In Vitro Silencing of MHC-I in Keratinocytes by Herpesvirus US11 Protein to Model Alloreactive Suppression
Abstract
1. Introduction
2. Materials and Methods
2.1. Cell Culture
2.2. Recombinant US11 Proteins and Stimulation Procedure
2.3. Western Blotting
2.4. IFN-γ Enzyme-Linked Immunosorbent Assay (ELISA)
2.5. Flow Cytometry
2.6. Skin Tissue Sampling and Immunofluorescence
3. Results
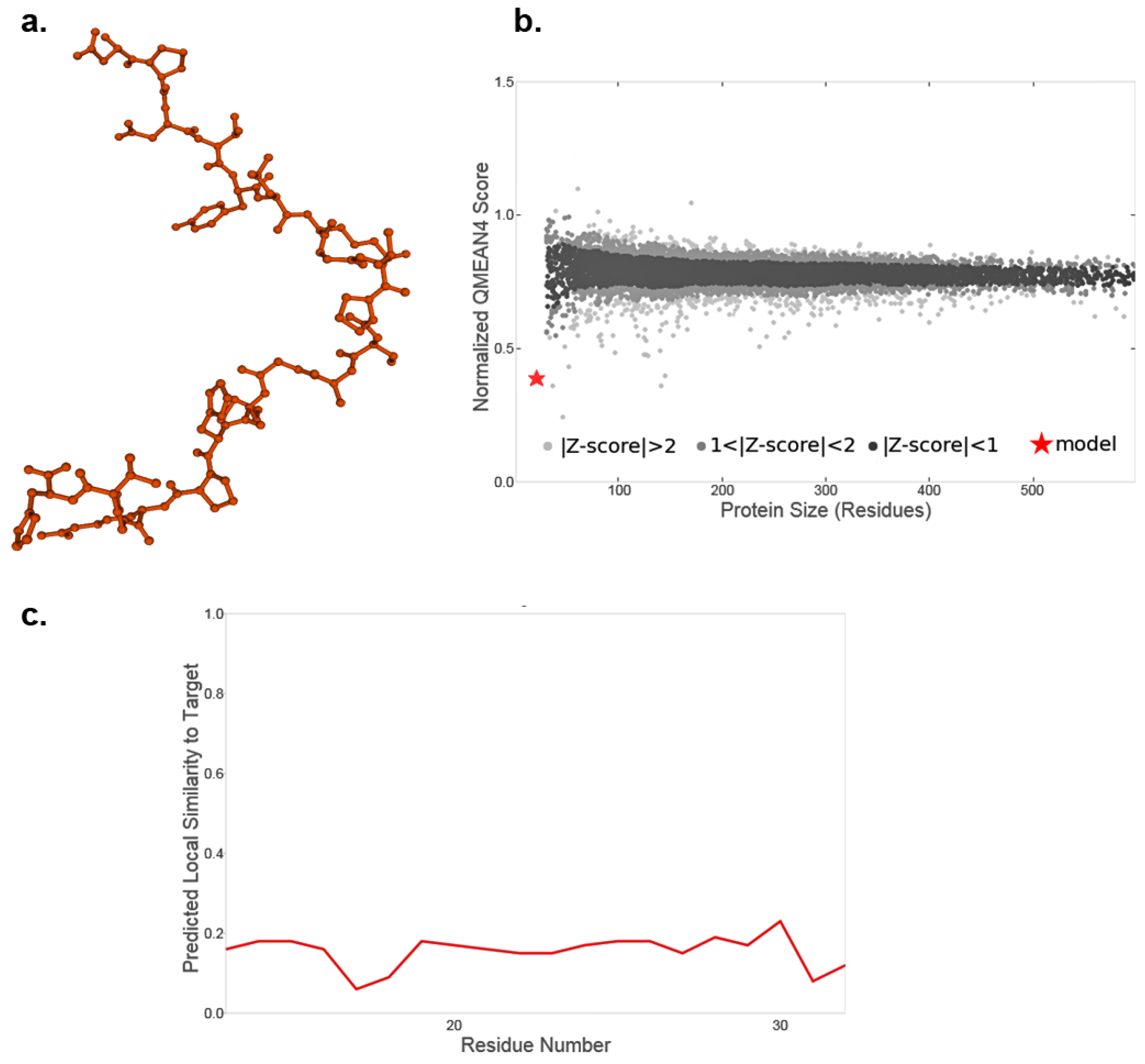
3.1. Recombinant US11 Proteins and Potential Three-Dimensional Protein Structure
3.2. Stimulation of Primary Keratinocytes with Recombinant US11 Protein
3.3. Treatment of Human Allogeneic Skin Tissue with US11 Protein
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| β2m | β2-microglobulin |
| CD | Cluster of differentiation |
| DAPI | 4′,6-Diamidino-2-phenylindol |
| ELISA | Enzyme-linked immunosorbent assay |
| ELiSpot | Enzyme-linked immunosorbent spot |
| GMQE | Global model quality estimate |
| HCMV | Human cytomegalovirus |
| HHV | Human herpes virus |
| HLA | Human leukocyte antigen |
| IgG | Immunoglobulin G |
| IFN-γ | Interferon gamma |
| MHC-I | Major histocompatibility complex class I |
| NK | Natural killer cells |
| PBMC | Peripheral blood mononuclear cells |
| PBS | Phosphate-buffered saline |
| PVDF | Polyvinylidene fluoride membrane |
| RNA | Ribonucleic acid |
| SDS-PAGE | Sodium dodecyl sulfate-polyacrylamide gel electrophoresis |
| US | Unique short glycoprotein |
| QMEAN | Qualitative model energy analysis |
References
- Smolle, C.; Cambiaso-Daniel, J.; Forbes, A.A.; Wurzer, P.; Hundeshagen, G.; Branski, L.K.; Huss, F.; Kamolz, L.-P. Recent Trends in Burn Epidemiology Worldwide: A Systematic Review. Burns 2017, 43, 249–257. [Google Scholar] [CrossRef] [PubMed]
- Rosenbaum, A.J.; Banerjee, S.; Rezak, K.M.; Uhl, R.L. Advances in Wound Management. J. Am. Acad. Orthop. Surg. 2018, 26, 833–843. [Google Scholar] [CrossRef] [PubMed]
- Janis, J.E.; Kwon, R.K.; Attinger, C.E. The New Reconstructive Ladder: Modifications to the Traditional Model. Plast. Reconstr. Surg. 2011, 127, 205S–212S. [Google Scholar] [CrossRef] [PubMed]
- Schlottmann, F.; Bucan, V.; Vogt, P.M.; Krezdorn, N. A Short History of Skin Grafting in Burns: From the Gold Standard of Autologous Skin Grafting to the Possibilities of Allogeneic Skin Grafting with Immunomodulatory Approaches. Medicina 2021, 57, 225. [Google Scholar] [CrossRef]
- Guogienė, I.; Kievišas, M.; Grigaitė, A.; Braziulis, K.; Rimdeika, R. Split-Thickness Skin Grafting: Early Outcomes of a Clinical Trial Using Different Graft Thickness. J. Wound Care 2018, 27, 5–13. [Google Scholar] [CrossRef]
- Wang, C.; Zhang, F.; Lineaweaver, W.C. Clinical Applications of Allograft Skin in Burn Care. Ann. Plast. Surg. 2020, 84, S158–S160. [Google Scholar] [CrossRef]
- Cronin, H.; Goldstein, G. Biologic Skin Substitutes and Their Applications in Dermatology. Dermatol. Surg. 2013, 39, 30–34. [Google Scholar] [CrossRef]
- Horch, R.; Stark, G.B.; Kopp, J.; Spilker, G. Cologne Burn Centre Experiences with Glycerol-Preserved Allogeneic Skin: Part I: Clinical Experiences and Histological Findings (Overgraft and Sandwich Technique). Burns 1994, 20 (Suppl. S1), S23–S26. [Google Scholar] [CrossRef]
- Horner, B.M.; Randolph, M.A.; Huang, C.A.; Butler, P.E.M. Skin Tolerance: In Search of the Holy Grail. Transpl. Int. 2008, 21, 101–112. [Google Scholar] [CrossRef]
- Benichou, G.; Yamada, Y.; Yun, S.-H.; Lin, C.; Fray, M.; Tocco, G. Immune Recognition and Rejection of Allogeneic Skin Grafts. Immunotherapy 2011, 3, 757–770. [Google Scholar] [CrossRef]
- Benichou, G.; Takizawa, P.A.; Olson, C.A.; McMillan, M.; Sercarz, E.E. Donor Major Histocompatibility Complex (MHC) Peptides Are Presented by Recipient MHC Molecules during Graft Rejection. J. Exp. Med. 1992, 175, 305–308. [Google Scholar] [CrossRef] [PubMed]
- Natarajan, K.; Li, H.; Mariuzza, R.A.; Margulies, D.H. MHC Class I Molecules, Structure and Function. Rev. Immunogenet. 1999, 1, 32–46. Available online: http://www.ncbi.nlm.nih.gov/pubmed/11256571 (accessed on 19 December 2022). [PubMed]
- Lanza, R.; Russell, D.W.; Nagy, A. Engineering Universal Cells That Evade Immune Detection. Nat. Rev. Immunol. 2019, 19, 723–733. [Google Scholar] [CrossRef] [PubMed]
- Tavakoli, S.; Klar, A.S. Bioengineered Skin Substitutes: Advances and Future Trends. Appl. Sci. 2021, 11, 1493. [Google Scholar] [CrossRef]
- Schlottmann, F.; Strauss, S.; Hake, K.; Vogt, P.M.; Bucan, V. Down-Regulation of MHC Class I Expression in Human Keratinocytes Using Viral Vectors Containing US11 Gene of Human Cytomegalovirus and Cultivation on Bovine Collagen-Elastin Matrix (Matriderm®): Potential Approach for an Immune-Privileged Skin Substitute. Int. J. Mol. Sci. 2019, 20, 2056. [Google Scholar] [CrossRef]
- Figueiredo, C.; Blasczyk, R. A Future with Less HLA: Potential Clinical Applications of HLA-Universal Cells. Tissue Antigens 2015, 85, 443–449. [Google Scholar] [CrossRef]
- Opelz, G.; Wujciak, T. The Influence of HLA Compatibility on Graft Survival after Heart Transplantation. The Collaborative Transplant Study. N. Engl. J. Med. 1994, 330, 816–819. [Google Scholar] [CrossRef]
- Opelz, G. Factors Influencing Long-Term Graft Loss. Transpl. Proc. 2000, 32, 647–649. [Google Scholar] [CrossRef]
- Opelz, G.; Wujciak, T.; Back, D.; Mytilineos, J.; Schwarz, V.; Albrecht, G. Effect of HLA Compatibility on Kidney Transplantation. Infusionsther. Transfusionsmed. 1994, 21, 198–202. Available online: http://www.ncbi.nlm.nih.gov/pubmed/7919908 (accessed on 19 December 2022).
- Hou, L.; Enriquez, E.; Persaud, M.; Steiner, N.; Oudshoorn, M.; Hurley, C.K. Next Generation Sequencing Characterizes HLA Diversity in a Registry Population from the Netherlands. HLA 2019, 93, 474–483. [Google Scholar] [CrossRef]
- Christinck, E.R.; Luscher, M.A.; Barber, B.H.; Williams, D.B. Peptide Binding to Class I MHC on Living Cells and Quantitation of Complexes Required for CTL Lysis. Nature 1991, 352, 67–70. [Google Scholar] [CrossRef] [PubMed]
- Sykulev, Y.; Joo, M.; Vturina, I.; Tsomides, T.J.; Eisen, H.N. Evidence That a Single Peptide–MHC Complex on a Target Cell Can Elicit a Cytolytic T Cell Response. Immunity 1996, 4, 565–571. [Google Scholar] [CrossRef] [PubMed]
- Timonen, T.; Helander, T.S. Natural Killer Cell—Target Cell Interactions. Curr. Opin. Cell Biol. 1997, 9, 667–673. [Google Scholar] [CrossRef] [PubMed]
- Shimizu, Y.; DeMars, R. Demonstration by Class I Gene Transfer That Reduced Susceptibility of Human Cells to Natural Killer Cell-Mediated Lysis Is Inversely Correlated with HLA Class I Antigen Expression. Eur. J. Immunol. 1989, 19, 447–451. [Google Scholar] [CrossRef]
- Fisher, J.D.; Acharya, A.P.; Little, S.R. Micro and Nanoparticle Drug Delivery Systems for Preventing Allotransplant Rejection. Clin. Immunol. 2015, 160, 24–35. [Google Scholar] [CrossRef]
- Weimer, R.; Melk, A.; Daniel, V.; Friemann, S.; Padberg, W.; Opelz, G. Switch from Cyclosporine A to Tacrolimus in Renal Transplant Recipients: Impact on Th1, Th2, and Monokine Responses. Hum. Immunol. 2000, 61, 884–897. [Google Scholar] [CrossRef]
- Vogt, P.M.; Thompson, S.; Andree, C.; Liu, P.; Breuing, K.; Hatzis, D.; Brown, H.; Mulligan, R.C.; Eriksson, E. Genetically Modified Keratinocytes Transplanted to Wounds Reconstitute the Epidermis. Proc. Natl. Acad. Sci. USA 1994, 91, 9307–9311. [Google Scholar] [CrossRef]
- Vogt, P.M.; Eriksson, E. Current Aspects of Epidermal Wound Healing. Handchir. Mikrochir. Plast. Chir. 1992, 24, 259–266. Available online: http://www.ncbi.nlm.nih.gov/pubmed/1427467 (accessed on 19 December 2022).
- Rong, Z.; Wang, M.; Hu, Z.; Stradner, M.; Zhu, S.; Kong, H.; Yi, H.; Goldrath, A.; Yang, Y.-G.; Xu, Y.; et al. An Effective Approach to Prevent Immune Rejection of Human ESC-Derived Allografts. Cell Stem Cell 2014, 14, 121–130. [Google Scholar] [CrossRef]
- Ljunggren, H.-G.; Van Kaer, L.; Sabatine, M.S.; Auchincloss, H.; Tonegawa, S.; Ploegh, H.L. MHC Class I Expression and CD8 + T Cell Development in TAP1/β 2 -Microglobulin Double Mutant Mice. Int. Immunol. 1995, 7, 975–984. [Google Scholar] [CrossRef]
- Zijlstra, M.; Auchincloss, H.; Loring, J.M.; Chase, C.M.; Russell, P.S.; Jaenisch, R. Skin Graft Rejection by Beta 2-Microglobulin-Deficient Mice. J. Exp. Med. 1992, 175, 885–893. [Google Scholar] [CrossRef]
- Markmann, J.F.; Bassiri, H.; Desai, N.M.; Odorico, J.S.; Kim, J.I.; Koller, B.H.; Smithies, O.; Barker, C.F. Indefinite Survival of MHC Class I-Deficient Murine Pancreatic Islet Allografts. Transplantation 1992, 54, 1085–1089. [Google Scholar] [CrossRef]
- Qian, S.; Fu, F.; Li, Y.; Lu, L.; Rao, A.S.; Strazl, T.E.; Thomson, A.W.; Fung, J.J. Impact of Donor MHC Class I or Class II Antigen Deficiency on First- and Second-set Rejection of Mouse Heart or Liver Allografts. Immunology 1996, 88, 124–129. [Google Scholar] [CrossRef]
- Mhashilkar, A.; Doebis, C.; Seifert, M.; Busch, A.; Zani, C.; Hoo, J.S.; Nagy, M.; Ritter, T.; Volk, H.-D.; Marasco, W. Intrabody-Mediated Phenotypic Knockout of Major Histocompatibility Complex Class I Expression in Human and Monkey Cell Lines and in Primary Human Keratinocytes. Gene Ther. 2002, 9, 307–319. [Google Scholar] [CrossRef][Green Version]
- Loenen, W.A.; Bruggeman, C.A.; Wiertz, E.J. Immune Evasion by Human Cytomegalovirus: Lessons in Immunology and Cell Biology. Semin. Immunol. 2001, 13, 41–49. [Google Scholar] [CrossRef]
- Jackson, S.E.; Mason, G.M.; Wills, M.R. Human Cytomegalovirus Immunity and Immune Evasion. Virus Res. 2011, 157, 151–160. [Google Scholar] [CrossRef] [PubMed]
- Jones, T.R.; Hanson, L.K.; Sun, L.; Slater, J.S.; Stenberg, R.M.; Campbell, A.E. Multiple Independent Loci within the Human Cytomegalovirus Unique Short Region Down-Regulate Expression of Major Histocompatibility Complex Class I Heavy Chains. J. Virol. 1995, 69, 4830–4841. [Google Scholar] [CrossRef] [PubMed]
- Wiertz, E.J.; Jones, T.R.; Sun, L.; Bogyo, M.; Geuze, H.J.; Ploegh, H.L. The Human Cytomegalovirus US11 Gene Product Dislocates MHC Class I Heavy Chains from the Endoplasmic Reticulum to the Cytosol. Cell 1996, 84, 769–779. [Google Scholar] [CrossRef] [PubMed]
- Shamu, C.E.; Story, C.M.; Rapoport, T.A.; Ploegh, H.L. The Pathway of US11-Dependent Degradation of MHC Class I Heavy Chains Involves a Ubiquitin-Conjugated Intermediate. J. Cell Biol. 1999, 147, 45–58. [Google Scholar] [CrossRef]
- Story, C.M.; Furman, M.H.; Ploegh, H.L. The Cytosolic Tail of Class I MHC Heavy Chain Is Required for Its Dislocation by the Human Cytomegalovirus US2 and US11 Gene Products. Proc. Natl. Acad. Sci. USA 1999, 96, 8516–8521. [Google Scholar] [CrossRef]
- Basta, S.; Chen, W.; Bennink, J.R.; Yewdell, J.W. Inhibitory Effects of Cytomegalovirus Proteins US2 and US11 Point to Contributions from Direct Priming and Cross-Priming in Induction of Vaccinia Virus-Specific CD8+ T Cells. J. Immunol. 2002, 168, 5403–5408. [Google Scholar] [CrossRef] [PubMed]
- Farrell, H.E.; Vally, H.; Lynch, D.M.; Fleming, P.; Shellam, G.R.; Scalzo, A.A.; Davis-Poynter, N.J. Inhibition of Natural Killer Cells by a Cytomegalovirus MHC Class I Homologue in Vivo. Nature 1997, 386, 510–514. [Google Scholar] [CrossRef] [PubMed]
- Falk, C.S.; Mach, M.; Schendel, D.J.; Weiss, E.H.; Hilgert, I.; Hahn, G. NK Cell Activity During Human Cytomegalovirus Infection Is Dominated by US2–11-Mediated HLA Class I Down-Regulation. J. Immunol. 2002, 169, 3257–3266. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.; Li, W.; Ma, N.; Steinhoff, G. Non-Viral Gene Delivery Methods. Curr. Pharm. Biotechnol. 2013, 14, 46–60. [Google Scholar] [CrossRef]
- Overton, T.W. Recombinant Protein Production in Bacterial Hosts. Drug Discov. Today 2014, 19, 590–601. [Google Scholar] [CrossRef]
- Couto, L.B.; High, K.A. Viral Vector-Mediated RNA Interference. Curr. Opin. Pharmacol. 2010, 10, 534–542. [Google Scholar] [CrossRef]
- Sanchez-Garcia, L.; Martín, L.; Mangues, R.; Ferrer-Miralles, N.; Vázquez, E.; Villaverde, A. Recombinant Pharmaceuticals from Microbial Cells: A 2015 Update. Microb. Cell Factories 2016, 15, 33. [Google Scholar] [CrossRef]
- Bienert, S.; Waterhouse, A.; de Beer, T.A.P.; Tauriello, G.; Studer, G.; Bordoli, L.; Schwede, T. The SWISS-MODEL Repository—New Features and Functionality. Nucleic Acids Res. 2017, 45, D313–D319. [Google Scholar] [CrossRef]
- Waterhouse, A.; Bertoni, M.; Bienert, S.; Studer, G.; Tauriello, G.; Gumienny, R.; Heer, F.T.; de Beer, T.A.P.; Rempfer, C.; Bordoli, L.; et al. SWISS-MODEL: Homology Modelling of Protein Structures and Complexes. Nucleic Acids Res. 2018, 46, W296–W303. [Google Scholar] [CrossRef]
- Huang, Y.; Wuyunerdeni; Yao, S. Expression, Purification of Herpes Simplex Virus Type 1 US11 Protein, and Production of US11 Polyclonal Antibody. Virol. J. 2011, 8, 490. [Google Scholar] [CrossRef]
- Schaerer-Uthurralt, N.; Erard, M.; Kindbeiter, K.; Madjar, J.J.; Diaz, J.J. Distinct Domains in Herpes Simplex Virus Type 1 US11 Protein Mediate Post-Transcriptional Transactivation of Human T-Lymphotropic Virus Type I Envelope Glycoprotein Gene Expression and Specific Binding to the Rex Responsive Element. J. Gen. Virol. 1998, 79 Pt 7, 1593–1602. [Google Scholar] [CrossRef] [PubMed]
- Attrill, H.L.; Cumming, S.A.; Clements, J.B.; Graham, S.V. The Herpes Simplex Virus Type 1 US11 Protein Binds the Coterminal UL12, UL13, and UL14 RNAs and Regulates UL13 Expression In Vivo. J. Virol. 2002, 76, 8090–8100. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Bryant, K.F. Binding of Herpes Simplex Virus-1 US11 to Specific RNA Sequences. Nucleic Acids Res. 2005, 33, 6090–6100. [Google Scholar] [CrossRef] [PubMed]
- Diaz, J.J.; Dodon, M.D.; Schaerer-Uthurralt, N.; Simonin, D.; Kindbeiter, K.; Gazzolo, L.; Madjar, J.J. Post-Transcriptional Transactivation of Human Retroviral Envelope Glycoprotein Expression by Herpes Simplex Virus Us11 Protein. Nature 1996, 379, 273–277. [Google Scholar] [CrossRef]
- AlQuraishi, M. AlphaFold at CASP13. Bioinformatics 2019, 35, 4862–4865. [Google Scholar] [CrossRef]
- Schwede, T. SWISS-MODEL: An Automated Protein Homology-Modeling Server. Nucleic Acids Res. 2003, 31, 3381–3385. [Google Scholar] [CrossRef]
- Biasini, M.; Bienert, S.; Waterhouse, A.; Arnold, K.; Studer, G.; Schmidt, T.; Kiefer, F.; Cassarino, T.G.; Bertoni, M.; Bordoli, L.; et al. SWISS-MODEL: Modelling Protein Tertiary and Quaternary Structure Using Evolutionary Information. Nucleic Acids Res. 2014, 42, W252–W258. [Google Scholar] [CrossRef]
- Fenjves, E.S. Approaches to Gene Transfer in Keratinocytes. J. Investig. Dermatol. 1994, 103, 70S–75S. [Google Scholar] [CrossRef][Green Version]
- Thomas, C.E.; Ehrhardt, A.; Kay, M.A. Progress and Problems with the Use of Viral Vectors for Gene Therapy. Nat. Rev. Genet. 2003, 4, 346–358. [Google Scholar] [CrossRef]
- Lee, E.M.; Kim, J.Y.; Cho, B.R.; Chung, W.K.; Yoon, B.W.; Kim, S.U.; Lee, B.C.; Hwang, W.S.; Moon, S.Y.; Lee, J.S.; et al. Down-Regulation of MHC Class I Expression in Human Neuronal Stem Cells Using Viral Stealth Mechanism. Biochem. Biophys. Res. Commun. 2005, 326, 825–835. [Google Scholar] [CrossRef]
- Radosevich, T.J.; Seregina, T.; Link, C.J. Effective Suppression of Class I Major Histocompatibility Complex Expression by the US11 or ICP47 Genes Can Be Limited by Cell Type or Interferon- γ Exposure. Hum. Gene Ther. 2003, 14, 1765–1775. [Google Scholar] [CrossRef] [PubMed]
- Besold, K.; Wills, M.; Plachter, B. Immune Evasion Proteins GpUS2 and GpUS11 of Human Cytomegalovirus Incompletely Protect Infected Cells from CD8 T Cell Recognition. Virology 2009, 391, 5–19. [Google Scholar] [CrossRef] [PubMed]
- Raulf-Heimsoth, M. T Cell—Primary Culture from Peripheral Blood. Methods Mol. Med. 2008, 138, 17–30. [Google Scholar] [CrossRef] [PubMed]
- Slota, M.; Lim, J.-B.; Dang, Y.; Disis, M.L. ELISpot for Measuring Human Immune Responses to Vaccines. Expert Rev. Vaccines 2011, 10, 299–306. [Google Scholar] [CrossRef]
- Ji, N.; Forsthuber, T.G. ELISPOT Techniques. Methods Mol. Biol. 2016, 1304, 63–71. [Google Scholar] [CrossRef]
- Raval, A.; Puri, N.; Rath, P.C.; Saxena, R.K. Cytokine Regulation of Expression of Class I MHC Antigens. Exp. Mol. Med. 1998, 30, 1–13. [Google Scholar] [CrossRef]
- Kim, J.Y.; Kim, D.; Choi, I.; Yang, J.S.; Lee, D.-S.; Lee, J.R.; Kang, K.; Kim, S.; Hwang, W.S.; Lee, J.S.; et al. MHC Expression in a Human Adult Stem Cell Line and Its Down-Regulation by HCMV US Gene Transfection. Int. J. Biochem. Cell Biol. 2005, 37, 69–78. [Google Scholar] [CrossRef]
- Furman, M.H.; Ploegh, H.L.; Tortorella, D. Membrane-Specific, Host-Derived Factors Are Required for US2- and US11-Mediated Degradation of Major Histocompatibility Complex Class I Molecules. J. Biol. Chem. 2002, 277, 3258–3267. [Google Scholar] [CrossRef]
- Noriega, V.M.; Hesse, J.; Gardner, T.J.; Besold, K.; Plachter, B.; Tortorella, D. Human Cytomegalovirus US3 Modulates Destruction of MHC Class I Molecules. Mol. Immunol. 2012, 51, 245–253. [Google Scholar] [CrossRef]
- Tortorella, D.; Gewurz, B.; Schust, D.; Furman, M.; Ploegh, H. Down-Regulation of MHC Class I Antigen Presentation by HCMV; Lessons for Tumor Immunology. Immunol. Investig. 2000, 29, 97–100. [Google Scholar] [CrossRef]
- Barel, M.T.; Pizzato, N.; Le Bouteiller, P.; Wiertz, E.J.H.J.; Lenfant, F. Subtle Sequence Variation among MHC Class I Locus Products Greatly Influences Sensitivity to HCMV US2- and US11-Mediated Degradation. Int. Immunol. 2006, 18, 173–182. [Google Scholar] [CrossRef][Green Version]
- Fotoran, W.L.; Santangelo, R.; de Miranda, B.N.M.; Irvine, D.J.; Wunderlich, G. DNA-Loaded Cationic Liposomes Efficiently Function as a Vaccine against Malarial Proteins. Mol. Ther. Methods Clin. Dev. 2017, 7, 1–10. [Google Scholar] [CrossRef]
- Green, J.J. Immunoengineering Has Arrived. J. Biomed. Mater. Res. A 2021, 109, 397–403. [Google Scholar] [CrossRef]



Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Published by MDPI on behalf of the European Burns Association. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Schlottmann, F.; Strauß, S.; Vogt, P.M.; Bucan, V. In Vitro Silencing of MHC-I in Keratinocytes by Herpesvirus US11 Protein to Model Alloreactive Suppression. Eur. Burn J. 2025, 6, 47. https://doi.org/10.3390/ebj6030047
Schlottmann F, Strauß S, Vogt PM, Bucan V. In Vitro Silencing of MHC-I in Keratinocytes by Herpesvirus US11 Protein to Model Alloreactive Suppression. European Burn Journal. 2025; 6(3):47. https://doi.org/10.3390/ebj6030047
Chicago/Turabian StyleSchlottmann, Frederik, Sarah Strauß, Peter Maria Vogt, and Vesna Bucan. 2025. "In Vitro Silencing of MHC-I in Keratinocytes by Herpesvirus US11 Protein to Model Alloreactive Suppression" European Burn Journal 6, no. 3: 47. https://doi.org/10.3390/ebj6030047
APA StyleSchlottmann, F., Strauß, S., Vogt, P. M., & Bucan, V. (2025). In Vitro Silencing of MHC-I in Keratinocytes by Herpesvirus US11 Protein to Model Alloreactive Suppression. European Burn Journal, 6(3), 47. https://doi.org/10.3390/ebj6030047






