1. Introduction
Nitrogen is an important nutrient for the growth and development of plants. Naturally occurring nitrogen compounds absorbed by plants are not sufficiently available. Hence, farmers apply nitrogen fertilizer to crops to enhance nitrogen absorption and increase crop production. According to the study by Ritchie [
1], 115 million tonnes of nitrogen fertilizer are annually applied to crops but only 35% is utilized by the crops. The excessive amount causes nitrogen pollution including the acidification of soil, water contamination, and nitrous oxide emissions that contribute to global warming. China is the largest contributor to this pollution, causing one third of the total pollution, followed by India with 18%, and USA with 11% of the total.
Nitrogen use efficiency (NUE) measures how much of the applied nitrogen is utilized by crops as opposed to being lost to the environment [
2]. The overapplication of nitrogen fertilizer can reduce NUE. By improving farming practices using the latest technology advancements, we can work towards increasing NUE, which will increase crop yield with reduced fertilizer application. Nitrogen fertilization is necessary for better yield and crop production, but the excessive use of fertilizers decreases NUE, increases overall fertilization costs without any improvement in crop yield, and has an undesirable impact on the environment. Therefore, it is necessary to prevent the overuse of nitrogen fertilizer by the targeted application of fertilizer and its rate adjustment.
Nitrogen is critical for chlorophyll formation in plant leaves, which produces the green color of vegetation. Nitrogen-deficient plants lack sufficient chlorophyll content, so the early signs of nitrogen deficiency appear in mature leaves as slight discoloration to a lighter green color in early stages, turning leaves to yellow later and further causing the premature fading of older leaves. Apart from this, thin and weak vegetation, slow growth of stem, and a purple hue are also signs of nitrogen deficiency. In later stages, it can cause premature fading and necrosis, resulting in early plant death and crop loss. Different crops show different symptoms of nitrogen deficiency. In maize, visual symptoms include a V-shaped yellowing pattern on the leaves, smaller or poorly filled maize ears, glossy maize kernels indicating low protein content, and premature browning of stover in the late season [
3]. These visual symptoms can be used to identify nitrogen deficiency in maize crop images using state-of-the-art computer vision techniques that are non-destructive and can be easily automated for faster response.
In this study, we have developed a custom-built vision transformer (ViT) model specifically designed for the classification of nitrogen stress levels in corn crops using RGB (red, green, and blue) image data. We also utilize the transfer learning approach to fine-tune some widely used classifiers for our application. Our approach focuses on leveraging the power of deep learning and vision transformers to accurately classify images into three distinct levels of nitrogen fertilization: no nitrogen applied, 75 kg nitrogen applied, and 136 kg nitrogen applied. Our contributions can be summarized as follows:
We have designed and implemented a vision transformer model tailored for the agricultural domain, specifically for nitrogen stress detection in corn crops.
Our developed vision and CNN models exclusively use RGB image data, making these models accessible and practical for widespread agricultural applications.
We have classified the nitrogen stress levels into three categories, providing detailed insights into the nitrogen status of the crops. For this purpose, we have used a publicly available dataset collected under field conditions.
We have utilized a transfer learning-based approach to fine-tune widely used vision transformer and CNN models for nitrogen stress level classification. This involved adapting pre-trained models to our specific application, enhancing their performance.
We have performed a detailed comparative analysis using two different image resolutions (100 × 100 and 224 × 224) to evaluate the impact of image resolution on classifier accuracy and identify the best-performing classifier.
Our research focuses on nitrogen-level classification from RGB images of maize crop, which is a foundational step towards achieving the goal of developing a precision agriculture system that can accurately determine the nitrogen fertilizer requirements for specific areas within a maize crop field. By detecting different nitrogen levels in crops, detailed nitrogen deficiency maps can be generated, which can then be used to divide the field into management zones, each with specific fertilizer requirements. With the precise identification of nitrogen-deficient areas, farmers can apply fertilizers only where needed, thereby increasing NUE and reducing waste.
2. Related Works
Traditional nitrogen status analysis in plants is performed using destructive chemical testing; therefore, the research community has been focused on exploring visual analysis techniques based on optical properties and machine learning models to approach the problem of nitrogen stress detection in crops. Hyperspectral imaging is a widely used method for stress detection in crops. Being a non-destructive method, it can capture detailed spectral information across various narrow spectral bands. Nitrogen affects the overall health of plants, which is reflected in the spectral properties of their leaves. Hyperspectral imaging is sensitive to these changes related to nitrogen deficiency, and therefore, many researchers have utilized hyperspectral imaging and vegetation indices associated with spectral properties of leaves for nitrogen stress detection. Authors in [
4,
5,
6,
7] used hyperspectral remote sensing to extract nitrogen information in cucumber, hemp, potato and tea leaves. Wang et al. [
8] used hyperspectral imagery data to determine nitrogen deficiency and its relation with crop yield in maize crop. The authors in [
9] determined nitrogen status in maize for variable rate fertilizer application using spectral vegetation indices. Wu et al. [
10] used a combination of spectral and texture-based features for estimating maize nitrogen content.
Even though hyperspectral imaging sensors have improved with time, due to the large volume of data requiring exhaustive processing and specialized expertise as well as high cost, hyperspectral imaging is not very feasible for real-time processing as compared to RGB imaging [
11]. Multispectral imaging, on the other hand, is relatively feasible for real-time applications due to the lower data volume and lesser computational cost. Burns et al. [
12] gave an overview of commonly used vegetation indices for assessing plant nitrogen stress. They used multispectral data to study the relationship between various vegetation indices with nitrogen stress and fertilizer rate application in maize. According to their study, chlorophyll index green (
), the green normalized difference vegetation index (GNDVI), and the red edge normalized difference vegetation index (RENDVI) are the best vegetation indices for the detection of nitrogen deficiency in maize. Zheng et al. [
13] used a five-band multispectral camera to obtain images of winter wheat using a drone during five growth stages of winter wheat. They compared various parametric and non-parametric modeling algorithms for nitrogen content determination, where parametric modeling is based on 19 vegetation indices. The best vegetation index for nitrogen content detection was reported to be the modified renormalized difference vegetation index (RDVI) with a coefficient of determination (
) of 0.73 and root mean squared error (RMSE) of 0.38. For non-parametric models, random forest performed best with
of 0.79 and RMSE of 0.33. The authors in [
14] used a combination of hyperspectral and multispectral data for nitrogen content estimation in tea plants and reported an
and RMSE of 0.9186 and 0.0560, respectively.
In contrast to the multispectral and hyperspectral imaging techniques, many recent studies have explored the use of advanced machine learning algorithms, particularly convolutional neural networks (CNNs), applied to RGB imaging data for nitrogen stress detection in crops. This approach utilizes the valuable information provided by RGB images, which, when combined with the deep learning capabilities of CNNs, has shown promising results in accurately identifying nitrogen stress indicators in plants. Moreover, RGB images are more amenable for real-time processing compared to other approaches. Azimi, Kaur, and Gandhi [
15] proposed a CNN for nitrogen stress indication in sorghum plant shoot images. The dataset they used for their study comprises RGB images of three classes: a healthy plants class representing 100% nitrogen treatment available, a semi-stressed class with 50% nitrogen treatment, and a severely stressed class with 10% nitrogen treatment available to the plants. They used a publicly available dataset obtained under laboratory conditions, consisting of images of plant shoots captured against a white background. They proposed a 23-layered CNN model with 5 convolutional layers, respectively, each followed by batch normalization and ReLU activation, where the first four convolutional layers are followed by max pooling layers. The fifth convolutional layer is followed by fully connected, softmax, and classification output layers. They reported a maximum accuracy of 84%.
Zermas et al. [
16] proposed an annotation assistant tool, which identifies plant areas demonstrating nitrogen deficiency. The tool provides a recommendation to the annotator to create a training set that is then used to train a deep learning model. The images in their work were collected through a drone. The annotator tool uses k-means clustering and support vector machine (SVM) models in various steps to divide pixels in green (healthy), yellow (stressed), and brown (soil) pixels. This step generates a training set with bounding boxes representing nitrogen-stressed areas in images. This set is then used to train a Faster RCNN architecture with a ResNet50 feature extractor for detection. They reported a mean average precision of 82.3% and an intersection over union percentage of 50%. Zhang et al. [
17] used RGB (red, green, and blue) imaging from a drone for nitrogen stress detection in rapeseed leaves during seedling stage. They used a U-Net model for purple rapeseed leaf segmentation and to establish a relationship between leaf purple area and nitrogen deficit. They reported that the purple area increases as the nitrogen application level decreases, suggesting a greater stress on the plants due to lower nitrogen levels. They determined the relationship between the purple area and nitrogen content to be negative exponential with an
of 0.858, indicating a strong correlation between these factors.
Haider et al. [
18] also proposed a vision-based analysis technique for nitrogen content estimation in spinach leaves. They used a specially designed board with white background and two reference colors that are represented by green and yellow circles.The first step of their methodology involves using Otsu’s thresholding and the bounding box method to extract the region of interest, which includes the reference circles and the leaf. The leaf and reference circles are then distinguished from each other using the width-to-height ratio of their corresponding bounding boxes. The next step of their methodology uses the Gaussian mixture model technique for background segmentation of the leaf. Their methodology then computes the mean color value of the reference circles and leaf. The decision is made on the basis of distance of leaf mean color from the mean color of the reference circles. They reported an
of 0.92 and RMSE of 0.0845.
3. Materials and Methods
Figure 1 depicts the flowchart of our methodology. The dataset utilized in this research is presented by Salaić et al. [
19] and is publicly available through the Mendeley data repository [
20]. The dataset is composed of 1200 images of maize canopy annotated by an agricultural expert. The images were collected in July 2023 over a 5-day period encompassing the flowering stage of maize. They were captured randomly along the rows between 7:30 a.m. and 11:00 a.m., with each image sized at 2400 × 1600 pixels. The images are categorized into three distinct classes, each representing a different nitrogen fertilization level. The classes include N0 (no fertilization), N75 (75 kg of nitrogen fertilizer applied), and NFull (136 kg of fertilizer applied), with 400 images per class. The images are captured using a digital single-lens reflex (DSLR) camera positioned at a 45° angle relative to the plots.
3.1. Pre-Processing
Pre-processing is a critical step in preparing data for classifier training. This process involves several key steps, including resizing the images to a uniform resolution. We performed experimentation using two image resolutions: 100 × 100 pixels and 224 × 224 pixels. Additionally, segmentation is performed to isolate the plant region. To achieve this, a threshold value of 30 is applied to the inverted blue image channel in the RGB color space to segment the sky. For removing the soil part, thresholding on the hue image is applied in the HSV color space. Two threshold values are selected based on the lighting and shadowing in the images. Images with higher exposure to sunlight are segmented using a threshold of 75, while images with lower exposure use a threshold of 40. Following thresholding, morphological closing is applied to refine the segmentation mask using a cross-shaped 5 × 5 kernel. All the values were determined empirically after extensive experimentation.
Figure 2 shows sample images from the dataset before and after segmentation.
3.2. Data Augmentation
Data augmentation is an important technique in expanding small datasets. Machine learning models require a large number of training samples to learn from, which are challenging to obtain in real-world settings. Data augmentation works by generating new data from the original data by introducing small variations, thereby artificially creating a bigger dataset to enhance the model’s generalization and performance. To increase the robustness of our models and prevent overfitting, we apply various transformations including rotations, shifts, shear, zooms, and flips to augment the input images. We apply rotation and flip transformations to both the original and segmented images, increasing the total number of images in our dataset from 1200 to 4800. We use a random flip set to horizontal and apply a random rotation range of 20%. During the training process, we use the ImageDataGenerator class in TensorFlow to apply shift, shear, and zoom transformations. Width and height shift values are set to 15% of the image width and height, respectively. The shear angle is set to 15 degrees, and the zoom range is set to 15%. In this way, we are able to generate a diverse set of examples for our deep learning models to learn from. This is crucial to generate an extensive and varied dataset that could train robust models for the precise classification of images based on nitrogen deficiency levels.
3.3. Classifier Selection
We use supervised machine learning to classify three levels of nitrogen fertilization (i.e., N0, N75, and NFull). Our study focuses on comparing the performance of ViTs and CNNs in detecting nitrogen deficiency in maize images. ViT is a revolutionary deep learning model introduced by Dosovitskiy et al. [
21] that extends the transformer architecture from natural language processing to image classification tasks. While traditional CNNs use a hierarchical approach to process images, ViTs use a sequential approach, treating images as a sequence of small tokens in a similar manner to words being processed in natural language processing. This enables ViTs to capture long-range dependencies more effectively in images, resulting in strong performance in image classification tasks. While CNNs have long been the dominant architecture for computer vision, delivering high effectiveness across various applications, ViTs have demonstrated remarkable performance in numerous image classification tasks, often matching or surpassing the accuracy of CNNs. Our research aims to examine the performance of vision transformers, particularly in identifying nitrogen stress in plants, by comparing their effectiveness against state-of-the-art neural networks.
4. Implementation
We use the Tensorflow library with Keras API to develop our framework. We compare the performance of state-of-the-art ViTs with widely used CNNs. We conduct two sets of experiments. In the first set, we implement a ViT model built from scratch and trained entirely on our dataset. In the second set, we implement a transfer learning approach, where we select various pre-trained deep learning architectures as the base models and fine-tune them on our dataset. The base models include a pre-trained vision transformer model, as well as four widely known CNN models: EfficientNetB0 [
22], DenseNet121 [
23], InceptionV3 [
24], and ResNet50 [
25]. The details for both sets of experiments are presented below.
4.1. Vision Transformer Custom-Built Model
In the first set of our experiments, we develop a custom vision transformer model tailored to our specific application. The vision transformer model is constructed in several key steps. The first step is to augment the input images and divide into patches. For this model, we perform experimentation using two image resolutions: image size of 100 × 100 with a patch size of 10 and image size of 224 × 224 with a patch size of 16. These patches are then fed into a dense layer, which learns positional embeddings for each patch. This step is crucial for the model to understand the spatial relationships between different parts of the image. The output of the dense layer is then passed into the transformer network. The transformer network consists of several blocks. The block diagram showing the transformer structure is given in
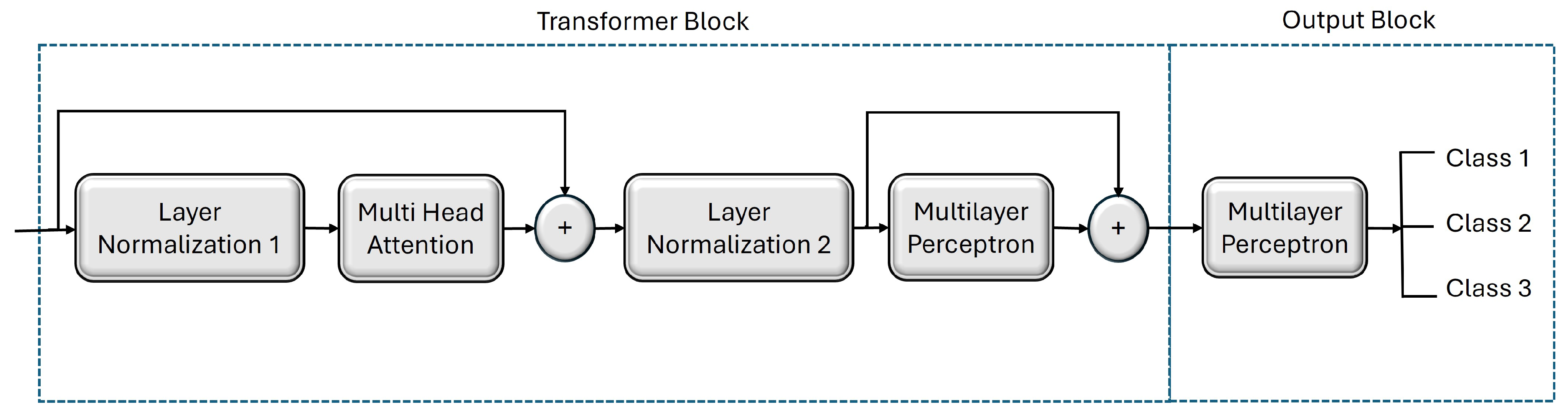
Figure 3. The transformer block contains a normalization layer followed by a multihead attention layer. The output of this attention layer is added to the original input and then fed into the next part of the block, which is composed of another normalization layer followed by a multilayer perceptron (MLP) that processes the intermediate features. The MLP in the transformer block comprises two hidden layers with 128 and 64 neurons, respectively. The output of this MLP is then passed to the MLP head in the output block for further processing. In our model, the MLP head unit has two hidden layers with 2048 and 1024 neurons, respectively. Finally, the output of the MLP head is passed to a dense layer that computes the class probabilities, determining the predicted class for each input image.
4.2. Transfer Learning and Fine-Tuned Models
In the second set of our experiments, we employ a transfer learning-based approach. In this set of experiments, we further compare a pre-trained vision transformer model with selected CNN models.
4.2.1. Fine-Tuned Vision Transformer Model
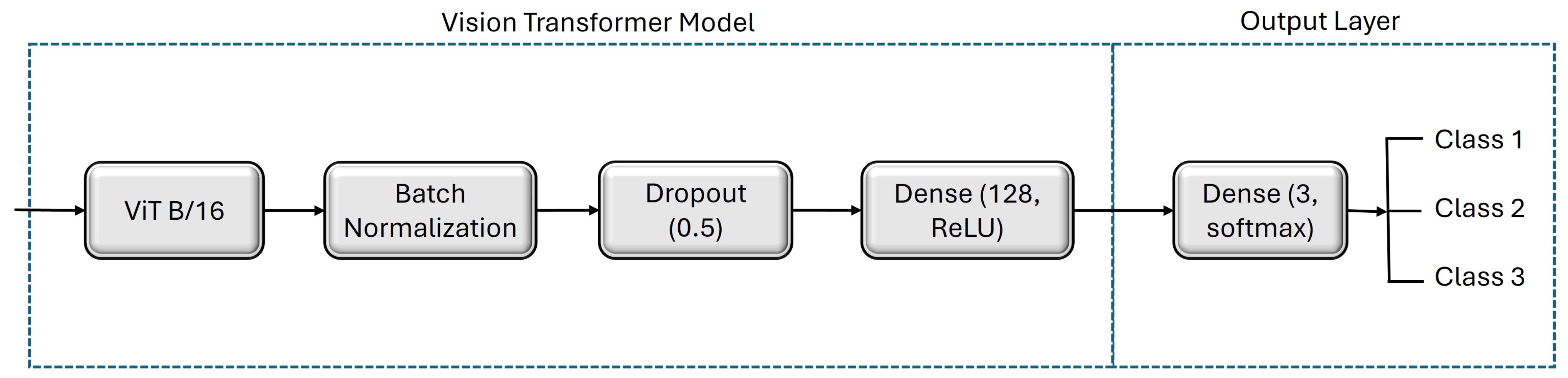
We select ViT-B/16 as the base model for fine-tuning on our dataset, which comprises 12 layers and 12 head units. This model configuration is known for its ability to capture intricate patterns in image data, making it a suitable choice for our classification task. Since this ViT base model accepts an image resolution of 224 × 224, we use only this resolution for training and testing in this part of the experiments. Following the base model, we incorporate a batch normalization layer, succeeded by an intermediate dense layer with 64 units and L2 regularization. Finally, the output layer, consisting of three units with a softmax activation function, completes the model.
Figure 4 shows the detailed architecture of the model.
4.2.2. Fine-Tuned Neural Network Models
The neural network architectures chosen as the base model for the second set of experiments are EfficientNetB0, DenseNet121, InceptionV3, and ResNet50. Similar to the custom-built ViT model, we perform experimentation using two image resolutions of 100 × 100 and 224 × 224. The base model’s output is fed into a global average pooling 2D layer, followed by a dropout layer with a 50% dropout rate to prevent overfitting. Subsequently, an intermediate dense layer with 128 units, ReLU activation, and L2 regularization is employed. Finally, the output layer consists of three units with a softmax activation function.
Figure 5 presents the details of this architecture.
4.3. Training Hyperparameters
We incorporate the Adam optimizer in our models with an initial learning rate of 0.001 and sparse categorical cross-entropy as the loss function. We implement several training callbacks to enhance model performance and prevent overfitting. Early stopping is employed to halt training if there is no improvement in performance for 10 consecutive epochs, thus promoting generalization. The Model Checkpoint callback saves the best-performing model based on validation accuracy, ensuring that the model with the highest predictive capability is retained. The ReduceLROnPlateau callback reduces the learning rate by a factor of 0.2 if the validation loss fails to improve for 5 epochs, with a lower bound set at 0.00001, allowing for fine-tuning of the model training. Additionally, a Learning Rate Scheduler adjusts the learning rate based on the epoch number to control the speed of model convergence, with an exponential decay scheduler employed in this instance. Each model has been trained for 50 epochs, and the results are detailed in the subsequent section.
5. Results and Discussion
For our experiments, we split the dataset into train and test sets, with 90% of the data allocated to the train set and 10% to the test set. We conduct the experiments using three-fold cross-validation for each classifier, with the validation ratio set to 20%. The best model configuration is selected based on the cross-validation results and evaluated on the test set. Accuracy, precision and recall are selected as the performance parameters. Additionally, training and validation loss curves are utilized to assess the performance of the classifiers.
Accuracy is defined as the ratio of correctly predicted instances to the total number of instances evaluated. Precision is defined as the ratio of true positive predictions to the total number of positive predictions made by the model. Recall is the ratio of true positive predictions to the total number of actual positive samples in the class. The training loss measures the error between the model’s predicted output and the actual target during training. Training aims at minimizing training loss through optimization techniques such as gradient descent. Decreasing training loss indicates improved fitting of the model to the training data. Validation loss, on the other hand, measures the error on a separate validation dataset that the model has not encountered during training, providing insight into its performance on unseen data. Validation loss evaluates the model’s generalization capability, with lower validation loss indicating effective generalization and higher validation loss suggesting potential overfitting.
5.1. Custom ViT Results
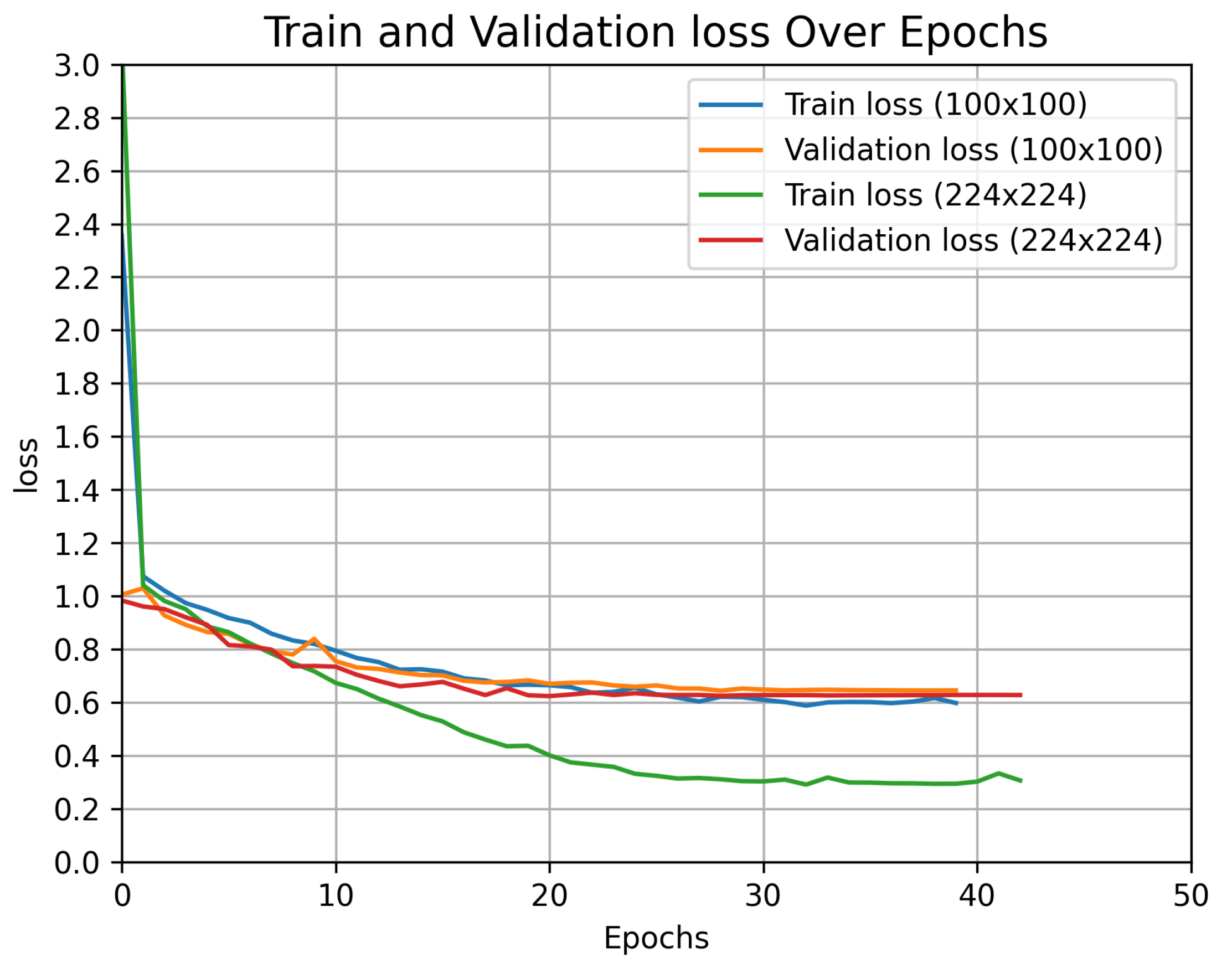
Figure 6 shows the training and validation loss curves for the custom ViT model for both image resolutions. For both cases, the training loss continues to decrease gradually, reaching a steady value by the end of the training. This gradual decrease suggests that the model continues to learn at a slower pace. The validation loss decreases at a relatively faster pace until epoch 20, after which it decreases very slowly and smoothly until the last epoch. This behavior indicates that while the model continues to improve its generalization, the rate of improvement becomes minimal after epoch 20. Overall, the model reaches a convergence point around epoch 30, where both training and validation losses stabilize, indicating effective learning and parameter stabilization.
From
Figure 6, it can be observed that even though the final training loss for the image resolution 224 × 224 is much lower (0.3) compared to the image resolution 100 × 100 (0.6), the validation loss is the same for both the cases (0.6). This indicates that the model with a larger image size might be overfitting to the training data. However, the test accuracy with the large-image resolution is higher (76%) as compared to the small-image resolution (70%). The improved test accuracy with the larger image size suggests that the model benefits from the additional information provided by larger images which contain more detail and spatial information than smaller images, making them the preferred choice for vision transformer classifier.
5.2. Transfer Learning and Fine-Tuned Models Results
5.2.1. Fine-Tuned ViT Results
Figure 7 shows the graphs of training and validation losses over 50 epochs for our fine-tuned model with the pre-trained ViT base. The graph demonstrates that the training loss gradually decreases, indicating that the model is effectively learning from the data. Initially, there are fluctuations in the validation loss, suggesting that the model is adjusting to the data and fine-tuning its parameters to find an optimal configuration. After epoch 25, the loss curve smooths out and remains relatively constant. This stability and similarity between the training and validation losses suggest good generalization. The curves indicate that by 50 epochs, the model has likely reached convergence, with its weights stabilized, and further training does not result in significant changes to the loss values. The pre-trained ViT model attains the highest training accuracy of 70% for the best configuration, and the best test accuracy of 64%.
Both ViT-based models exhibit similar performance, with the custom-built model showing better performance. Both models achieve their best loss values within 30 epochs, although the pre-trained model displays more fluctuations at the beginning of training. The stability of the loss curves and the consistent values of both training and validation losses after 30 epochs suggest that the models have likely reached a point of convergence. Further training is unlikely to lead to significant improvements, as it can be inferred from the training curve that the models have already learned fully from the data given the current architecture and hyperparameters.
5.2.2. Fine-Tuned CNN Models Results
Table 1 gives a comparison of train and test accuracies for all CNN-based models for both image resolutions (100 × 100 and 224 × 224). It has been observed that all base models perform better with a higher image resolution of 224 × 224 as compared to a lower resolution of 100 × 100. Among all the models, the largest difference in test accuracy is observed with InceptionV3, which achieves a test accuracy of 74% for smaller images and 91% for larger images. ResNet50 shows slightly better performance, with 78% test accuracy for smaller images and 84.5% for larger images. The DenseNet121-based model performs reasonably well, with 90% and 93% test accuracy for small and large images, respectively. The best performance is obtained with the EfficientNetB0 base model, achieving a test accuracy of 92% for smaller images (100 × 100), and 97% for larger images (224 × 224), making it the most suitable model for our application.
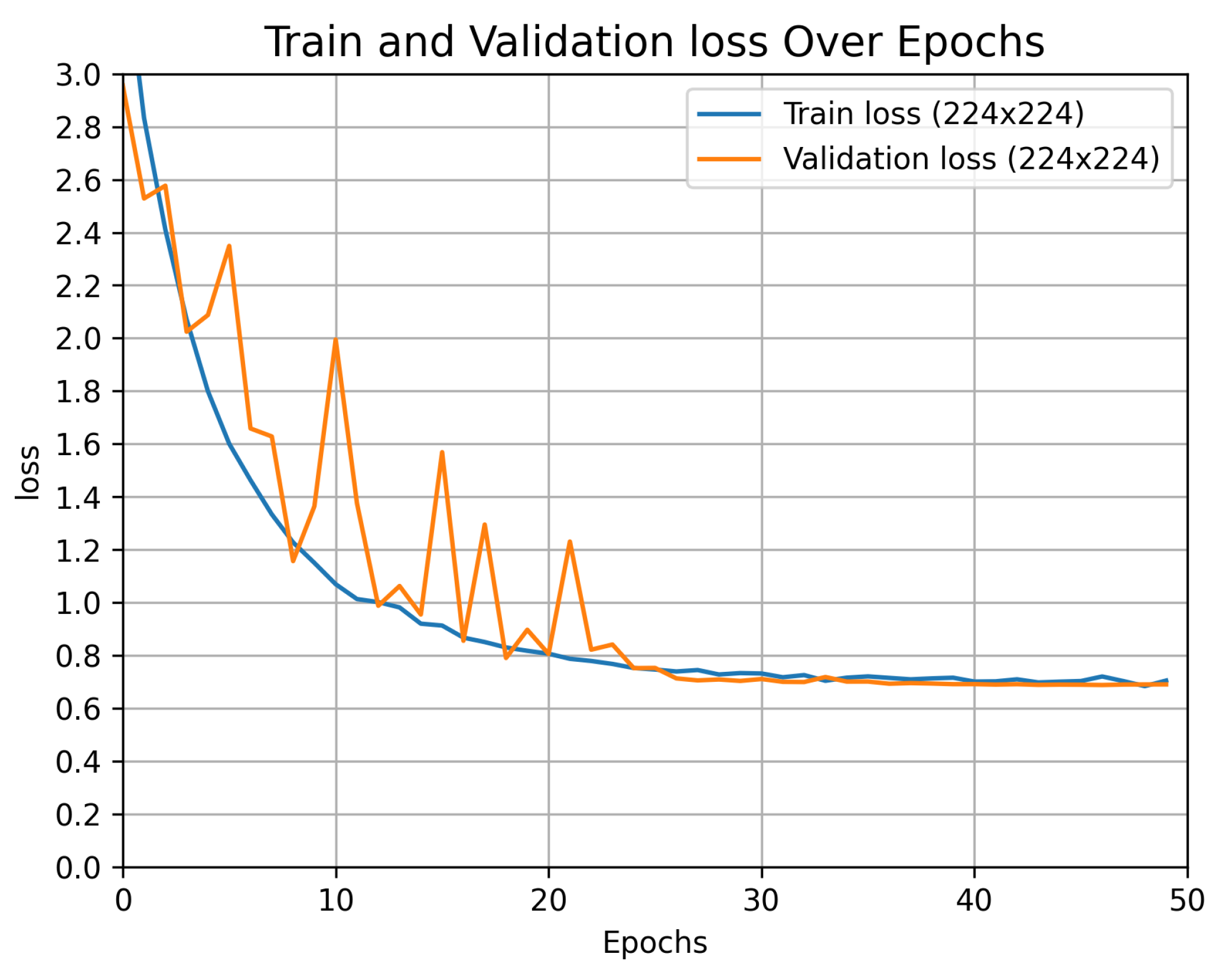
Figure 8 gives the training and validation loss curves for the fine-tuned EfficientNetB0 model for both image resolutions. A gradually decreasing and smooth training curve in both cases indicates that the model’s optimization process is stable and effective. The fluctuations in the validation loss early in the training are due to the model making adjustments to the weights and learning the data. Steady values of training and validation losses indicate that the model has converged. Training loss values are similar for both resolutions and become steady after 30 epochs. However, the validation loss for the small-image resolution is higher (0.23) compared to the large-image resolution (0.08). The significant difference in validation loss suggests that higher resolution images provide more detailed information, enabling the model to make more accurate predictions. The test accuracy values are 92% for small images and 97% for large images, indicating better performance with higher image resolution.
The training curves of both ViT and CNN models indicate that CNN-based fine-tuned models outperform ViT models for detecting nitrogen stress in maize crop images. While ViT models demonstrate good generalization, they tend to converge prematurely, ceasing to learn once a stable loss curve is achieved. Despite their generalization ability, ViT models exhibit higher loss values, indicating potential for improvement.
Table 2 gives a comparison of train and test accuracies for the best CNN and vision transformer models for the image size 224 × 224. It can be seen that the fine-tuned CNN with EfficientNetB0 as the base model achieves higher accuracy as compared to ViT models.
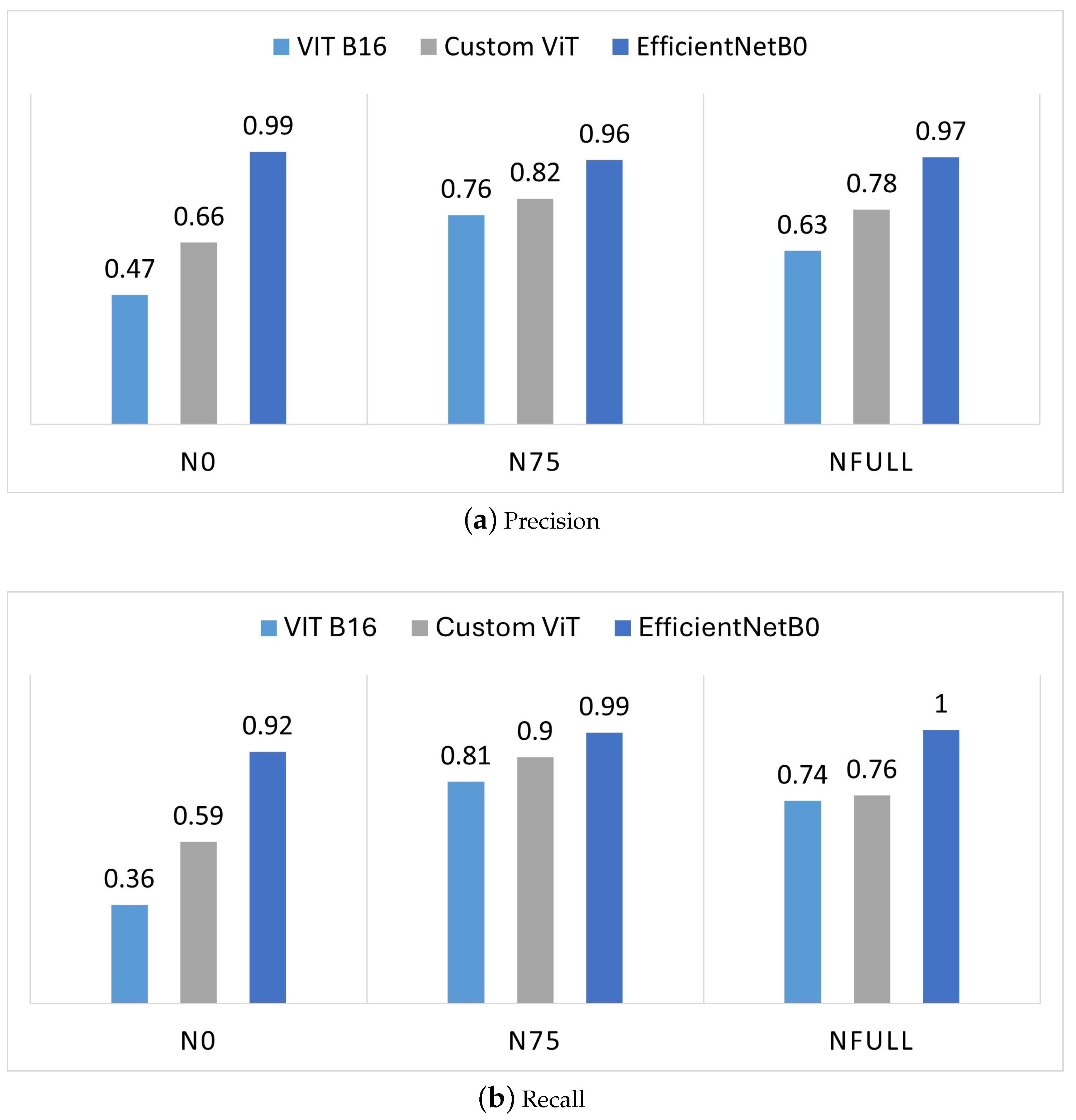
The individual class results (i.e., fertilization-levels N0, N75, and NFull) are presented in
Figure 9.
Figure 9 only presents the results for the CNN model with EfficientNetB0 as the base model, as it outperforms other CNN models (as depicted in
Table 1). For class N0, although the recall value for the CNN model (EfficientNetB0) is higher compared to the ViT models, it remains the lowest when compared to the other two classes, which have recall values of 0.99 and 1. This indicates that the CNN fine-tuned model performs better for N75 and NFull than for N0. However, the recall value for class N0 for the CNN model is still the best performing among the three classifiers for the N0 class. The fine-tuned ViT model shows both precision and recall values of less than 50% for the N0 class, indicating poor performance in identifying N0 samples. However, the fine-tuned ViT model demonstrates better recall for the N75 class as compared to the other classes, with the highest value of 0.81. The custom ViT model has moderate recall and precision values for the N0 class, suggesting there is room for improvement as compared to the other classes, where it performs relatively better. For N75, the custom ViT shows the best performance with a recall of 0.9 and precision of 0.82, both of which are higher than the values for the other two classes (N0 and NFull). This indicates that the custom ViT is particularly effective at classifying the N75 class compared to the other classes. Overall, the custom ViT outperforms the fine-tuned ViT for all the three classes; however, the fine-tuned CNN model with EfficientNetB0 as the base outperforms both ViTs for all the classes.
6. Conclusions
The current research uses computer vision-based analysis techniques and deep learning models to assess the nitrogen status of maize crops. In this study, we have employed two distinct approaches to develop models for nitrogen fertilization-level classification task. The first approach involves constructing a vision transformer (ViT) model from scratch. This custom-built model has been designed and trained entirely on our dataset, allowing us to tailor the architecture and training process specifically to our needs. The second approach utilizes transfer learning, leveraging a pre-trained ViT model and four widely implemented CNN models. These pre-trained models have been fine-tuned on our dataset, allowing us to benefit from the rich feature representations learned from large-scale datasets like ImageNet. By adding custom layers on top of the pre-trained base and fine-tuning the models, we have enhanced the models’ effectiveness by combining the generalization capabilities of the pre-trained models with the specific characteristics of our dataset. Additionally, we have tested two image resolutions, 100 × 100 and 224 × 224, to observe the relationship between image resolution and model performance. Our findings indicate that the performance of all the models is significantly affected by image size, with larger images (224 × 224) yielding better results as compared to smaller images (100 × 100). Furthermore, through a comparative analysis of fine-tuned and custom-built ViT models against fine-tuned convolutional neural networks (CNNs), we observe that CNN models outperform ViT models at both image resolutions. Specifically, the CNN model with EfficientNetB0 as the base successfully classifies the crops into stressed, non-stressed, and semi-stressed classes, achieving a best test accuracy of 97%. Even though the final training and validation losses for vision transformer models are consistent, the overall loss values are large. These results indicate that while vision transformers slowly learn and generalize well to the data, to reduce loss values and increase accuracy, further optimization is needed, which might be achieved with a larger number of data samples.
The current research on nitrogen deficiency detection using RGB images is a critical step towards the long-term goal of developing a real-time precision agriculture system for targeted nitrogen fertilizer application. Our research focuses on using only RGB images as compared to hyperspectral imagery, which is costly and less suitable for real-time implementation. The superior performance of EfficientNetB0 suggests its potential for real-time decision-making in precision agriculture applications. Vision transformers require a large number of data samples and significantly more training time and parameters, suggesting their somewhat lower suitability for typical agricultural datasets (particularly for nitrogen stress identification), which are usually smaller. After a detailed comparison and analysis, it is concluded that CNNs are more practical for real-time agricultural applications, and the vision transformers need further refinement for agricultural datasets.
Author Contributions
Conceptualization, A.M.; methodology, S.G., N.K. and A.M.; software, S.G. and N.K.; validation, S.G. and N.K.; formal analysis, S.G. and N.K.; investigation, A.M.; resources, A.M.; data curation, S.G. and N.K.; writing—original draft preparation, S.G. and N.K.; writing—review and editing, A.M.; visualization, S.G. and N.K.; supervision, A.M.; project administration, A.M.; funding acquisition, A.M. All authors have read and agreed to the published version of the manuscript.
Funding
This work was supported in part by the United States Department of Agriculture (USDA) National Institute of Food and Agriculture (NIFA), Award Number 2023-67021-40614. Any opinions, findings, and conclusions or recommendations expressed in this material are those of the author(s) and do not necessarily reflect the views of the USDA NIFA.
Institutional Review Board Statement
Not Applicable.
Informed Consent Statement
Not Applicable.
Data Availability Statement
Conflicts of Interest
The authors declare no conflicts of interest.
Abbreviations
| RGB | Red, green, blue |
| NUE | Nitrogen use efficiency |
| Chlorophyll index green |
| GNDVI | Green normalized difference vegetation index |
| RENDVI | Red edge normalized difference vegetation index |
| RDVI | Renormalized difference vegetation index |
| Coefficient of determination |
| RMSE | Root mean squared error |
| ReLU | Rectified linear unit |
| CNN | Convolutional neural network |
| SVM | Support vector machine |
| RCNN | Region-based convolutional neural networks |
| DSLR | Digital single-lens reflex |
| ViT | Vision transformer |
| MLP | Multilayer perceptron |
References
- Ritchie, H. Excess Fertilizer Use: Which Countries Cause Environmental Damage by Overapplying Fertilizers? 2021. Available online: https://ourworldindata.org/excess-fertilizer (accessed on 12 July 2024).
- Sainju, U.M.; Ghimire, R.; Pradhan, G.P. Nitrogen Fertilization I: Impact on Crop, Soil, and Environment. In Nitrogen Fixation; Rigobelo, E.C., Serra, A.P., Eds.; IntechOpen: Rijeka, Croatia, 2019; Chapter 5. [Google Scholar] [CrossRef]
- Nitrogen Deficiency in Crops: How to Detect & Fix It. 2021. Available online: https://eos.com/blog/nitrogen-deficiency/ (accessed on 31 May 2024).
- Shi, J.-Y.; Zou, X.-B.; Zhao, J.-W.; Wang, K.-L.; Chen, Z.-W.; Huang, X.-W.; Zhang, D.-T.; Holmes, M. Nondestructive diagnostics of nitrogen deficiency by cucumber leaf chlorophyll distribution map based on near infrared hyperspectral imaging. Sci. Hortic. 2012, 138, 190–197. [Google Scholar] [CrossRef]
- Sanaeifar, A.; Yang, C.; Min, A.; Jones, C.R.; Michaels, T.E.; Krueger, Q.J.; Barnes, R.; Velte, T.J. Noninvasive Early Detection of Nutrient Deficiencies in Greenhouse-Grown Industrial Hemp Using Hyperspectral Imaging. Remote Sens. 2024, 16, 187. [Google Scholar] [CrossRef]
- Liu, N.; Townsend, P.A.; Naber, M.R.; Bethke, P.C.; Hills, W.B.; Wang, Y. Hyperspectral imagery to monitor crop nutrient status within and across growing seasons. Remote Sens. Environ. 2021, 255, 112303. [Google Scholar] [CrossRef]
- Yamashita, H.; Sonobe, R.; Hirono, Y.; Morita, A.; Ikka, T. Dissection of hyperspectral reflectance to estimate nitrogen and chlorophyll contents in tea leaves based on machine learning algorithms. Sci. Rep. 2020, 10, 17360. [Google Scholar] [CrossRef]
- Wang, S.; Guan, K.; Wang, Z.; Ainsworth, E.A.; Zheng, T.; Townsend, P.A.; Liu, N.; Nafziger, E.; Masters, M.D.; Li, K.; et al. Airborne hyperspectral imaging of nitrogen deficiency on crop traits and yield of maize by machine learning and radiative transfer modeling. Int. J. Appl. Earth Obs. Geoinf. 2021, 105, 102617. [Google Scholar] [CrossRef]
- Cilia, C.; Panigada, C.; Rossini, M.; Meroni, M.; Busetto, L.; Amaducci, S.; Boschetti, M.; Picchi, V.; Colombo, R. Nitrogen Status Assessment for Variable Rate Fertilization in Maize through Hyperspectral Imagery. Remote Sens. 2014, 6, 6549–6565. [Google Scholar] [CrossRef]
- Wu, L.; Gong, Y.; Bai, X.; Wang, W.; Wang, Z. Nondestructive Determination of Leaf Nitrogen Content in Corn by Hyperspectral Imaging Using Spectral and Texture Fusion. Appl. Sci. 2023, 13, 1910. [Google Scholar] [CrossRef]
- Ram, B.G.; Oduor, P.; Igathinathane, C.; Howatt, K.; Sun, X. A systematic review of hyperspectral imaging in precision agriculture: Analysis of its current state and future prospects. Comput. Electron. Agric. 2024, 222, 109037. [Google Scholar] [CrossRef]
- Burns, B.W.; Green, V.S.; Hashem, A.A.; Massey, J.H.; Shew, A.M.; Adviento-Borbe, M.A.A.; Milad, M. Determining nitrogen deficiencies for maize using various remote sensing indices. Precis. Agric. 2022, 23, 791–811. [Google Scholar] [CrossRef]
- Zheng, H.; Li, W.; Jiang, J.; Liu, Y.; Cheng, T.; Tian, Y.; Zhu, Y.; Cao, W.; Zhang, Y.; Yao, X. A Comparative Assessment of Different Modeling Algorithms for Estimating Leaf Nitrogen Content in Winter Wheat Using Multispectral Images from an Unmanned Aerial Vehicle. Remote Sens. 2018, 10, 2026. [Google Scholar] [CrossRef]
- Cao, Q.; Yang, G.; Duan, D.; Chen, L.; Wang, F.; Xu, B.; Zhao, C.; Niu, F. Combining multispectral and hyperspectral data to estimate nitrogen status of tea plants (Camellia sinensis (L.) O. Kuntze) under field conditions. Comput. Electron. Agric. 2022, 198, 107084. [Google Scholar] [CrossRef]
- Azimi, S.; Kaur, T.; Gandhi, T.K. A deep learning approach to measure stress level in plants due to Nitrogen deficiency. Measurement 2021, 173, 108650. [Google Scholar] [CrossRef]
- Zermas, D.; Nelson, H.J.; Stanitsas, P.; Morellas, V.; Mulla, D.J.; Papanikolopoulos, N. A Methodology for the Detection of Nitrogen Deficiency in Corn Fields Using High-Resolution RGB Imagery. IEEE Trans. Autom. Sci. Eng. 2021, 18, 1879–1891. [Google Scholar] [CrossRef]
- Zhang, J.; Xie, T.; Yang, C.; Song, H.; Jiang, Z.; Zhou, G.; Zhang, D.; Feng, H.; Xie, J. Segmenting Purple Rapeseed Leaves in the Field from UAV RGB Imagery Using Deep Learning as an Auxiliary Means for Nitrogen Stress Detection. Remote Sens. 2020, 12, 1403. [Google Scholar] [CrossRef]
- Haider, T.; Farid, M.S.; Mahmood, R.; Ilyas, A.; Khan, M.H.; Haider, S.T.A.; Chaudhry, M.H.; Gul, M. A Computer-Vision-Based Approach for Nitrogen Content Estimation in Plant Leaves. Agriculture 2021, 11, 766. [Google Scholar] [CrossRef]
- Salaić, M.; Novoselnik, F.; Žarko, I.P.; Galić, V. Nitrogen deficiency in maize: Annotated image classification dataset. Data Brief 2023, 50, 109625. [Google Scholar] [CrossRef] [PubMed]
- Galic, V.; Podnar Žarko, I.; Novoselnik, F.; Salaic, M. Nitrogen deficiency in maize: Annotated image classification dataset. Mendeley Data 2023. [Google Scholar] [CrossRef]
- Dosovitskiy, A.; Beyer, L.; Kolesnikov, A.; Weissenborn, D.; Zhai, X.; Unterthiner, T.; Dehghani, M.; Minderer, M.; Heigold, G.; Gelly, S.; et al. An Image is Worth 16 × 16 Words: Transformers for Image Recognition at Scale. In Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition, New Orleans, LA, USA, 18–24 June 2021; pp. 3156–3164. [Google Scholar]
- Tan, M.; Le, Q.V. EfficientNet: Rethinking Model Scaling for Convolutional Neural Networks. arXiv 2019, arXiv:1905.11946. [Google Scholar]
- Huang, G.; Liu, Z.; van der Maaten, L.; Weinberger, K.Q. Densely Connected Convolutional Networks. arXiv 2018, arXiv:1608.06993. [Google Scholar]
- Szegedy, C.; Vanhoucke, V.; Ioffe, S.; Shlens, J.; Wojna, Z. Rethinking the Inception Architecture for Computer Vision. arXiv 2015, arXiv:1512.00567. [Google Scholar]
- He, K.; Zhang, X.; Ren, S.; Sun, J. Identity Mappings in Deep Residual Networks. In Proceedings of the Computer Vision—ECCV 2016, Amsterdam, The Netherlands, 11–14 October 2016; Leibe, B., Matas, J., Sebe, N., Welling, M., Eds.; Springer: Cham, Switzerland, 2016; pp. 630–645. [Google Scholar]
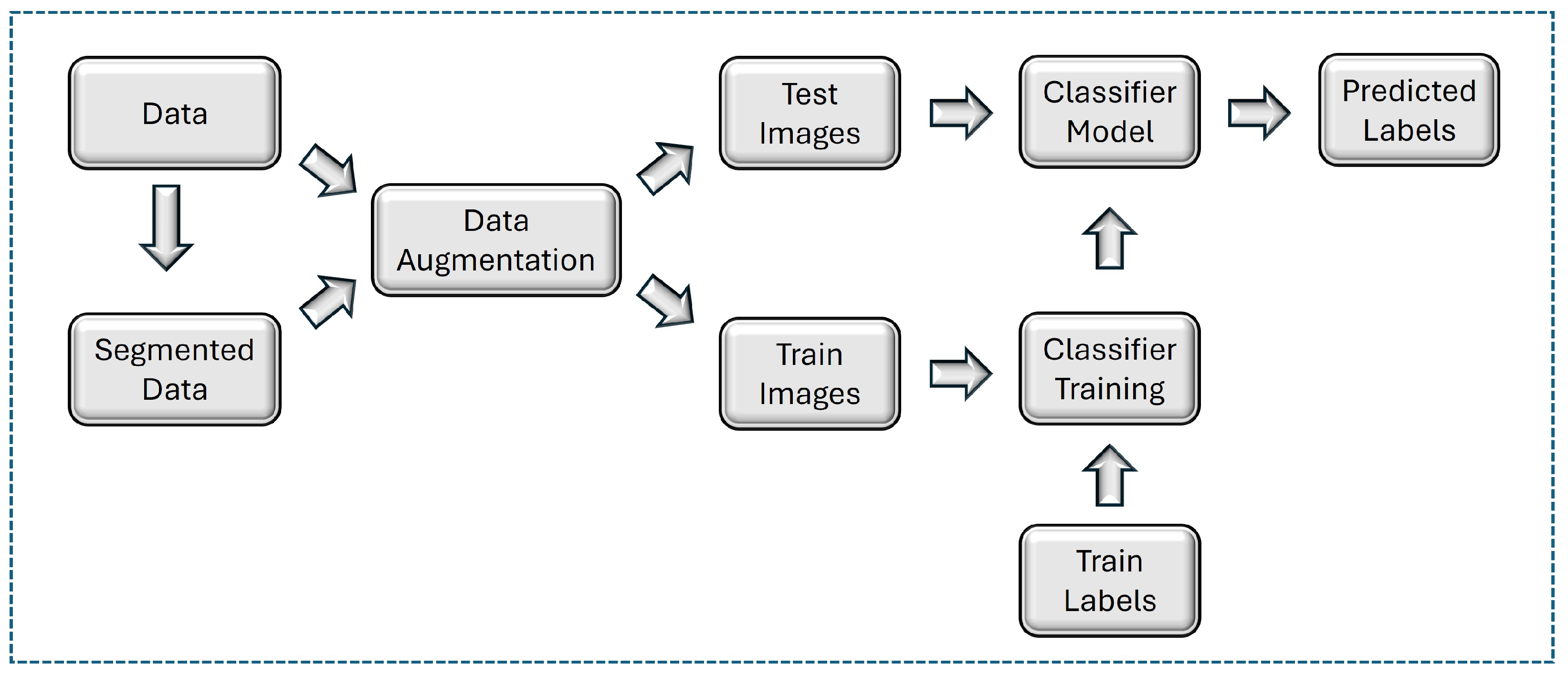
Figure 1.
Flowchart of proposed methodology for nitrogen stress detection.
Figure 1.
Flowchart of proposed methodology for nitrogen stress detection.
Figure 2.
Samples of maize images: (a) sample image from class N0 with no fertilization, (b) sample image from class N75 with 75 kg of fertilizer applied, (c) sample image from class NFull with 136 kg of fertilizer applied, (d–f) same images as (a–c) after segmentation.
Figure 2.
Samples of maize images: (a) sample image from class N0 with no fertilization, (b) sample image from class N75 with 75 kg of fertilizer applied, (c) sample image from class NFull with 136 kg of fertilizer applied, (d–f) same images as (a–c) after segmentation.
Figure 3.
Architecture of our custom-built vision transformer model to classify three levels of nitrogen fertilization (i.e., N0, N75, and NFull).
Figure 3.
Architecture of our custom-built vision transformer model to classify three levels of nitrogen fertilization (i.e., N0, N75, and NFull).
Figure 4.
Architecture of the fine-tuned vision transformer model to classify three levels of nitrogen fertilization (i.e., N0, N75, and NFull).
Figure 4.
Architecture of the fine-tuned vision transformer model to classify three levels of nitrogen fertilization (i.e., N0, N75, and NFull).
Figure 5.
Architecture of fine-tuned CNN-based model to classify three levels of nitrogen fertilization (i.e., N0, N75, and NFull).
Figure 5.
Architecture of fine-tuned CNN-based model to classify three levels of nitrogen fertilization (i.e., N0, N75, and NFull).
Figure 6.
Custom-built vision transformer model results for train and validation losses for two image resolutions (100 × 100 and 224 × 224) for nitrogen fertilization-level prediction.
Figure 6.
Custom-built vision transformer model results for train and validation losses for two image resolutions (100 × 100 and 224 × 224) for nitrogen fertilization-level prediction.
Figure 7.
Fine-tuned vision transformer model results for train and validation losses for image resolution 224 × 224 for nitrogen fertilization-level prediction.
Figure 7.
Fine-tuned vision transformer model results for train and validation losses for image resolution 224 × 224 for nitrogen fertilization-level prediction.
Figure 8.
Fine-tuned CNN best model results for train and validation losses for two image resolutions for nitrogen fertilization-level prediction.
Figure 8.
Fine-tuned CNN best model results for train and validation losses for two image resolutions for nitrogen fertilization-level prediction.
Figure 9.
Classification results for individual fertilization-level classes for image resolution 224 × 224.
Figure 9.
Classification results for individual fertilization-level classes for image resolution 224 × 224.
Table 1.
Comparison of test accuracies of fine-tuned CNN classifier models for two image resolutions (100 × 100 and 224 × 224) for nitrogen fertilization-level prediction.
Table 1.
Comparison of test accuracies of fine-tuned CNN classifier models for two image resolutions (100 × 100 and 224 × 224) for nitrogen fertilization-level prediction.
| CNN-Based Model | Train Accuracy (%) | Test Accuracy (%) |
|---|
| 100 × 100 | 224 × 224 | 100 × 100 | 224 × 224 |
|---|
| EfficientNetB0 | 99 | 99.7 | 92 | 97 |
| DenseNet121 | 98 | 98 | 90 | 93 |
| InceptionV3 | 76 | 99 | 74 | 91 |
| ResNet50 | 84 | 90 | 78 | 84.5 |
Table 2.
Comparison of train and test accuracies of vision transformer models and CNN best model for image resolution 224×224 for nitrogen fertilization-level prediction.
Table 2.
Comparison of train and test accuracies of vision transformer models and CNN best model for image resolution 224×224 for nitrogen fertilization-level prediction.
| Architecture | Train Accuracy (%) | Test Accuracy (%) |
|---|
| ViT (Custom) | 87 | 76 |
| Fine-tuned ViT (VIT-b/16) | 71 | 69 |
| CNN (EfficientNetB0) | 99.7 | 97 |
| Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).