Abstract
In this paper, we report the relationship between the anti-MERS-CoV activities of the HKU4 derived peptides for some peptidomimetic compounds and various descriptors using the quantitative structure activity relationships (QSAR) methods. The used descriptors were computed using ChemSketch, Marvin Sketch and ChemOffice software. The principal components analysis (PCA) and the multiple linear regression (MLR) methods were used to propose a model with reliable predictive capacity. The original data set of 41 peptidomimetic derivatives was randomly divided into training and test sets of 34 and 7 compounds, respectively. The predictive ability of the best MLR model was assessed by determination coefficient R2 = 0.691, cross-validation parameter Q2cv = 0.528 and the external validation parameter R2test = 0.794.
1. Introduction
Middle East Respiratory Syndrome (MERS) is a respiratory infection disease that emerged in Saudi Arabia in 2012 [1,2]. In addition to Saudi Arabia, Egypt, Oman and Qatar were affected by this outbreak, with a high percentage of cases (>85%) [3,4,5]. The outbreak continued its spread until 2015 to affect 27 countries in Asia. Among these countries, South Korea was the most affected with 186 confirmed cases including 38 deaths. Approximately 35% of patients with MERS have died, but this may be an overestimate of the true mortality rate [6]. MERS-CoV is a zoonotic virus, which was transmitted from animals to human reservoirs [7,8]. The virus appears to cause more severe disease in older people, people with weakened immune systems, and those with chronic diseases such as renal disease, cancer, chronic lung disease and diabetes. In 2019, 203 new cases of MERS-CoV were reported. So far, neither vaccine nor effective treatment is available for this disease. Several efforts have been made by researchers throughout the world to develop an effective therapy against MERS-CoV infection. Many previous studies have shown that the MERS-CoV possesses a single-stranded positive-sense RNA genome with 2 open reading frames (ORFs) and encodes two polyprotein precursors [9,10,11,12] which are cleaved by 3CLPro and a papain-like cysteine protease (PLPro) to generate 16 nonstructural proteins (NSP1−16) [13,14,15,16]. Thus, it represents a potential target for antiviral drug development. Nowadays, very few data are available on MERS-CoV 3CLpro inhibition by active molecules. Furthermore, HKU4-CoV 3CLpro shares a high sequence identity (81%) with the MERS-CoV enzyme and thus represents a potential surrogate model for anti-MERS drug discovery [17].
A quantitative structure−activity relationship approach attempts to explore the relationship between molecular descriptors that describe the unique physicochemical properties of the studied compounds and their respective biological activity [18]. It encodes the chemical structure through a variety of molecular descriptors, such as constitutional, topological, thermodynamic, electronic, geometrical. The development of new cheminformatics software allows the calculation of a thousand molecular descriptors [19].
This study aims to build QSAR models, which explain the relationship between anti-MERS-CoV activity and the structure of 41 peptidomimetic based on physicochemical descriptors using statistical methods. Multiple linear regression (MLR) was used for numerical characterization of the compounds based on the selected descriptors by PCA. The quality of the developed QSAR model was checked using statistical parameters and several validation methods.
2. Material and Methods
2.1. Data Set
A series of 41 peptidomimetic derivatives was studied for their anti-HKU4 activity [13]. Table 1 presents the structure and the activity values for these compounds (pIC50 = −log (IC50)). The compounds of this series were drawn using the ChemDraw, available in ChemOffice software, as shown in Table 1, and the descriptors were calculated using ChemSketch, ChemOffice and Marvin Sketch software. The studied compounds were randomly divided into a training set used to build QSAR models and a test set used to evaluate the predictive power of models, consisting of 34 and 7 compounds, respectively.

Table 1.
Chemical structures and activity experiment of 41 peptidomimetic compounds.
In QSAR studies, it is recommended that the dataset is divided into several training and test sets (5:1 ratio) [20]. In the present study, QSAR models have been built following the OECD principles for acceptable QSAR models. This approach led to the generation of QSAR models possessing excellent statistical performance. Therefore, the whole dataset was randomly split into training and test sets by a good number of MLR models with the same size of training and test sets. Of the chemicals in the dataset, 35 compounds were selected for the training set used to build QSAR models and the remaining (7 compounds) were considered as the test set used to evaluate the predictive power of the models [21,22].
ChemSketch software was used to calculate formula weight (FW), percentage of carbon, hydrogen, nitrogen, oxygen and sulfur atoms (% C, % H, % N, % O and % S), molar volume (MV (cm3)), parachor (Pa (cm3)), refractive index (RI), surface tension (ST (dyne/cm)), density (D (g/cm3)), polarizability (Po (cm3)), ring double bond equivalents (RDBE), and nominal mass (NM (Da)) (Table S1).
MarvinSketch and ChemOffice have been used to build-in structure to calculate the following descriptors: partition coefficient octanol-water (Log P), hydrophilic-lipophilic balance (HLB kcal/mol)), MMFF94 energy (ME (kcal/mol)), polar surface area (PSA), Van Der Waals surface area (VDWSA), Van Der Waals volume (VDWV), refractivity (R), number of H-bond acceptors (NHA), number of H-bond donors (NHD), molar refractivity (MR), partition coefficient (PC), topological diameter (TD), winner index (WI), Balaban index (BI), molecular topological index (MTI), number of rotator band (NRB), and number of oxygen atoms (NO) (Tables S2 and S3).
2.2. Statistical Analysis
In this study, XLSTAT [23] was used to accomplish both principal component analysis (PCA) and multiple linear regression (MLR). The method allows us to reduce the number of descriptors and keeps only those that are closely related to the activity. It also relies on studying the correlation matrix by removing those involving a large correlation. The MLR was initiated, with the aim to establish a mathematical relationship between inhibitory activity and a set of molecular descriptors. In other words, these two statistical methods depend on the assumption that there is a relationship that combines both the dependent variable (activity) and a series of independent variables (descriptors).
2.3. Validation of the QSAR Model
The predictive power of the built QSAR models was checked using internal and external validations.
We have used the leave-one-out (LOO) cross-validation for the internal validation. The cross validation parameter Q2cv was calculated. However, several previous studies have suggested that the only way to estimate the true predictive power of a QSAR model is to compare the predicted and observed activities for an external test set of compounds that were not used in the model’s development [24,25,26,27,28,29]. The quality of the QSAR model is mostly determined by its ability to make predictions for things not included in the training set. The external validation parameter R2test was calculated.
The y-randomization test was used to validate the developed QSAR models, whereby the performance of the original model in data description (R2) was compared to that of the built models. In other words, in this test, the random MLR models were generated by randomly shuffling the dependent variable while keeping the independent variables as they were. The newly established QSAR models were expected to have significantly low R2 and Q2 values for several trials, which confirmed that the developed QSAR models were robust. Another parameter, CRp2 was also calculated which should be more than 0.5 [24].
3. Results and Discussion
3.1. Principal Components Analysis (PCA)
Thirty descriptors were calculated using ChemSketch, MarvinSketch and ChemOffice software (Tables S1–S3). The correlation matrix obtained by the ACP was analyzed to extract important information from a multivariate spreadsheet and to express this information as a set of a few new variables called the main components. Therefore, PCA was a very important stage for reducing descriptors while ensuring a minimum level of information loss.
The descriptors that remained after the PCA for the rest of this study were: % C, % H, % N, % O, % S, RI, ST, D, RDBE, Log P, HLB, PSA, R, NHA, NHD, MR, PC, VDWSA, VDWV, BI, NRB, TD and NO.
3.2. Multiple Linear Regression (MLR)
Those descriptors remaining after PCA were used as an input for establishing MLR models. The best model obtained using MLR with the best statistical keys is represented by the following equation:
where R2 is the coefficient of determination; R2test is the coefficient of determination of the external test; R2adj is the adjusted coefficient of determination; MSE is the means of the square errors of the model; RMSE is root mean square error, F the coefficient of Fischer (Fisher statistics F) and P-value is the significance level.
pIC50 = 1.017 + 0.699 O% + 0.364 PC + 0.065 VDWV − 0.037 VDWSA − 2.158 NO
R2 = 0.691; R2test = 0.794; R2adj = 0.636; MSE = 0.108; RMSE = 0.328; F = 12.549; Pr < 0.0001.
R2 = 0.691; R2test = 0.794; R2adj = 0.636; MSE = 0.108; RMSE = 0.328; F = 12.549; Pr < 0.0001.
From the model found we deduce that the activity depends on the following descriptors: PC, VDWV, VDWSA, NO and O%.
The high values obtained for the coefficient of determination, the coefficient of determination of the external test and the adjusted coefficient of determination, which exceeded 0.6, as well as the low value of mean squared errors and root mean square error, confirmed that the established model had reliable predictive power.
On the other hand, the Fisher test associated with the p-value indicates that we would take less than 0.01% of the risk assuming the null hypothesis was false and the regression equation was statistically significant.
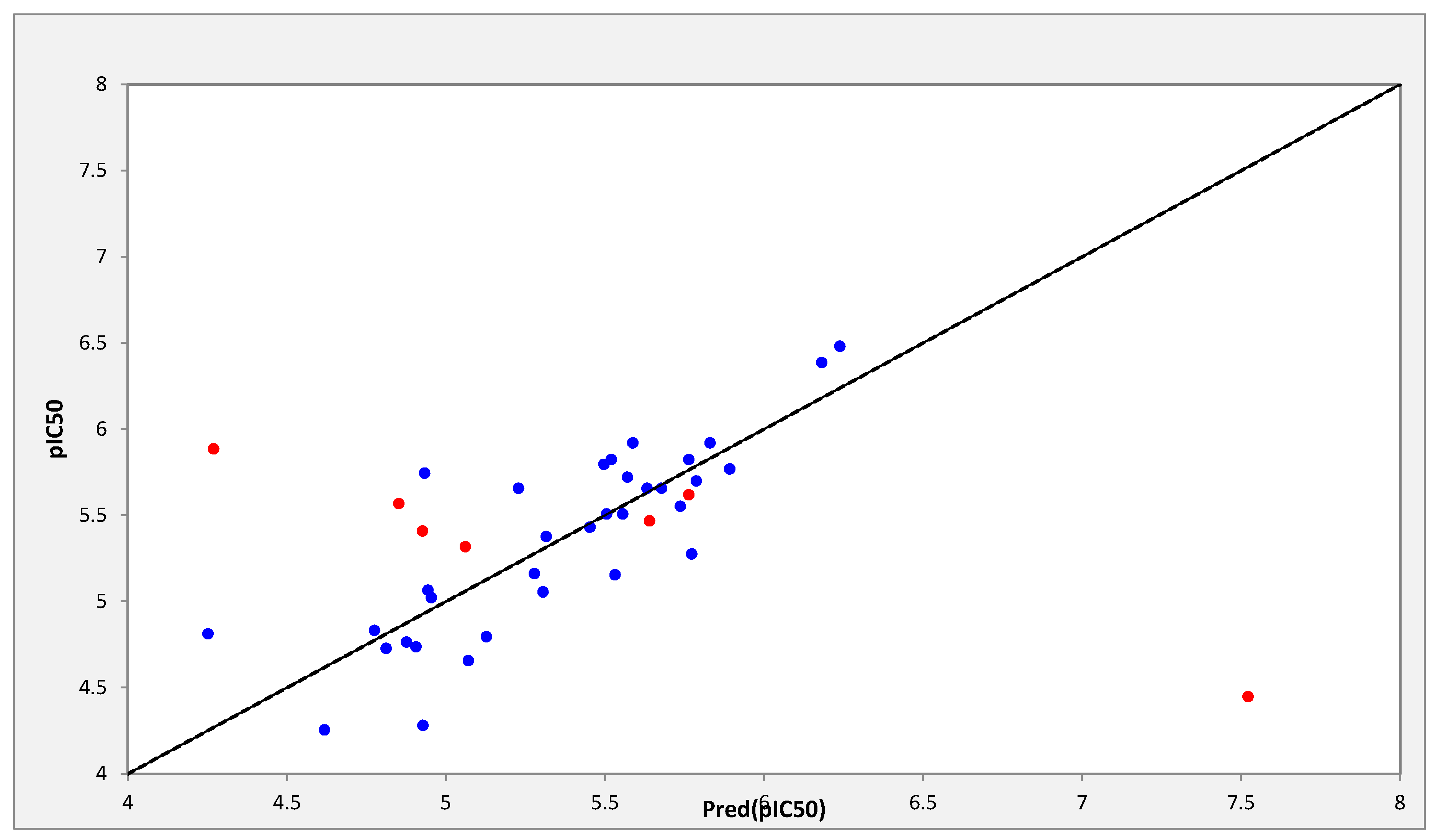
The correlations between the predicted and observed activities are represented in Table 2 and illustrated in Figure 1.

Table 2.
Experimental and predicted activities (pIC50) and residual values, according to MLR model.
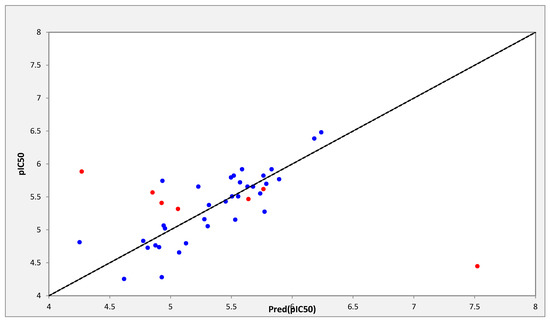
Figure 1.
Representation of observed and predicted activities values (pIC50).
3.3. Y-Randomization
The y-randomization test was applied to verify the validity and robustness of the built model. The obtained outcomes (Table 3) confirmed that the model was not obtained by chance.

Table 3.
Various values obtained after testing of y-randomization.
Based on all these results obtained by MLR, we can conclude that the built model has a good predictive power.
4. Conclusions
In this study, we have used thirty predefined descriptors for 41 peptidomimetic derivatives using ChemSketch, MarvinSketch and ChemOffice software. These descriptors are subjected to a statistical study using PCA analysis. In fact, the PCA was used to analyze and visualize the dataset, as well as to group the data into principal components. A linear model that combined five descriptors was found using the MLR method to predict the pIC50 activity. The proposed QSAR model by the MLR in this study was statistically significant and has sufficient capacity to predict the anti-MERS-CoV activity.
Supplementary Materials
The following are available online at https://www.mdpi.com/2624-8549/3/1/29/s1, Table S1: Chemical descriptors calculated by ChemSketch, Table S2: Chemical descriptors calculated by Marvin Sketch, Table S3: Chemical descriptors calculated by ChemOffice.
Author Contributions
Conceptualization, S.M. and I.H.; methodology, S.C.; software, S.C.; validation, S.C., S.B. and M.B.; formal analysis, S.C., S.B. and M.B.; investigation, S.C.; resources, S.C.; data curation, S.C.; writing—original draft preparation, S.M. and I.H.; writing—review and editing, S.C., S.B. and M.B.; visualization, S.C., S.B. and M.B.; supervision, S.C., S.B. and M.B.; project administration, S.C.; funding acquisition, S.C. All authors have read and agreed to the published version of the manuscript.
Funding
The authors are thankful to the “Agence Universitaire de la Francophone (AUF)” for financial support under the project AUF- 463/2020.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
The data presented in this study are available in Supplementary Materials.
Conflicts of Interest
The authors declare that there is no conflict of interest.
References
- Memish, Z.A.; Zumla, A.I.; Al-Hakeem, R.F.; AlRabeeah, A.A.; Stephens, G.M. Family cluster of Middle East respiratory syndrome coronavirus infections. N. Engl. J. Med. 2013, 368, 2487–2494. [Google Scholar] [CrossRef]
- Zaki, A.M.; Boheemen, S.V.; Bestebroer, T.M.; Osterhaus, A.D.; Fouchier, R.A. Isolation of a novel coronavirus from a man with pneumonia in Saudi Arabia. N. Engl. J. Med. 2012, 367, 1814–1820. [Google Scholar] [CrossRef] [PubMed]
- Bermingham, A.; Chand, M.A.; Brown, C.S.; Aarons, E.; Tong, C.; Langrish, C.; Hoschler, K.; Brown, K.; Galiano, M.; Myers, R.; et al. Severe respiratory illness caused by a novel coronavirus, in a patient transferred to the United Kingdom from the Middle East, September 2012. Euro. Surveill 2012, 17, 20290. [Google Scholar] [PubMed]
- Sridhar, S.; Brouqui, P.; Parola, P.; Gautret, P. Imported cases of Middle East respiratory syndrome: An update. Travel Med. Infect Dis. 2015, 13, 106–109. [Google Scholar] [CrossRef] [PubMed]
- Bialek, R.; Allen, D.; Alvarado-Ramy, F.; Arthur, R.; Balajee, A.; Bell, D.; Best, S.; Blackmore, C.; Breakwell, L.; Cannons, A.; et al. First confirmed cases of Middle East respiratory syndrome coronavirus (MERS-CoV) infection in the United States, updated information on the epidemiology of MERS-CoV infection, and guidance for the public, clinicians, and public health authorities—May 2014. MMWR. Morb. Mortal. Wkly. Rep. 2014, 63, 431–436. [Google Scholar]
- World Health Organization WHO. Middle East Respiratory Syndrome Coronavirus (MERS-CoV). 2019. Available online: https://www.who.int/health-topics/middle-east-respiratory-syndrome-coronavirus-merscom (accessed on 9 March 2021).
- Haagmans, B.L.; Al Dhahiry, S.H.S.; Reusken, C.B.E.M.; Raj, V.S.; Galiano, M.; Myers, R.; Godeke, G.J.; Jonges, M.; Farag, E.; Diab, A.; et al. Middle East respiratory syndrome coronavirus in dromedary camels: An outbreak investigation. Lancet Infect. Dis. 2014, 14, 140–145. [Google Scholar] [CrossRef]
- Sabir, J.S.M.; Lam, T.T.-Y.; Ahmed, M.M.M.; Li, L.; Shen, Y.; Abo-Aba, S.E.M.; Qureshi, M.I.; Abu-Zeid, M.; Zhang, Y.; Khiyami, M.A.; et al. Co-circulation of three camel coronavirus species and recombination of MERS-CoVs in Saudi Arabia. Science 2016, 351, 81–84. [Google Scholar] [CrossRef]
- Van den Brand, J.M.; Smits, S.L.; Haagmans, B.L. Pathogenesis of Middle East Respiratory Syndrome Coronavirus. J. Pathol. 2015, 235, 175–184. [Google Scholar] [CrossRef] [PubMed]
- Lee, H.-J.; Shieh, C.-K.; Gorbalenya, A.E.; Koonin, E.V.; La Monica, N.; Tuler, J.; Bagdzhadzhyan, A.; Lai, M.M.C. The Complete Sequence (22 kilobases) of Murine Coronavirus Gene 1 Encoding the Putative Proteases and RNA Polymerase. Virology 1991, 180, 567–582. [Google Scholar] [CrossRef]
- Marra, M.A.; Jones, S.J.M.; Astell, C.R.; Holt, R.A.; Angela, B.W.; Butterfield, Y.S.N.; Jaswinder, K.; Asano, J.K.; Barber, S.A.; Chan, S.Y. The Genome Sequence of the SARS-Associated Coronavirus. Science 2003, 300, 1399–1404. [Google Scholar] [CrossRef]
- Woo, P.C.Y.; Huang, Y.; Lau, S.K.P.; Yuen, K.-Y. Coronavirus Genomics and Bioinformatics Analysis. Viruses 2010, 2, 1804–1820. [Google Scholar] [CrossRef] [PubMed]
- St John, S.E.; Tomar, S.; Stauffer, S.R.; Mesecar, A.D. Targeting Zoonotic Viruses: Structure-Based Inhibition of the 3C-Like Protease from Bat Coronavirus HKU4-The Likely Reservoir Host to the Human Coronavirus that Causes Middle East Respiratory Syndrome (MERS). Bioorg. Med. Chem. 2015, 23, 6036–6048. [Google Scholar] [CrossRef]
- Ratia, K.; Saikatendu, K.S.; Santarsiero, B.D.; Barretto, N.; Baker, S.C.; Stevens, R.C.; Mesecar, A.D. Severe Acute Respiratory Syndrome Coronavirus Papain-Like Protease: Structure of a Viral Deubiquitinating Enzyme. Proc. Natl. Acad. Sci. USA 2006, 103, 5717–5722. [Google Scholar] [CrossRef]
- Chen, S.; Chen, L.; Tan, J.; Chen, J.; Du, L.; Sun, T.; Shen, J.; Chen, K.; Jiang, H.; Shen, X. Severe Acute Respiratory Syndrome Coronavirus 3C-Like Proteinase N Terminus is Indispensable for Proteolytic Activity but not for Enzyme Dimerization. Biochemical and Thermodynamic Investigation in Conjunction with Molecular Dynamics Simulations. J. Biol. Chem. 2005, 280, 164–173. [Google Scholar] [CrossRef] [PubMed]
- Wojdyla, J.A.; Manolaridis, I.; van Kasteren, P.B.; Kikkert, M.; Snijder, E.J.; Gorbalenya, A.E.; Tucker, P.A. Papain-Like Protease 1 from Transmissible Gastroenteritis Virus: Crystal Structure and Enzymatic Activity toward Viral and Cellular Substrates. J. Virol. 2010, 84, 10063–10073. [Google Scholar] [CrossRef]
- Abuhammad, A.; Al-Aqtash, R.A.; Anson, B.J.; Mesecar, A.D.; Taha, M.O. Computational modeling of the bat HKU4 coronavirus 3CLpro inhibitors as a tool for the development of antivirals against the emerging Middle East respiratory syndrome (MERS) coronavirus. J. Mol. Recognit. 2017, 30, e2644. [Google Scholar] [CrossRef]
- Nantasenamat, C.; Isarankura-Na-Ayudhya, C.; Naenna, T.; Prachayasittikul, V. A practical overview of quantitative structure-activity relationship. J. Excli. 2009, 8, 74–88. [Google Scholar]
- Khan, A.U. Descriptors and their selection methods in QSAR analysis: Paradigm for drug design. Drug Discov. Today 2016, 21, 1291–1302. [Google Scholar]
- Toropova, A.P.; Toropov, A.A.; Benfenatia, E.; Leszczynska, D.; Leszczynski, J. QSAR model as a random event: A case of rat toxicity. Bioorg. Med. Chem. 2015, 23, 1223–1230. [Google Scholar] [CrossRef]
- Chtita, S.; Belhassan, A.; Bakhouch, M.; Taourat, A.I.; Aouidate, A.; Belaidi, S.; Moutaabbid, M.; Belaaouad, S.; Bouachrine, M.; Lakhlifi, T. QSAR study of unsymmetrical aromatic disulfides as potent avian SARS-CoV main protease inhibitors using quantum chemical descriptors and statistical methods. Chemo. Intel. Lab. Syst. 2021, 210, 104266. [Google Scholar] [CrossRef]
- Chtita, S.; Aouidate, A.; Belhassan, A.; Ousaa, A.; Taourati, A.I.; Elidrissi, B.; Ghamali, M.; Bouachrine, M.; Lakhlifi, T. QSAR study of N -substituted Oseltamivir derivatives as potent avian influenza virus H5N1 inhibitors using quantum chemical descriptors and statistical methods. New J. Chem. 2020, 44, 1747–1760. [Google Scholar] [CrossRef]
- XLSTAT Software. 2020. Available online: http://www.xlstat.com (accessed on 9 March 2021).
- Veerasamy, R.; Rajak, H.; Jain, A.; Sivadasan, S.; Varghese, C.; Agrawal, R. Validation of QSAR Models—Strategies and Importance. Int. J. Drug Disc. 2011, 2, 511–519. [Google Scholar]
- Muhammad, U.; Uzairu, A.; Arthur, D.E. Review on: Quantitative structure activity relationship (QSAR) modeling. J. Anal Pharm. Res. 2018, 7, 240–242. [Google Scholar] [CrossRef]
- Roy, K.; Mitra, I.; Kar, S.; Ojha, P.K.; Das, R.N.; Kabir, H. Comparative Studies on some metrics for external validation of QSAR model. J. Chem. Inf. Mdel. 2012, 52, 396–408. [Google Scholar] [CrossRef]
- Rücker, C.; Rücker, G.; Meringer, M. Y-Randomization and Its Variants in QSPR/QSAR. J. Chem. Inf. Model. 2007, 47, 2345–2357. [Google Scholar] [CrossRef] [PubMed]
- Pravin Ambure of Drug Theoretics & Cheminformatics (DTC) Laboratory; Jadavpur University: Kolkata, India, 2013.
- Chtita, S.; Belhassan, A.; Aouidate, A.; Belaidi, S.; Bouachrine, M.; Lakhlifi, T. Discovery of Potent SARS-CoV-2 Inhibitors from Approved Antiviral Drugs via Docking Screening. Comb. Chem. High Throughput Screen. 2020, 23, 441–454. [Google Scholar]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).