comprehensive collection of the disease’s progression. In total, the dataset comprised 1061 images. This included 418 images from plants inoculated with B. cinerea and 355 images from mock-inoculated plants, both sets using mycelial plugs. Additionally, there were 210 images from plants inoculated with B. cinerea and 78 from mock-inoculated plants, both inoculated using the spraying technique. Figure 2 illustrates the spectra captured at various wavelengths.
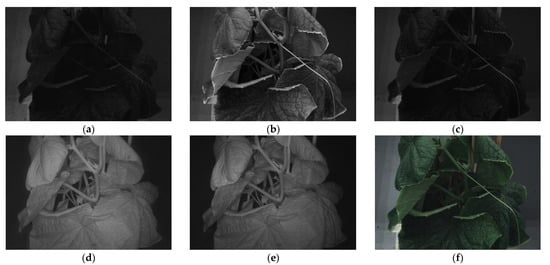
Figure 2.
Spectral images at different wavelengths: (a) 460 nm, (b) 540 nm, (c) 640 nm, (d) 775 nm, and (e) 875 nm, accompanied by (f) the RGB image.
Each leaf in the dataset was annotated, using Roboflow [45] by marking its area with a polygon and then assigning a class label to it based on the disease’s progress. This annotation process involved classifying the leaves into different stages, depending on the percentage of leaf area showing visible symptoms of B. cinerea infection. Leaves from mock-inoculated plants received the label “control” if they had mock inoculation, while leaves without any inoculation were categorized as “healthy”. For B. cinerea-inoculated plants, labels were assigned based on the degree of visible symptoms observed on the leaves. Two different annotation systems were generated for this dataset, one for six infection stages and another for three infection stages. For the six-class system, the categories were defined as follows: leaves with 0.1–2% symptoms were labeled as invisible-early, those with 2–5% as invisible-late, denoting hardly visible by naked eye symptoms, leaves with 5–10% symptoms as visible-light, 10–20% as visible-moderate, and leaves with 20–100% symptoms as visible-heavy. For the simplified three-class system, leaves with 0.1–5% symptoms were categorized as “invisible” indicating symptoms that are not easily seen, while those with 5–100% affected leaf area were labeled as “visible”. In the initial annotation phase, leaves were categorized based on clear symptoms, ensuring precise classification into their respective categories. However, as the model predictions extended to unannotated leaves, the annotation process was expanded to encompass all leaves. Consequently, for both class systems, all leaves were annotated, including those showing minimal (0–0.1%) or no symptoms, which were assigned under the healthy class, similar to the leaves from mock-inoculated plants. This study focuses on creating a segmentation model for the three-class system, while the six-class system is used for a better and more comprehensive visualization of disease progression.
Of the total 1061 images, 773 resulted from using the inoculation method A, while 288 were from method B. Artificial inoculated plants were numbered 1 to 20, whereas mock-inoculated plants were numbered 1 to 9. Specifically, plants inoculated with B. cinerea and designated for training included those numbered 1, 2, 3, 4, 5, 6, 7, 9, 11, 12, 14, 15, 17, 19, and 20, while validation samples consisted of those numbered 8, 10, 13, 16, and 18. For control plants, training samples included those numbered 4, 5, 6, 8, 9, 10, 11, 12, 13, 14, 15, 16, 17, 19, 20, 21, 22, 23, 24, 26, 27, 28, and 29, while validation samples comprised those numbered 1, 2, 3, 7, and 18. Following the annotation process, the dataset was divided into training and validation sets using a 78–22% split. Comprehensive statistics on the annotated class distribution for both the training and validation datasets for the six-class and the three-class categories are shown in Table 1 and Table 2, respectively.

Table 1.
Dataset’s class distribution statistics for the six-class system.

Table 2.
Dataset’s class distribution statistics for the three-class system.
Utilizing the six-class system to offer a more thorough visualization, Figure 3 and Figure 4 collectively provide a detailed visual narrative of B. cinerea infection dynamics on a specific plant. Figure 3 captures the entire temporal span of the experiment, displaying the progression of infection at various dpi timepoints. This visualization enables a chronological examination of the plant’s response to B. cinerea, showcasing the transition from the initial stages of invisibility to the development of visible symptoms.

Figure 3.
Evolution of B. cinerea captured at different dpi for a particular plant during the experiment presented in varied colors on top of the 460 nm wavelength grayscale image adjusted to 1024 × 1024 and overlaid with the ground truth masks for the six-class label system: (a) dpi 0, (b) dpi 2, (c) dpi 13, (d) dpi 20, (e) dpi 30, and (f) dpi 33. Leaves in healthy condition are indicated in green, “invisible-early” stage in yellow, “invisible-late” stage in orange, “visible-light” infection in red, “visible-moderate” infection in a deeper red, and “visible-heavy” infection in the deepest red. This figure shows plant number 2, which was infected with inoculation method B (B_Bc_2).
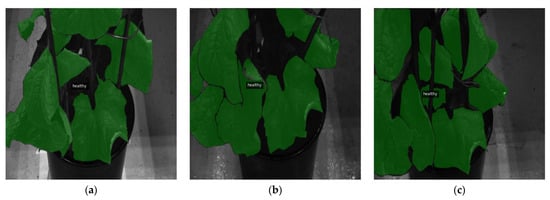
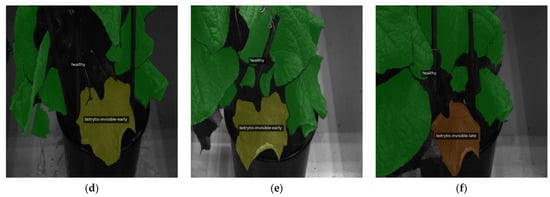
Figure 4.
Evolution of B. cinerea captured in the early dpi for a particular plant during the experiment presented in varied colors on top of the 460 nm wavelength grayscale image adjusted to 1024 × 1024 and overlaid with the ground truth masks for the six-class label system: (a) dpi 0, (b) dpi 1, (c) dpi 2, (d) dpi 3, (e) dpi 5, and (f) dpi 6. Annotations follow the same color-coding scheme as Figure 2. This figure shows plant number 2, which was infected with inoculation method B (B_Bc_2).
In contrast, Figure 4 zooms in on the critical early dpi, presenting a more focused view of the plant’s response. The six subfigures offer a close-up look at the initial stages of infection. This detailed view provides an understanding of the rapid changes in leaf conditions during the first days of the experiment.
2.3. Deep Learning Segmentation Models
For this study, four state-of-the-art DL segmentation architectures were employed: U-Net++ [46], PAN [47], MA-Net [48], and DeepLabV3+ [49]. U-Net++ stands out as an evolved version of the original U-Net [50], tailored for biomedical image segmentation. It introduces an encoder–decoder structure with nested and skip pathway enhancements. Path Aggregation Network (PAN) leverages advanced techniques to capture precise, dense features, enhancing the model’s capacity to catch finer details. Multi-Attention Network (MA-Net) further extends these capabilities by incorporating self-attention mechanisms for adaptive local–global feature integration and segmentation precision. DeepLabV3+ extends the capabilities of DeepLabV3 [51], combining atrous convolutions with an enhanced encoder–decoder architecture.
To boost the feature extraction capabilities of these architectures and conduct an in-depth examination of ViT’s effectiveness, they were paired with diverse encoders, i.e., MobileViT-S [52], MobileViTv2-1.25 [53]. MobileViT-S combines the strengths of CNNs and ViTs, offering an efficient and powerful approach to computer vision tasks. On the other hand, MobileViTv2-1.25 further improves efficiency by making self-attention operations simpler.
The models were adjusted to process multi-spectral images by modifying their input channels to accept five different spectral channels. This allows the models to fully use each spectral band for image segmentation, enhancing the use of multi-spectral data.
2.4. Model Training
The DL segmentation models underwent training on the three-class system. Initially, each backbone received random initialization. U-Net++ and MA-Net utilized a combination of Dice loss [54] and Binary Cross-Entropy (BCE) loss, while PAN and DeepLabV3+ employed Cross-Entropy (CE). Weighted loss functions were applied, with normalized weights derived from the inverse logarithm of class frequencies. The weights were adjusted to a range of 0.1 to 1.0, preventing the logarithm of zero by adding one. Different optimizers were assigned to each architecture: AdamW [55] for U-Net++ and DeepLabV3+, Adam [56] for MA-Net, and SGD for PAN.
U-Net++ was set with a base learning rate of 2 × 10−4, whereas MA-Net, PAN, and DeepLabV3+ were assigned a higher rate of 2 × 10−3. A warm-up phase spanning 5 epochs used a learning rate set to ten times the base rate. Following this, a cosine scheduler gradually decreased the learning rate to one hundredth of the base rate over 100 training epochs. A consistent weight decay of 10−2 was applied to all models. The batch size varied between 2 and 8, based on the computational resources.
During training, various geometric data augmentation techniques were employed for enhancing dataset diversity and equipping the models for different visual contexts [57]. The training images underwent resizing to dimensions of 1024 × 1024 pixels, with an additional step of resizing them to 11/7 times the original size before applying a random crop to the desired dimensions. This resizing strategy, coupled with a random crop, aims to maintain crucial image details while introducing variability in the training data. Following the resizing, each image had a 50% probability of undergoing a horizontal flip and a 5% probability of a vertical flip. The above augmentation techniques were designed for bolstering the resilience to typical geometric alterations and prevent overfitting for the model. Conversely, the validation set was subjected solely to resizing.
2.5. Evaluation Metrics
This research evaluated DL segmentation models using various common metrics. These metrics were selected to highlight different aspects of the model’s accuracy and effectiveness in detecting and segmenting the progression of B. cinerea infection. Pixel accuracy was used as a basic measure to quantify the proportion of correctly classified pixels. The Intersection over Union (IoU) measured the overlap between predicted and actual labels, providing an understanding of the model’s effectiveness in differentiating classes. The model’s sensitivity was measured by recall and its specificity by precision. Additionally, the Dice Similarity Coefficient (DSC) was employed to give a comprehensive view of the model’s segmentation ability, which combines both precision and recall. Similarly, mean Intersection over Union (mIoU) and mean Dice Similarity Coefficient (mDSC) are used when aggregating all classes.
2.6. Cut-and-Paste Augmentation Technique
As evident in Table 2, the class distribution statistics for the three classes are not distributed equally, which is expected since the dataset and its images are based on the progression of the fungal pathogen in cucumber plants. More specifically, the healthy leaves annotated in the dataset far outnumber the corresponding leaves of the classes with invisible and visible B. cinerea symptoms. This makes the dataset imbalanced and complicates the process of correct identification of all classes by the models. The initial training contained only geometric data augmentations, as described in Section 2.4, a method that was not sufficient to give satisfactory results. Therefore, to further expand on the augmentation process and improve the results, a Cut-and-Paste [58,59,60] implementation was utilized. Essentially, Cut-and-Paste is a technique where objects are copied from one image and pasted onto another, forming a new image, thereby virtually multiplying the training dataset. In this study, separate leaves from classes of interest were extracted from original images and pasted dynamically to others during the training process to augment the original dataset. A two-step approach was followed to apply this augmentation technique.
The first step involves the creation of the object dataset. This is a preprocessing step that needs to be completed before the model training can be conducted. To achieve this, the original dataset images are used with their corresponding annotation masks to create a new image and a new mask for each object (leaf) in the images. Each new image contains the object, surrounded by fully transparent pixels for the rest of the image. Similarly, each new mask contains the masking information for that object, while the rest of the mask is set to be background. For this dataset, each leaf annotation was extracted from all the wavelengths used in this study in order to paste them properly later. Figure 5 shows an example of the extraction process of a leaf belonging to the class with visible B. cinerea symptoms. Figure 5a displays the corresponding RGB image of the specific plant, while Figure 5b–f show the extracted leaf in all available multi-spectral wavelengths.
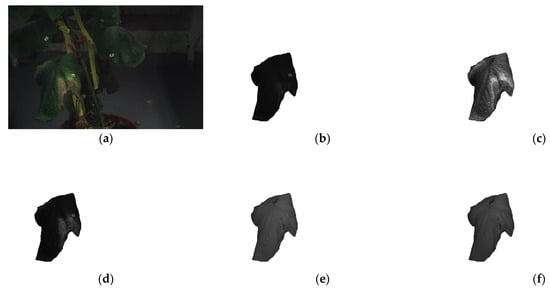
Figure 5.
Leaf extraction process used for the Cut-and-Paste augmentation method: (a) original RGB plant image, before being resized to 1024 × 1024, (b) extracted leaf at 460 nm, (c) extracted leaf at 540 nm, (d) extracted leaf at 640 nm, (e) extracted leaf at 775 nm, and (f) extracted leaf at 875 nm.
The second step occurs during the data loading in training, where the images fed into the model undergo the Cut-and-Paste augmentation. This step determines the number and class of objects (leaves) to paste into the new image. After experimentation and considering the relatively large size of the leaves compared to the image, the distribution of pasted leaves was established: 25% probability to paste one leaf, 40% to paste two leaves, 15% to paste three leaves, and 20% to leave the original image unchanged. To address the issue of an imbalanced dataset, the classes with invisible and visible symptoms of B. cinerea were prioritized with higher weights during the process of pasting new leaves. Initial training without the Cut-and-Paste augmentation confirmed this approach. The underlined line of Table A1 shows the model’s performance before implementing the new augmentation, where the healthy class (with numerous leaf annotations) achieved the highest IoU, while the invisible and visible classes were significantly lower.
Based on these IoU values, the probability of selecting which class to paste into a new image was determined using the following formula:
where denotes the likelihood of a leaf being assigned to class , refers to the IoU result for class from the initial experiment that lacked Cut-and-Paste augmentation, and represents the number of classes. For this study, the IoU values outlined in the underlined line of Table A1 were utilized. The probability of adding a leaf from the healthy class ( = 0.658) was 25.8%, from the invisible class ( = 0.429) was 39.5%, and from the visible class ( = 0.490) was 34.6%. Figure 6 demonstrates the operation of the Cut-and-Paste augmentation technique. Figure 6a,c showcase in different wavelengths the same plant that initially contained only healthy annotated leaves. Figure 6a contains a newly pasted leaf from the visible class, while Figure 6c includes both an invisible class leaf and a visible class leaf. Similarly, Figure 6b,d display the new masks created and provided to the model as input.
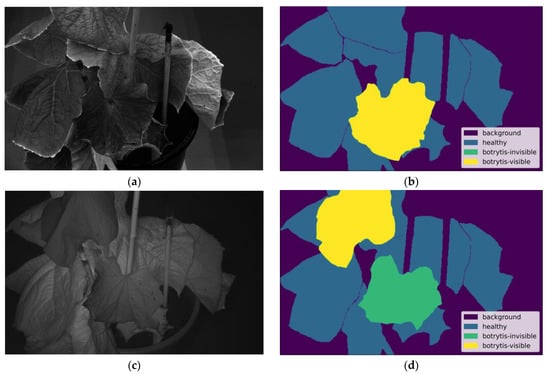
Figure 6.
Demonstration of the Cut-and-Paste technique: (a) augmented image at 540 nm, before being resized to 1024 × 1024, with an added leaf from the botrytis-visible class, (b) corresponding mask of (a) with a class legend to differentiate each class, (c) augmented image at 875 nm, before being resized to 1024 × 1024, with two added leaves, one from the botrytis-invisible class and one from the botrytis-visible class, (d) corresponding mask of (a) with a class legend to differentiate each class.
The Cut-and-Paste augmentation method enabled the newly trained model to reach higher IoU values across all three classes and substantially addressed the imbalanced dataset issue by dynamically creating new images for each training epoch. Specifically, as outlined below in the results section, this technique improved IoU values by 14.29% for the healthy class, 56.45% for the invisible class, and 23.27% for the visible B. cinerea symptoms class.
3. Results and Discussion
3.1. Assessment of Deep Learning Segmentation Models
Evaluating the performance of DL segmentation models for categorizing B. cinerea severity indicates varying degrees of efficacy among the models, with a specific focus on the mDSC and mIoU metrics, as detailed in Table 3. More specifically, Table 3 contains the model performances for the three-class system, while Table 4 presents a comprehensive breakdown of segmentation results for all classes for the best-performing model before and after applying the Cut-and-Paste augmentation technique.

Table 3.
Overall performance of DL segmentation models for three-class system (best-performing model indicated in bold).

Table 4.
Class-specific performance of DL segmentation models for three-class system before and after applying the Cut-and-Paste augmentation technique.
The underlined row of Table 3 shows that the U-Net++ architecture, when paired with the MobileViTV2-1.25, reports the highest mDSC score at the three-class system before applying the Cut-and-Paste augmentation technique. With a parameter count of 17.84 M, this combination achieves 90.6% accuracy, an mDSC of 0.771, an mIoU of 0.750, and a recall rate reaching 0.848 by the 8th epoch. MobileViTV2-1.25 features, with a higher parameter count, achieve an effective balance between complexity and performance, allowing accurate segmentation by capturing critical information. U-Net++ stands out for its U-shaped design and skip connections, which help it capture details of B. cinerea symptoms on cucumber effectively. Other architectures, such as MA-Net, PAN, and DeepLabV3+, despite employing the MobileViTV2-1.25 encoder, exhibit comparatively lower mDSC values, indicating potential challenges in extracting critical features for accurate segmentation. PAN’s introduction of a Feature Pyramid Attention module and a Global Attention Upsample module emphasizes a unique approach to feature extraction, potentially impacting effectiveness in B. cinerea segmentation. DeepLabV3+ is known for using atrous convolutions and spatial pyramid pooling to handle object scale and detail. However, these methods might face challenges when dealing with the complexities of the B. cinerea segmentation task. As for MA-Net, Table A1 in Appendix A reveals that this architecture struggles significantly to detect the key invisible class of B. cinerea with either encoder, as evidenced by its low IoU and recall scores. This may be due to MA-Net’s Multi-scale Attention Net design, which focuses on rich contextual dependencies through self-attention mechanisms, potentially offering a broader perspective in feature representation, but may not align well with the specifics of the invisible class.
Comparing the results of MA-Net and U-Net++ with MobileViT-S versus MobileViTV2-1.25 emphasizes the relationship between architecture and encoder. Using ViT encoders with more parameters resulted in an mDSC score increase of 3.41% for MA-Net and 18.07% for U-Net++. Interestingly, a further increase in parameters for the PAN architecture using the MobileViTV2-1.25 encoder did not yield further improvements, resulting in an 11.02% decrease in mDSC score, demonstrating the complex interaction within this particular architecture–encoder pairing. This underscores the critical need for choosing the appropriate combination of architecture and encoder for optimal segmentation outcomes.
The architecture–encoder combination that achieved the highest mDSC score with only geometric data augmentations was used to develop a refined model utilizing the Cut-and-Paste method. As indicated by the bold line in Table 3, the improved model utilizes U-Net++ architecture with the MobileViTV2-1.25 encoder, reaching almost 92% accuracy, while also increases the mDSC score by 2.72% from 0.771 to 0.792 and mIoU by 8.8% from 0.750 to 0.816, at epoch 89. Table 4 details the class-specific performance of the two DL segmentation models before and after applying the Cut-and-Paste augmentation method. The enhancement is substantial across all three classes, as reflected in both the IoU and recall metrics. The most notable improvement is seen in the invisible class of B. cinerea symptoms, where IoU increases significantly from 42.9% to 67.1%. Similar positive trends are observed in the recall metric for this class, as well as for both metrics in the healthy and visible classes.
The use of the Cut-and-Paste augmentation method significantly improved the segmentation model by effectively expanding the training dataset and addressing the imbalanced dataset that originally existed. By copying separate leaves from classes with higher weights from one image and pasting them onto another, the method creates new images containing more leaves with the classes of interest, thereby virtually multiplying the available training data. This enriched variety in the training dataset enhances the model’s capability to learn various contextual environments and complex scenarios.
Appendix A details comprehensive segmentation results per class for three-class systems, along with IoU and recall metrics. Table A1 presents detailed information for the three-class system, while Figure A1 provides a clear representation of IoU progression across epochs. Figure A1a shows the model’s performance with only geometric data augmentations, while Figure A1b demonstrates the performance boost after incorporating the Cut-and-Paste augmentation technique. These analyses offer a thorough breakdown of the segmentation performance, providing a detailed breakdown of accuracy, recall, and IoU metrics for each disease severity category, thus giving valuable perspectives on the model’s performance across various classes.
3.2. Early-Stage Evaluation
To evaluate the effectiveness of early detection and track the progression of infection in the initial week, the model’s performance was assessed using IoU on validation data from dpi 1 to dpi 6. As shown in Figure 7, the model demonstrated a strong ability to recognize early-stage invisible symptoms of B. cinerea, while also delivering highly encouraging results in detecting visible B. cinerea symptoms very early for both inoculation methods.
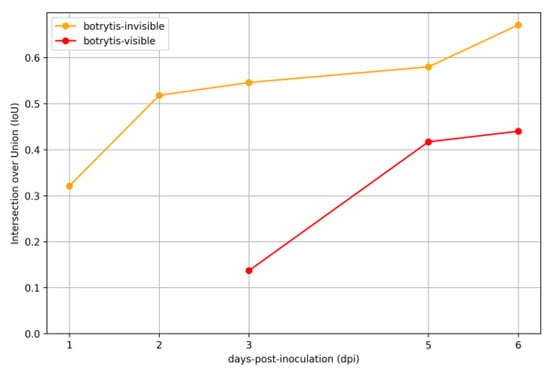
Figure 7.
IoU for B. cinerea invisible and visible classes from dpi 1 to dpi 6 across the two inoculation methods. The model’s IoU scores for the B. cinerea invisible class were 0.321 on dpi 1, 0.518 on dpi 2, 0.546 on day 3, 0.58 on dpi 5, and peaked at 0.671 on dpi 6. For B. cinerea visible symptoms, the IoU values were 0.137 on dpi 3, 0.417 on dpi 5, and 0.44 on dpi 6.
On dpi 1, the model detected the pathogen with an initial IoU of 0.321, indicating a developing sensitivity to its presence. This detection progressively intensified over the following days, with the model reaching an IoU of 0.518 on the 2nd day post-inoculation and 0.546 on the 3rd day post-inoculation. Identification of early signs of infection improved steadily, reaching an IoU of 0.58 on dpi 5. By dpi 6, the model had reached its peak performance, recording a further increase of 15.69% to reach an IoU of 0.671, showcasing the solid proficiency of the model in identifying the pathogen’s initial stages. For detecting visible B. cinerea symptoms, the model’s performance improved significantly over the observation period. On dpi 3, an IoU of 0.137 was recorded for visible symptoms. This initial IoU value was lower due to misclassification during the early stages, when B. cinerea was primarily invisible and visible symptoms were minimal. By dpi 5, the IoU showed a significant increase of 204.38%, rising to 0.417, and then further up to 0.44 on dpi 6. These results demonstrate the model’s growing capability in detecting visible symptoms as the disease progresses.
The ability of the model to accurately recognize both invisible and visible classes from dpi 2 onward, with its accuracy reaching its highest point on dpi 6, reinforces its effectiveness in early-stage detection. This skill is vital since it detects symptoms before they become visible, providing an essential opportunity for prompt intervention and effective management practices in farming.
3.3. Qualitative Results of Disease Severity Levels
Precisely identifying and segmenting disease severity in B. cinerea infections on cucumber is essential for understating plant pathology and implementing effective agricultural practices. The challenge lies in recognizing the progression of the disease, spanning from hidden to obvious symptoms. The current section reviews the performance of different models tested and highlights the top-performing model’s ability to identify and segment various levels of B. cinerea.
Figure 8 provides a visual comparison of each model’s performance, corresponding to the metrics presented in Table 3, using the true mask annotation in Figure 8a as a reference. Figure 8b,c display the predicted segmentation masks from the PAN architecture combined with the MobileViT-S and MobileViTV2-1.25 encoders, respectively, both showing suboptimal segmentation. Similarly, Figure 8d,e illustrate the results from the MA-Net architecture with the MobileViT-S and MobileViTV2-1.25 encoders, respectively, also exhibiting poor segmentation. Figure 8f presents the output of the DeepLabV3+ architecture with the MobileViTV2-1.25 encoder, showing improved performance over the previous models, while Figure 8g shows the U-Net++ and MobileViT-S combination, demonstrating similar segmentation performance. Figure 8h represents the U-Net++ architecture with the MobileViTV2-1.25 encoder prior to applying the Cut-and-Paste augmentation technique. Finally, the best segmentation results are shown in Figure 8i, where the U-Net++ architecture with the MobileViTV2-1.25 encoder, enhanced by the Cut-and-Paste augmentation technique, delivers the most accurate segmentation of all the previous models, consistent with the findings in Table 3.
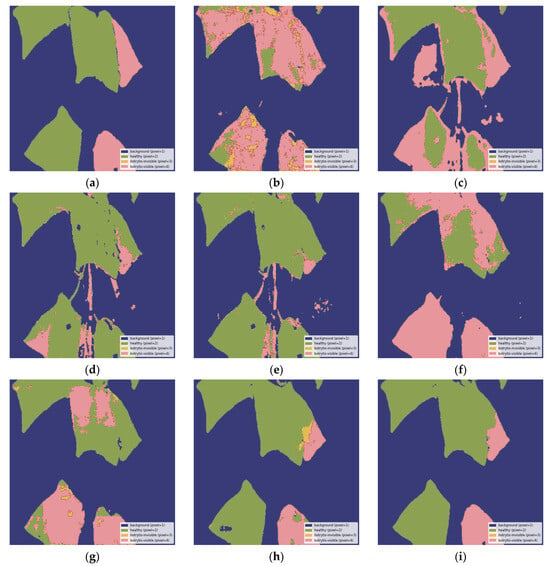
Figure 8.
Visual comparison of each model’s performance: (a) true segmentation mask, (b) segmentation output from the PAN—MobileViT-S model; (c) segmentation output from the PAN—MobileViTV2-1.25 model; (d) segmentation output from the MA-Net—MobileViT-S model; (e) segmentation output from the MA-Net—MobileViT-S model; (f) segmentation output from the DeepLabV3+—MobileViTV2-1.25 model; (g) segmentation output from the U-Net++—MobileViT-S model; (h) segmentation output from the U-Net++—MobileViTV2-1.25 model before applying the Cut-and-Paste augmentation technique; (i) segmentation output from the U-Net++—MobileViTV2-1.25 model after applying the Cut-and-Paste augmentation method. The above images have been resized to 1024 × 1024 and displayed with the standard colormap.
The comprehensive qualitative evaluation, as depicted in Figure 9, reveals both the capabilities and weaknesses of the best-performing model. The combination of U-Net++ and MobileViTV2-1.25 demonstrates notable improvements following the application of the Cut-and-Paste augmentation technique. Figure 9a–c present images with overlaid ground truth masks, while Figure 9d–f exhibit the model-generated output masks. Figure 9d aligns accurately with the ground truth mask in Figure 9a, showcasing the adeptness that the model has in precisely outlining healthy regions and invisible B. cinerea. Figure 9e successfully segments most healthy and visible areas of the leaf with B. cinerea, with minor errors misclassifying some spots of visible symptoms as healthy. Similarly, Figure 9f highlights a discrepancy, indicating misinterpretation of healthy areas as containing visible symptoms.
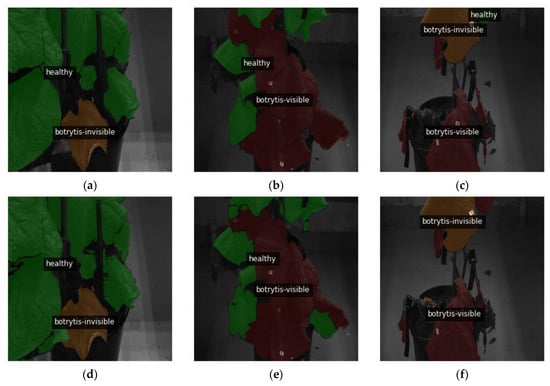
Figure 9.
Qualitative evaluation of segmentation performance: (a–c) showcase the original images, overlaid with ground truth labels, (d–f) display the images overlaid with the output masks generated by the top-performing segmentation model. Specifically, image (a) is the ground truth for the accurate prediction in (d). Image (b) corresponds to the ground truth for the prediction in (e), in which most healthy and visible parts of the leaf affected by B. cinerea are accurately segmented, with a few minor mistakes classifying certain visible symptom spots as healthy. Image (c) represents the ground truth for prediction (f), revealing an inconsistency where the model confuses a healthy leaf with visible B. cinerea symptoms. All of the above images have been resized to 1024 × 1024.
In Figure A4 and Figure A5 (Appendix B), the model encounters challenges in accurately classifying certain symptoms of B. cinerea infection. The misclassification of leaves with visible B. cinerea symptoms as invisible (Figure A4) and the misclassification of parts of healthy leaves as both invisible and visible symptoms (Figure A5) highlight the complexities associated with the diverse symptoms caused by B. cinerea. The difficulties in distinguishing between healthy and symptomatic leaves can be attributed to the intricate nature of the symptoms, especially when the symptomatic areas are relatively minor. Additionally, the wide range of symptoms induced by B. cinerea across various plant organs and tissues, coupled with the variability in symptom manifestation at the early stages of infection, contributes to the inherent difficulty in achieving precise classifications for the model.
3.4. Qualitative Results of Biotic and Abiotic Plant Stress Factors
Abiotic stresses, such as sunburn, often display signs similar to B. cinerea [61]. The visual resemblance presents a major challenge for distinguishing between plant stressors with comparable appearances. This section explores the ability to differentiate between biotic stress factors, specifically B. cinerea infections in inoculated plants, and abiotic stresses such as sunburn and/or aging in healthy control plants. The assessment emphasizes the precise differentiation of responses attributed to B. cinerea infection from those influenced by environmental factors. In a particular instance from the validation dataset, symptoms unrelated to gray mold led to a misclassification, as depicted in Figure 10a–c. Specifically, Figure 10b illustrates a slight misclassification of a region which is erroneously marked as displaying visible symptoms of B. cinerea, a disparity evident when contrasted with its ground truth mask in Figure 10a. However, beyond this specific case, the model accurately segments the leaves. For instance, Figure 10e aligns closely with the ground truth mask in Figure 10d, showcasing the model’s ability to recognize areas with symptoms arising from abiotic disorders.
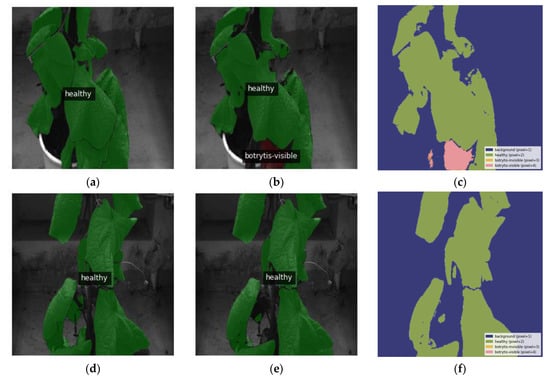
Figure 10.
Qualitative assessment of segmentation performance: (a,d) present the original images overlaid with ground truth labels; (b,e) depict the images overlaid with the output masks; (c,f) show the output segmentation mask by the top-performing model along with the standard colormap. Precisely, image (a) is the ground truth mask of the prediction as seen in (b,c), in which a small region is incorrectly classified as a visible symptom of B. cinerea with an mDSC of 0.941, while image (d) is the ground truth mask for the correct output seen in (e,f) with an mDSC of 0.963. All of the above images have been resized to 1024 × 1024.
4. Conclusions
A wide array of biotic and abiotic stressors that exist in the natural environment, like pests and diseases, are the driving force behind plant symptom manifestation. Since similar symptoms can be caused by different factors, relying solely on the symptoms’ characteristics for disease identification is not ideal. On the other hand, rigorous and extensive testing for precise metrics requires time, labor, and specialized skills and equipment that are often not available. The integration of DL techniques in agricultural practices holds promise for enhancing crop quality by assisting farmers in prompt identification and effective management of diseases, particularly in extensive operations.
This study focuses on B. cinerea disease and specifically on the early and late detection of its gray mold symptom. The experiment was conducted on cucumber plants grown in greenhouse-controlled conditions. The assessment of the U-Net++ model combined with the MobileViTV2-1.25 encoder and Cut-and-Paste technique yielded an mDSC of 0.792, an mIoU of 0.816, and a recall rate of 0.885, achieving an accuracy of 91.9%. Additionally, it successfully identified the early stages of B. cinerea, achieving an IoU of 0.518 at dpi 2, while it reached its peak at dpi 6 with an IoU of 0.615.
The Cut-and-Paste augmentation technique was crucial in improving the effectiveness of the DL models by addressing the issue of dataset imbalance. This method involved extracting leaves from images and dynamically pasting them onto new images during training, effectively multiplying the training dataset. By prioritizing the underrepresented classes with invisible and visible symptoms of B. cinerea, the Cut-and-Paste technique ensured a more balanced dataset. This approach led to substantial improvements in the model’s capability in correctly segmenting and classifying the various stages of infection, with substantial increases in IoU by 14.29% for the healthy class, 56.45% for the invisible class, and 23.27% for the visible B. cinerea symptoms class. The success of this augmentation method underscores the critical importance of employing advanced data augmentation techniques to maximize model performance and effectively address dataset imbalance in developing robust and effective plant disease identification models.
By qualitatively interpreting the study, it is determined that the integration of ViT capabilities with traditional convolutional architectures provides notable improvements in plant disease segmentation and classification. This successful method for prompt identification of diseases underscores the promise of advanced DL methods in precision farming and proactive greenhouse health management. Combined with multi-spectral imaging, this approach provides an effective solution for real-time disease monitoring, allowing farmers to take faster, well-informed actions for improved plant health management. Such DL models can be seamlessly integrated with other digital systems, like self-navigating robots and mobile apps, providing precise measurements and visual insights for farmers. These technological advancements represent a promising step toward optimizing resources in smart agriculture, minimizing chemical inputs, and enhancing efficiency across the farm-to-fork sector.
Future steps should include experiments using the existing six-class annotation system for this dataset, as well as examining the DL model’s potential for early detection of other plant diseases.
Author Contributions
Conceptualization, E.-M.P. and N.G.; methodology, N.G. and P.C.; software, P.C.; validation, D.K. and N.G.; formal analysis, P.C.; investigation, P.C. and D.K.; resources, E.-M.P.; data curation, P.C.; writing—original draft preparation, P.C., N.G. and D.K.; writing—review and editing, E.-M.P.; visualization, P.C.; supervision, D.T. and E.-M.P.; project administration, E.-M.P.; funding acquisition, E.-M.P. All authors have read and agreed to the published version of the manuscript.
Funding
This work is supported by the Green Deal PestNu project, funded by European Union’s Horizon 2020 research and innovation program under grant agreement No. 101037128. The APC was funded by the Centre for Research and Technology Hellas.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Data available on request.
Acknowledgments
We thank our project partner iKnowHow S.A. for supporting the creation of the imagery dataset and their subcontractor Benaki Phytopathological Institute for the bioassays.
Conflicts of Interest
The authors declare no conflicts of interest.
Appendix A
This appendix presents a comprehensive breakdown of segmentation results for various classes, encompassing the three-class annotation system. The detailed IoU and recall metrics in Table A1 offer a thorough assessment of each model’s performance across different classes. This class-specific evaluation is crucial for assessing model precision in distinguishing between disease stages, emphasizing their capabilities, and identifying potential areas for improvement across different model–encoder combinations. Figure A1 visualizes the IoU progression across epochs for the top-performing architecture–encoder model on the validation dataset.

Table A1.
Performance of all three classes of DL segmentation models (best-performing model highlighted in bold).
Table A1.
Performance of all three classes of DL segmentation models (best-performing model highlighted in bold).
| Architecture | Encoder | IoU (Healthy) | IoU (Invisible) | IoU (Visible) | Recall (Healthy) | Recall (Invisible) | Recall (Visible) |
|---|---|---|---|---|---|---|---|
| PAN | MobileViT-S | 0.742 | 0.247 | 0.360 | 0.848 | 0.435 | 0.612 |
| PAN | MobileViTV2-125 | 0.600 | 0.135 | 0.197 | 0.710 | 0.253 | 0.391 |
| MA-Net | MobileViT-S | 0.711 | 9.69 × 10−16 | 0.146 | 0.905 | 9.69 × 10−16 | 0.310 |
| MA-Net | MobileViTV2-125 | 0.692 | 9.69 × 10−16 | 0.213 | 0.827 | 9.69 × 10−16 | 0.504 |
| DeepLabV3+ | MobileViT-S | 0.790 | 0.279 | 0.470 | 0.883 | 0.370 | 0.761 |
| U-Net++ | MobileViT-S | 0.778 | 0.293 | 0.470 | 0.880 | 0.303 | 0.635 |
| U-Net++ 1 | MobileViTV2-125 | 0.658 | 0.429 | 0.490 | 0.770 | 0.503 | 0.782 |
| U-Net++ 2 | MobileViTV2-125 | 0.752 | 0.671 | 0.604 | 0.865 | 0.795 | 0.781 |
1 Underlined row indicates the best-performing model before applying the Cut-and-Paste augmentation technique. 2 Bold row indicates the best-performing model after applying the Cut-and-Paste augmentation technique.
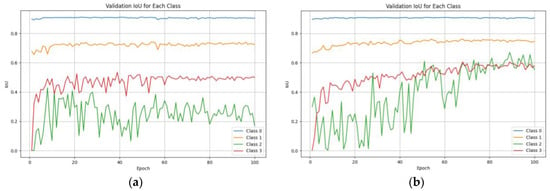
Figure A1.
IoU per epoch for the best-performing architecture–encoder model for the validation dataset: (a) only with geometric data augmentations, (b) after applying the Cut-and-Paste augmentation technique.
Appendix B
This appendix offers qualitative evaluations of segmentation outputs, demonstrating the capabilities of the best model, U-Net++ with the MobileViTV2-125 encoder, for the three-class system. Figure A2 illustrates the accurate detection of healthy areas, highlighting the precise identification of asymptomatic regions. Similarly, Figure A3 successfully identifies healthy leaves and perfectly segments the leaf exhibiting invisible gray mold symptoms. In contrast, Figure A4 and Figure A5 present examples of classification vagueness: Figure A4 depicts a case in which a very small patch of healthy leaf is erroneously segmented as having invisible B. cinerea symptoms, while larger areas of visible symptoms are misclassified as invisible B. cinerea symptoms and healthy tissue. Figure A5 illustrates a misclassification error, as healthy parts of leaves are identified as invisible and visible gray mold symptoms.
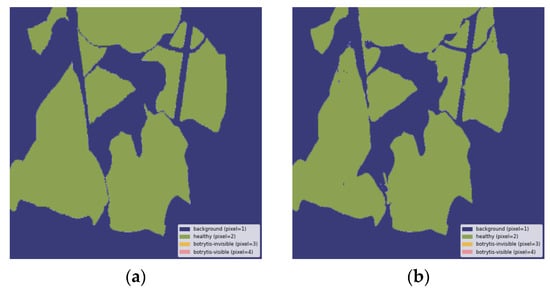
Figure A2.
Healthy leaves correctly segmented: (a) ground truth label, (b) output segmentation mask of the model. All images are resized to 1024 × 1024 and displayed with standard colormap.
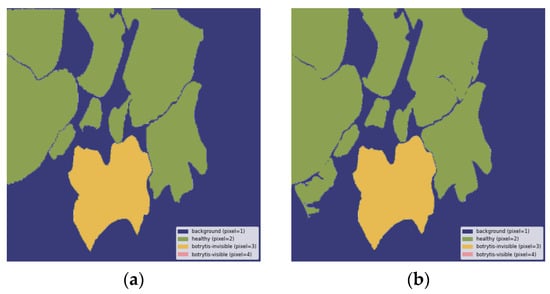
Figure A3.
Healthy and invisible B. cinerea leaves correctly segmented: (a) ground truth segmentation label, (b) output segmentation mask of the model. All images are resized to 1024 × 1024 and displayed with standard colormap.
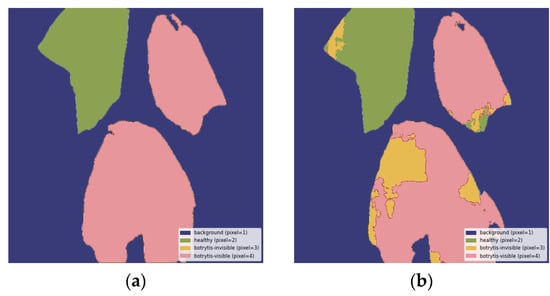
Figure A4.
Misclassification of a small healthy area as invisible and larger visible areas as a mix of invisible B. cinerea symptoms and healthy: (a) ground truth segmentation label, (b) output segmentation mask of the model. All images are resized to 1024 × 1024 and displayed with standard colormap.
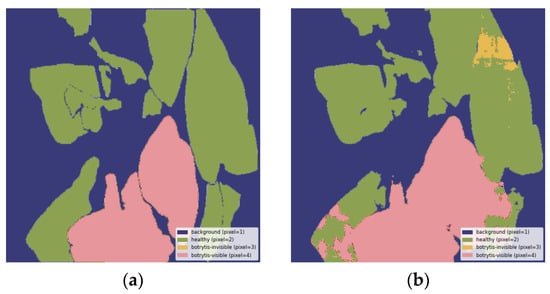
Figure A5.
Misclassification of healthy leaves as invisible and visible B. cinerea symptoms: (a) ground truth segmentation label, (b) output segmentation mask of the model. All images are resized to 1024 × 1024 and displayed with standard colormap.
References
- Williamson, B.; Tudzynski, B.; Tudzynski, P.; Van Kan, J.A.L. Botrytis cinerea: The cause of grey mould disease. Mol. Plant Pathol. 2007, 8, 561–580. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Chen, Y.; Zhang, Z.; Li, B.; Qin, G.; Tian, S. Pathogenic mechanisms and control strategies of Botrytis cinerea causing post-harvest decay in fruits and vegetables. Food Qual. Saf. 2018, 2, 111–119. [Google Scholar] [CrossRef]
- Latorre, B.A.; Elfar, K.; Ferrada, E.E. Gray mold caused by Botrytis cinerea limits grape production in Chile. Cienc. Investig. Agrar. 2015, 42, 305–330. [Google Scholar] [CrossRef]
- Al-Sarayreh, M.; Reis, M.M.; Yan, W.Q.; Klette, R. Potential of deep learning and snapshot hyperspectral imaging for classification of species in meat. Food Control 2020, 117, 107332. [Google Scholar] [CrossRef]
- Romanazzi, G.; Smilanick, J.L.; Feliziani, E.; Droby, S. Integrated management of postharvest gray mold on fruit crops. Postharvest Biol. Technol. 2016, 113, 69–76. [Google Scholar] [CrossRef]
- Leroux, P.; Fritz, R.; Debieu, D.; Albertini, C.; Lanen, C.; Bach, J.; Gredt, M.; Chapeland, F. Mechanisms of resistance to fungicides in field strains of Botrytis cinerea. Pest Manag. Sci. 2002, 58, 876–888. [Google Scholar] [CrossRef] [PubMed]
- Bilkiss, M.; Shiddiky, M.J.A.; Ford, R. Advanced Diagnostic Approaches for Necrotrophic Fungal Pathogens of Temperate Legumes with a Focus on Botrytis spp. Front. Microbiol. 2019, 10, 1889. [Google Scholar] [CrossRef] [PubMed]
- Rosslenbroich, H.-J.; Stuebler, D. Botrytis cinerea—History of chemical control and novel fungicides for its management. Crop Prot. 2000, 19, 557–561. [Google Scholar] [CrossRef]
- Wäldchen, J.; Mäder, P. Machine learning for image based species identification. Methods Ecol. Evol. 2018, 9, 2216–2225. [Google Scholar] [CrossRef]
- Giakoumoglou, N.; Pechlivani, E.M.; Tzovaras, D. Generate-Paste-Blend-Detect: Synthetic dataset for object detection in the agriculture domain. Smart Agric. Technol. 2023, 5, 100258. [Google Scholar] [CrossRef]
- Tsiakas, K.; Papadimitriou, A.; Pechlivani, E.M.; Giakoumis, D.; Frangakis, N.; Gasteratos, A.; Tzovaras, D. An Autonomous Navigation Framework for Holonomic Mobile Robots in Confined Agricultural Environments. Robotics 2023, 12, 146. [Google Scholar] [CrossRef]
- Pechlivani, E.M.; Gkogkos, G.; Giakoumoglou, N.; Hadjigeorgiou, I.; Tzovaras, D. Towards Sustainable Farming: A Robust Decision Support System’s Architecture for Agriculture 4.0. In Proceedings of the 2023 24th International Conference on Digital Signal Processing (DSP), Rhodes (Rodos), Greece, 11–13 June 2023; IEEE: New York, NY, USA, 2023; pp. 1–5. [Google Scholar] [CrossRef]
- Robertson, S.; Azizpour, H.; Smith, K.; Hartman, J. Digital image analysis in breast pathology—From image processing techniques to artificial intelligence. Transl. Res. 2018, 194, 19–35. [Google Scholar] [CrossRef] [PubMed]
- Voulodimos, A.; Doulamis, N.; Doulamis, A.; Protopapadakis, E. Deep Learning for Computer Vision: A Brief Review. Comput. Intell. Neurosci. 2018, 2018, 7068349. [Google Scholar] [CrossRef] [PubMed]
- Saleem, M.H.; Potgieter, J.; Arif, K.M. Plant Disease Detection and Classification by Deep Learning. Plants 2019, 8, 468. [Google Scholar] [CrossRef] [PubMed]
- Shoaib, M.; Shah, B.; Ei-Sappagh, S.; Ali, A.; Ullah, A.; Alenezi, F.; Gechev, T.; Hussain, T.; Ali, F. An advanced deep learning models-based plant disease detection: A review of recent research. Front. Plant Sci. 2023, 14, 1158933. [Google Scholar] [CrossRef]
- Khan, S.; Naseer, M.; Hayat, M.; Zamir, S.W.; Khan, F.S.; Shah, M. Transformers in Vision: A Survey. ACM Comput. Surv. 2022, 54, 1–41. [Google Scholar] [CrossRef]
- Bahdanau, D.; Cho, K.; Bengio, Y. Neural Machine Translation by Jointly Learning to Align and Translate. arXiv 2016, arXiv:1409.0473. [Google Scholar] [CrossRef]
- Jamil, S.; Piran, M.J.; Kwon, O.-J. A Comprehensive Survey of Transformers for Computer Vision. Drones 2023, 7, 287. [Google Scholar] [CrossRef]
- Sykes, J.; Denby, K.; Franks, D.W. Computer vision for plant pathology: A review with examples from cocoa agriculture. Appl. Plant Sci. 2023, 12, e11559. [Google Scholar] [CrossRef]
- Dhanya, V.G.; Subeesh, A.; Kushwaha, N.L.; Vishwakarma, D.K.; Kumar, T.N.; Ritika, G.; Singh, A.N. Deep learning based computer vision approaches for smart agricultural applications. Artif. Intell. Agric. 2022, 6, 211–229. [Google Scholar] [CrossRef]
- Thisanke, H.; Deshan, C.; Chamith, K.; Seneviratne, S.; Vidanaarachchi, R.; Herath, D. Semantic segmentation using Vision Transformers: A survey. Eng. Appl. Artif. Intell. 2023, 126, 106669. [Google Scholar] [CrossRef]
- Remez, T.; Huang, J.; Brown, M. Learning to Segment via Cut-and-Paste. arXiv 2018, arXiv:1803.06414. [Google Scholar] [CrossRef]
- Dirr, J.; Bauer, J.C.; Gebauer, D.; Daub, R. Cut-paste image generation for instance segmentation for robotic picking of industrial parts. Int. J. Adv. Manuf. Technol. 2024, 130, 191–201. [Google Scholar] [CrossRef]
- Omia, E.; Bae, H.; Park, E.; Kim, M.S.; Baek, I.; Kabenge, K.; Cho, B.-K. Remote Sensing in Field Crop Monitoring: A Comprehensive Review of Sensor Systems, Data Analyses and Recent Advances. Remote Sens. 2023, 15, 354. [Google Scholar] [CrossRef]
- Pechlivani, E.M.; Papadimitriou, A.; Pemas, S.; Giakoumoglou, N.; Tzovaras, D. Low-Cost Hyperspectral Imaging Device for Portable Remote Sensing. Instruments 2023, 7, 32. [Google Scholar] [CrossRef]
- Fahrentrapp, J.; Ria, F.; Geilhausen, M.; Panassiti, B. Detection of Gray Mold Leaf Infections Prior to Visual Symptom Appearance Using a Five-Band Multispectral Sensor. Front. Plant Sci. 2019, 10, 628. [Google Scholar] [CrossRef] [PubMed]
- Sahin, H.M.; Miftahushudur, T.; Grieve, B.; Yin, H. Segmentation of weeds and crops using multispectral imaging and CRF-enhanced U-Net. Comput. Electron. Agric. 2023, 211, 107956. [Google Scholar] [CrossRef]
- Giakoumoglou, N.; Pechlivani, E.M.; Katsoulas, N.; Tzovaras, D. White Flies and Black Aphids Detection in Field Vegetable Crops using Deep Learning. In Proceedings of the 2022 IEEE 5th International Conference on Image Processing Applications and Systems (IPAS), Genova, Italy, 5–7 December 2022; pp. 1–6. [Google Scholar] [CrossRef]
- Giakoumoglou, N.; Pechlivani, E.-M.; Frangakis, N.; Tzovaras, D. Enhancing Tuta absoluta Detection on Tomato Plants: Ensemble Techniques and Deep Learning. AI 2023, 4, 996–1009. [Google Scholar] [CrossRef]
- Giakoumoglou, N.; Pechlivani, E.M.; Sakelliou, A.; Klaridopoulos, C.; Frangakis, N.; Tzovaras, D. Deep learning-based multi-spectral identification of grey mould. Smart Agric. Technol. 2023, 4, 100174. [Google Scholar] [CrossRef]
- Bhujel, A.; Khan, F.; Basak, J.K.; Jaihuni, H.; Sihalath, T.; Moon, B.-E.; Park, J.; Kim, H.-T. Detection of gray mold disease and its severity on strawberry using deep learning networks. J. Plant Dis. Prot. 2022, 129, 579–592. [Google Scholar] [CrossRef]
- Sánchez, M.G.; Miramontes-Varo, V.; Chocoteco, J.A.; Vidal, V. Identification and Classification of Botrytis Disease in Pomegranate with Machine Learning. In Intelligent Computing 1229 (Advances in Intelligent Systems and Computing 1229); Arai, K., Kapoor, S., Bhatia, R., Eds.; Springer International Publishing: Cham, Switzerland, 2020; pp. 582–598. [Google Scholar] [CrossRef]
- Ilyas, T.; Khan, A.; Umraiz, M.; Jeong, Y.; Kim, H. Multi-Scale Context Aggregation for Strawberry Fruit Recognition and Disease Phenotyping. IEEE Access 2021, 9, 124491–124504. [Google Scholar] [CrossRef]
- Meng, L.; Audenaert, K.; Van Labeke, M.-C.; Höfte, M. Imaging Detection of Botrytis cinerea On Strawberry Leaves Upon Mycelial Infection. SSRN 2023, preprint. [Google Scholar] [CrossRef]
- Wang, C.; Du, P.; Wu, H.; Li, J.; Zhao, C.; Zhu, H. A cucumber leaf disease severity classification method based on the fusion of DeepLabV3+ and U-Net. Comput. Electron. Agric. 2021, 189, 106373. [Google Scholar] [CrossRef]
- Qasrawi, R.; Amro, M.; Zaghal, R.; Sawafteh, M.; Polo, S.V. Machine Learning Techniques for Tomato Plant Diseases Clustering, Prediction and Classification. In Proceedings of the 2021 International Conference on Promising Electronic Technologies (ICPET), Deir El-Balah, Palestine, 17–18 November 2021; IEEE: New York, NY, USA, 2021; pp. 40–45. [Google Scholar] [CrossRef]
- Giakoumoglou, N.; Kalogeropoulou, E.; Klaridopoulos, C.; Pechlivani, E.M.; Christakakis, P.; Markellou, E.; Frangakis, N.; Tzovaras, D. Early detection of Botrytis cinerea symptoms using deep learning multi-spectral image segmentation. Smart Agric. Technol. 2024, 8, 100481. [Google Scholar] [CrossRef]
- O’Sullivan, C. U-Net Explained: Understanding Its Image Segmentation Architecture. Medium. Available online: https://towardsdatascience.com/u-net-explained-understanding-its-image-segmentation-architecture-56e4842e313a (accessed on 24 May 2024).
- Decognet, V.; Bardin, M.; Trottin-Caudal, Y.; Nicot, P.C. Rapid Change in the Genetic Diversity of Botrytis cinerea Populations After the Introduction of Strains in a Tomato Glasshouse. Phytopathology 2009, 99, 185–193. [Google Scholar] [CrossRef] [PubMed]
- La Camera, S.; L’haridon, F.; Astier, J.; Zander, M.; Abou-Mansour, E.; Page, G.; Thurow, C.; Wendehenne, D.; Gatz, C.; Métraux, J.-P.; et al. The glutaredoxin ATGRXS13 is required to facilitate Botrytis cinerea infection of Arabidopsis thaliana plants: Role of ATGRXS13 during B. cinerea infection. Plant J. 2011, 68, 507–519. [Google Scholar] [CrossRef] [PubMed]
- De Meyer, G.; Bigirimana, J.; Elad, Y.; Höfte, M. Induced systemic resistance in Trichoderma harzianum T39 biocontrol of Botrytis cinerea. Eur. J. Plant Pathol. 1998, 104, 279–286. [Google Scholar] [CrossRef]
- Lee Campbell, C.; Madden, L.V. Introduction to Plant Disease Epidemiology, 1st ed.; Wiley-Interscience: New York, NY, USA, 1990. [Google Scholar]
- IBM SPSS Statistics for Windows, Version 27.0; IBM Corp.: Armonk, NY, USA, 2020.
- “Roboflow” (Version 1.0) [Software]. Available online: https://roboflow.com (accessed on 3 March 2024).
- Zhou, Z.; Siddiquee, M.M.R.; Tajbakhsh, N.; Liang, J. UNet++: A Nested U-Net Architecture for Medical Image Segmentation. arXiv 2018, arXiv:1807.10165. [Google Scholar] [CrossRef]
- Li, H.; Xiong, P.; An, J.; Wang, L. Pyramid Attention Network for Semantic Segmentation. arXiv 2018, arXiv:1805.10180. [Google Scholar] [CrossRef]
- Fan, T.; Wang, G.; Li, Y.; Wang, H. MA-Net: A Multi-Scale Attention Network for Liver and Tumor Segmentation. IEEE Access 2020, 8, 179656–179665. [Google Scholar] [CrossRef]
- Chen, L.-C.; Zhu, Y.; Papandreou, G.; Schroff, F.; Adam, H. Encoder-Decoder with Atrous Separable Convolution for Semantic Image Segmentation. arXiv 2018, arXiv:1802.02611. [Google Scholar] [CrossRef]
- Ronneberger, O.; Fischer, P.; Brox, T. U-Net: Convolutional Networks for Biomedical Image Segmentation. arXiv 2015, arXiv:1505.04597. [Google Scholar] [CrossRef]
- Chen, L.-C.; Papandreou, G.; Schroff, F.; Adam, H. Rethinking Atrous Convolution for Semantic Image Segmentation. arXiv 2017, arXiv:1706.05587. [Google Scholar] [CrossRef]
- Mehta, S.; Rastegari, M. MobileViT: Light-weight, General-purpose, and Mobile-friendly Vision Transformer arXiv 2022, arXiv:2110.02178. Available online: http://arxiv.org/abs/2110.02178 (accessed on 31 October 2023).
- Mehta, S.; Rastegari, M. Separable Self-attention for Mobile Vision Transformers. arXiv 2022, arXiv:2206.02680. Available online: http://arxiv.org/abs/2206.02680 (accessed on 31 October 2023).
- Sudre, C.H.; Li, W.; Vercauteren, T.; Ourselin, S.; Cardoso, M.J. Generalised Dice overlap as a deep learning loss function for highly unbalanced segmentations. arXiv 2017, arXiv:1707.03237. [Google Scholar] [CrossRef]
- Loshchilov, I.; Hutter, F. Decoupled Weight Decay Regularization. arXiv 2019, arXiv:1711.05101. Available online: http://arxiv.org/abs/1711.05101 (accessed on 31 October 2023).
- Kingma, D.P.; Ba, J. Adam: A Method for Stochastic Optimization. arXiv 2014, arXiv:1412.6980. [Google Scholar] [CrossRef]
- Krizhevsky, A.; Sutskever, I.; Hinton, G.E. ImageNet classification with deep convolutional neural networks. Commun. ACM 2017, 60, 88–90. [Google Scholar] [CrossRef]
- Ghiasi, G.; Cui, Y.; Srinivas, A.; Qian, R.; Lin, T.-Y.; Cubuk, E.D.; Le, Q.V.; Zoph, B. Simple Copy-Paste is a Strong Data Augmentation Method for Instance Segmentation. arXiv 2021, arXiv:2012.07177. [Google Scholar] [CrossRef]
- Dvornik, N.; Mairal, J.; Schmid, C. Modeling Visual Context is Key to Augmenting Object Detection Datasets. arXiv 2018, arXiv:1807.07428. [Google Scholar] [CrossRef]
- Dwibedi, D.; Misra, I.; Hebert, M. Cut, Paste and Learn: Surprisingly Easy Synthesis for Instance Detection. arXiv 2017, arXiv:1708.01642. [Google Scholar] [CrossRef]
- Gull, A.; Lone, A.A.; Wani, N.U.I. Biotic and abiotic stresses in plants. In Abiotic and Biotic Stress in Plants; IntechOpen: London, UK, 2019; pp. 1–19. [Google Scholar]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).