Dissecting the Inorganic Nanoparticle-Driven Interferences on Adhesome Dynamics
Abstract
1. Introduction
2. Adhesome Structure, Function, and Dynamics
3. Impact of Inorganic Nanoparticles on Adhesome Dynamics
3.1. Inorganic Nanoparticles and Focal Adhesions Dynamic
3.2. Nanopaterneting as a Platform for FA Study
3.3. Inorganic Nanoparticles and Invadosome Dynamic
4. Perspectives
Funding
Conflicts of Interest
References
- Habibi, N.; Quevedo, D.F.; Gregory, J.V.; Lahann, J. Emerging Methods in Therapeutics Using Multifunctional Nanoparticles. WIREs Nanomed. Nanobiotechnol. 2020, 12, e1625. [Google Scholar] [CrossRef] [PubMed]
- Khan, A.U.; Khan, M.; Cho, M.H.; Khan, M.M. Selected Nanotechnologies and Nanostructures for Drug Delivery, Nanomedicine and Cure. Bioprocess Biosyst. Eng. 2020, 43, 1339–1357. [Google Scholar] [CrossRef]
- Wicki, A.; Witzigmann, D.; Balasubramanian, V.; Huwyler, J. Nanomedicine in Cancer Therapy: Challenges, Opportunities, and Clinical Applications. J. Control. Release 2015, 200, 138–157. [Google Scholar] [CrossRef]
- Hare, J.I.; Lammers, T.; Ashford, M.B.; Puri, S.; Storm, G.; Barry, S.T. Challenges and Strategies in Anti-Cancer Nanomedicine Development: An Industry Perspective. Adv. Drug Deliv. Rev. 2017, 108, 25–38. [Google Scholar] [CrossRef]
- Gharagozloo, M.; Majewski, S.; Foldvari, M. Therapeutic Applications of Nanomedicine in Autoimmune Diseases: From Immunosuppression to Tolerance Induction. Nanomed. Nanotechnol. Biol. Med. 2015, 11, 1003–1018. [Google Scholar] [CrossRef]
- Qamar, N.; Arif, A.; Bhatti, A.; John, P. Nanomedicine: An Emerging Era of Theranostics and Therapeutics for Rheumatoid Arthritis. Rheumatology 2019, 58, 1715–1721. [Google Scholar] [CrossRef] [PubMed]
- Poovaiah, N.; Davoudi, Z.; Peng, H.; Schlichtmann, B.; Mallapragada, S.; Narasimhan, B.; Wang, Q. Treatment of Neurodegenerative Disorders through the Blood–Brain Barrier Using Nanocarriers. Nanoscale 2018, 10, 16962–16983. [Google Scholar] [CrossRef]
- Ramanathan, S.; Archunan, G.; Sivakumar, M.; Selvan, S.T.; Fred, A.L.; Kumar, S.; Gulyás, B.; Padmanabhan, P. Theranostic Applications of Nanoparticles in Neurodegenerative Disorders. Int. J. Nanomed. 2018, 13, 5561–5576. [Google Scholar] [CrossRef]
- Steichen, S.D.; Caldorera-Moore, M.; Peppas, N.A. A Review of Current Nanoparticle and Targeting Moieties for the Delivery of Cancer Therapeutics. Eur. J. Pharm. Sci. 2013, 48, 416–427. [Google Scholar] [CrossRef] [PubMed]
- Amreddy, N.; Babu, A.; Muralidharan, R.; Panneerselvam, J.; Srivastava, A.; Ahmed, R.; Mehta, M.; Munshi, A.; Ramesh, R. Chapter Five—Recent Advances in Nanoparticle-Based Cancer Drug and Gene Delivery. In Advances in Cancer Research; Tew, K.D., Fisher, P.B., Eds.; Academic Press: Cambridge, MA, USA, 2018; Volume 137, pp. 115–170. ISBN 0065-230X. [Google Scholar]
- Deirram, N.; Zhang, C.; Kermaniyan, S.S.; Johnston, A.P.R.; Such, G.K. PH-Responsive Polymer Nanoparticles for Drug Delivery. Macromol. Rapid Commun. 2019, 40, 1800917. [Google Scholar] [CrossRef] [PubMed]
- Karimi, M.; Eslami, M.; Sahandi-Zangabad, P.; Mirab, F.; Farajisafiloo, N.; Shafaei, Z.; Ghosh, D.; Bozorgomid, M.; Dashkhaneh, F.; Hamblin, M.R. PH-Sensitive Stimulus-Responsive Nanocarriers for Targeted Delivery of Therapeutic Agents. WIREs Nanomed. Nanobiotechnol. 2016, 8, 696–716. [Google Scholar] [CrossRef] [PubMed]
- Xu, X.; Saw, P.E.; Tao, W.; Li, Y.; Ji, X.; Bhasin, S.; Liu, Y.; Ayyash, D.; Rasmussen, J.; Huo, M.; et al. ROS-Responsive Polyprodrug Nanoparticles for Triggered Drug Delivery and Effective Cancer Therapy. Adv. Mater. 2017, 29, 1700141. [Google Scholar] [CrossRef]
- Lv, X.; Zhu, Y.; Ghandehari, H.; Yu, A.; Wang, Y. An ROS-Responsive and Self-Accelerating Drug Release Nanoplatform for Overcoming Multidrug Resistance. Chem. Commun. 2019, 55, 3383–3386. [Google Scholar] [CrossRef] [PubMed]
- Halappanavar, S.; Vogel, U.; Wallin, H.; Yauk, C.L. Promise and Peril in Nanomedicine: The Challenges and Needs for Integrated Systems Biology Approaches to Define Health Risk. WIREs Nanomed. Nanobiotechnol. 2018, 10, e1465. [Google Scholar] [CrossRef] [PubMed]
- Pelaz, B.; Charron, G.; Pfeiffer, C.; Zhao, Y.; de la Fuente, J.M.; Liang, X.-J.; Parak, W.J.; Del Pino, P. Interfacing Engineered Nanoparticles with Biological Systems: Anticipating Adverse Nano-Bio Interactions. Small 2013, 9, 1573–1584. [Google Scholar] [CrossRef]
- Su, H.; Wang, Y.; Gu, Y.; Bowman, L.; Zhao, J.; Ding, M. Potential Applications and Human Biosafety of Nanomaterials Used in Nanomedicine. J. Appl. Toxicol. 2018, 38, 3–24. [Google Scholar] [CrossRef]
- Tang, D.D.; Gerlach, B.D. The Roles and Regulation of the Actin Cytoskeleton, Intermediate Filaments and Microtubules in Smooth Muscle Cell Migration. Respir. Res. 2017, 18, 54. [Google Scholar] [CrossRef]
- Fife, C.M.; McCarroll, J.A.; Kavallaris, M. Movers and Shakers: Cell Cytoskeleton in Cancer Metastasis. Br. J. Pharmacol. 2014, 171, 5507–5523. [Google Scholar] [CrossRef] [PubMed]
- Burbage, M.; Keppler, S.J. Shaping the Humoral Immune Response: Actin Regulators Modulate Antigen Presentation and Influence B-T Interactions. Mol. Immunol. 2018, 101, 370–376. [Google Scholar] [CrossRef] [PubMed]
- Mostowy, S.; Shenoy, A.R. The Cytoskeleton in Cell-Autonomous Immunity: Structural Determinants of Host Defence. Nat. Rev. Immunol. 2015, 15, 559–573. [Google Scholar] [CrossRef]
- Nishimura, Y.; Kasahara, K.; Inagaki, M. Intermediate Filaments and IF-Associated Proteins: From Cell Architecture to Cell Proliferation. Proc. Jpn. Acad. Ser. B Phys. Biol. Sci. 2019, 95, 479–493. [Google Scholar] [CrossRef]
- Pawlik, A.; Szczepanski, M.A.; Klimaszewska-Wisniewska, A.; Gackowska, L.; Zuryn, A.; Grzanka, A. Cytoskeletal Reorganization and Cell Death in Mitoxantrone-Treated Lung Cancer Cells. Acta Histochem. 2016, 118, 784–796. [Google Scholar] [CrossRef]
- Zaidel-Bar, R.; Itzkovitz, S.; Ma’ayan, A.; Iyengar, R.; Geiger, B. Functional Atlas of the Integrin Adhesome. Nat. Cell Biol. 2007, 9, 858–867. [Google Scholar] [CrossRef]
- Winograd-Katz, S.E.; Fässler, R.; Geiger, B.; Legate, K.R. The Integrin Adhesome: From Genes and Proteins to Human Disease. Nat. Rev. Mol. Cell Biol. 2014, 15, 273–288. [Google Scholar] [CrossRef]
- Singer, I.I.; Scott, S.; Kawka, D.W.; Kazazis, D.M. Adhesomes: Specific Granules Containing Receptors for Laminin, C3bi/Fibrinogen, Fibronectin, and Vitronectin in Human Polymorphonuclear Leukocytes and Monocytes. J. Cell Biol. 1989, 109, 3169–3182. [Google Scholar] [CrossRef] [PubMed]
- Whittaker, C.A.; Bergeron, K.-F.; Whittle, J.; Brandhorst, B.P.; Burke, R.D.; Hynes, R.O. The Echinoderm Adhesome. Dev. Biol. 2006, 300, 252–266. [Google Scholar] [CrossRef] [PubMed]
- Jacquemet, G.; Hamidi, H.; Ivaska, J. Filopodia in Cell Adhesion, 3D Migration and Cancer Cell Invasion. Curr. Opin. Cell Biol. 2015, 36, 23–31. [Google Scholar] [CrossRef] [PubMed]
- Innocenti, M. New Insights into the Formation and the Function of Lamellipodia and Ruffles in Mesenchymal Cell Migration. Cell Adhes. Migr. 2018, 12, 401–416. [Google Scholar] [CrossRef] [PubMed]
- Burridge, K.; Guilluy, C. Focal Adhesions, Stress Fibers and Mechanical Tension. Exp. Cell Res. 2016, 343, 14–20. [Google Scholar] [CrossRef]
- Alonso, F.; Spuul, P.; Daubon, T.; Kramer, I.; Génot, E. Variations on the Theme of Podosomes: A Matter of Context. Biochim. Biophys. Acta Mol. Cell Res. 2019, 1866, 545–553. [Google Scholar] [CrossRef] [PubMed]
- Cmoch, A.; Groves, P.; Pikuła, S. Biogenesis of Invadopodia and Their Cellular Functions. Postepy Biochem. 2014, 60, 62–68. [Google Scholar]
- Phng, L.-K.; Gebala, V.; Bentley, K.; Philippides, A.; Wacker, A.; Mathivet, T.; Sauteur, L.; Stanchi, F.; Belting, H.-G.; Affolter, M.; et al. Formin-Mediated Actin Polymerization at Endothelial Junctions Is Required for Vessel Lumen Formation and Stabilization. Dev. Cell 2015, 32, 123–132. [Google Scholar] [CrossRef] [PubMed]
- Sahasrabudhe, A.; Ghate, K.; Mutalik, S.; Jacob, A.; Ghose, A. Formin 2 Regulates the Stabilization of Filopodial Tip Adhesions in Growth Cones and Affects Neuronal Outgrowth and Pathfinding In Vivo. Development 2016, 143, 449–460. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Grobe, H.; Wüstenhagen, A.; Baarlink, C.; Grosse, R.; Grikscheit, K. A Rac1-FMNL2 Signaling Module Affects Cell-Cell Contact Formation Independent of Cdc42 and Membrane Protrusions. PLoS ONE 2018, 13, e0194716. [Google Scholar] [CrossRef] [PubMed]
- Disanza, A.; Bisi, S.; Winterhoff, M.; Milanesi, F.; Ushakov, D.S.; Kast, D.; Marighetti, P.; Romet-Lemonne, G.; Müller, H.-M.; Nickel, W.; et al. CDC42 Switches IRSp53 from Inhibition of Actin Growth to Elongation by Clustering of VASP. EMBO J. 2013, 32, 2735–2750. [Google Scholar] [CrossRef]
- Chacón, M.R.; Navarro, A.I.; Cuesto, G.; del Pino, I.; Scott, R.; Morales, M.; Rico, B. Focal Adhesion Kinase Regulates Actin Nucleation and Neuronal Filopodia Formation during Axonal Growth. Development 2012, 139, 3200–3210. [Google Scholar] [CrossRef]
- Young, L.E.; Latario, C.J.; Higgs, H.N. Roles for Ena/VASP Proteins in FMNL3-Mediated Filopodial Assembly. J. Cell Sci. 2018, 131, jcs220814. [Google Scholar] [CrossRef] [PubMed]
- Barzik, M.; McClain, L.M.; Gupton, S.L.; Gertler, F.B. Ena/VASP Regulates MDia2-Initiated Filopodial Length, Dynamics, and Function. Mol. Biol. Cell 2014, 25, 2604–2619. [Google Scholar] [CrossRef]
- Alieva, N.O.; Efremov, A.K.; Hu, S.; Oh, D.; Chen, Z.; Natarajan, M.; Ong, H.T.; Jégou, A.; Romet-Lemonne, G.; Groves, J.T.; et al. Myosin IIA and Formin Dependent Mechanosensitivity of Filopodia Adhesion. Nat. Commun. 2019, 10, 3593. [Google Scholar] [CrossRef]
- Hogg, N.; Laschinger, M.; Giles, K.; McDowall, A. T-Cell Integrins: More than Just Sticking Points. J. Cell Sci. 2003, 116, 4695–4705. [Google Scholar] [CrossRef]
- Pröls, F.; Sagar; Scaal, M. Signaling Filopodia in Vertebrate Embryonic Development. Cell. Mol. Life Sci. 2016, 73, 961–974. [Google Scholar] [CrossRef] [PubMed]
- Romero, S.; Quatela, A.; Bornschlögl, T.; Guadagnini, S.; Bassereau, P.; Van Nhieu, G.T. Filopodium Retraction Is Controlled by Adhesion to Its Tip. J. Cell Sci. 2012, 125, 4999–5004. [Google Scholar] [CrossRef]
- Wong, S.; Guo, W.-H.; Wang, Y.-L. Fibroblasts Probe Substrate Rigidity with Filopodia Extensions before Occupying an Area. Proc. Natl. Acad. Sci. USA 2014, 111, 17176–17181. [Google Scholar] [CrossRef] [PubMed]
- Spinardi, L.; Rietdorf, J.; Nitsch, L.; Bono, M.; Tacchetti, C.; Way, M.; Marchisio, P.C. A Dynamic Podosome-like Structure of Epithelial Cells. Exp. Cell Res. 2004, 295, 360–374. [Google Scholar] [CrossRef]
- Wang, Y.; McNiven, M.A. Invasive Matrix Degradation at Focal Adhesions Occurs via Protease Recruitment by a FAK–P130Cas Complex. J. Cell Biol. 2012, 196, 375–385. [Google Scholar] [CrossRef] [PubMed]
- Luxenburg, C.; Geblinger, D.; Klein, E.; Anderson, K.; Hanein, D.; Geiger, B.; Addadi, L. The Architecture of the Adhesive Apparatus of Cultured Osteoclasts: From Podosome Formation to Sealing Zone Assembly. PLoS ONE 2007, 2, e179. [Google Scholar] [CrossRef]
- Sharma, V.P.; Eddy, R.; Entenberg, D.; Kai, M.; Gertler, F.B.; Condeelis, J. Tks5 and SHIP2 Regulate Invadopodium Maturation, but Not Initiation, in Breast Carcinoma Cells. Curr. Biol. 2013, 23, 2079–2089. [Google Scholar] [CrossRef]
- Fu, P.; Ebenezer, D.L.; Berdyshev, E.V.; Bronova, I.A.; Shaaya, M.; Harijith, A.; Natarajan, V. Role of Sphingosine Kinase 1 and S1P Transporter Spns2 in HGF-Mediated Lamellipodia Formation in Lung Endothelium. J. Biol. Chem. 2016, 291, 27187–27203. [Google Scholar] [CrossRef]
- Usatyuk, P.V.; Fu, P.; Mohan, V.; Epshtein, Y.; Jacobson, J.R.; Gomez-Cambronero, J.; Wary, K.K.; Bindokas, V.; Dudek, S.M.; Salgia, R.; et al. Role of C-Met/Phosphatidylinositol 3-Kinase (PI3k)/Akt Signaling in Hepatocyte Growth Factor (HGF)-Mediated Lamellipodia Formation, Reactive Oxygen Species (ROS) Generation, and Motility of Lung Endothelial Cells. J. Biol. Chem. 2014, 289, 13476–13491. [Google Scholar] [CrossRef] [PubMed]
- Buhrmann, C.; Yazdi, M.; Popper, B.; Kunnumakkara, A.B.; Aggarwal, B.B.; Shakibaei, M. Induction of the Epithelial-to-Mesenchymal Transition of Human Colorectal Cancer by Human TNF-β (Lymphotoxin) and Its Reversal by Resveratrol. Nutrients 2019, 11, 704. [Google Scholar] [CrossRef] [PubMed]
- Gazquez, E.; Watanabe, Y.; Broders-Bondon, F.; Paul-Gilloteaux, P.; Heysch, J.; Baral, V.; Bondurand, N.; Dufour, S. Endothelin-3 Stimulates Cell Adhesion and Cooperates with Β1-Integrins during Enteric Nervous System Ontogenesis. Sci. Rep. 2016, 6, 37877. [Google Scholar] [CrossRef] [PubMed]
- Savin, V.J.; Sharma, M.; Zhou, J.; Gennochi, D.; Fields, T.; Sharma, R.; McCarthy, E.T.; Srivastava, T.; Domen, J.; Tormo, A.; et al. Renal and Hematological Effects of CLCF-1, a B-Cell-Stimulating Cytokine of the IL-6 Family. J. Immunol. Res. 2015, 2015, 714964. [Google Scholar] [CrossRef] [PubMed]
- Kiso, M.; Tanaka, S.; Saji, S.; Toi, M.; Sato, F. Long Isoform of VEGF Stimulates Cell Migration of Breast Cancer by Filopodia Formation via NRP1/ARHGAP17/Cdc42 Regulatory Network. Int. J. Cancer 2018, 143, 2905–2918. [Google Scholar] [CrossRef]
- Feng, H.; Chen, L.; Wang, Q.; Shen, B.; Liu, L.; Zheng, P.; Xu, S.; Liu, X.; Chen, J.; Teng, J. Calumenin-15 Facilitates Filopodia Formation by Promoting TGF-β Superfamily Cytokine GDF-15 Transcription. Cell Death Dis. 2013, 4, e870. [Google Scholar] [CrossRef]
- Maisel, S.; Broka, D.; Schroeder, J. Intravesicular Epidermal Growth Factor Receptor Subject to Retrograde Trafficking Drives Epidermal Growth Factor-Dependent Migration. Oncotarget 2017, 9, 6463–6477. [Google Scholar] [CrossRef][Green Version]
- Shi, M.-D.; Liao, Y.-C.; Shih, Y.-W.; Tsai, L.-Y. Nobiletin Attenuates Metastasis via Both ERK and PI3K/Akt Pathways in HGF-Treated Liver Cancer HepG2 Cells. Phytomedicine 2013, 20, 743–752. [Google Scholar] [CrossRef]
- Marivin, A.; Berthelet, J.; Cartier, J.; Paul, C.; Gemble, S.; Morizot, A.; Boireau, W.; Saleh, M.; Bertoglio, J.; Solary, E.; et al. CIAP1 Regulates TNF-Mediated Cdc42 Activation and Filopodia Formation. Oncogene 2014, 33, 5534–5545. [Google Scholar] [CrossRef] [PubMed]
- Oswald, J.; Büttner, M.; Jasinski-Bergner, S.; Jacobs, R.; Rosenstock, P.; Kielstein, H. Leptin Affects Filopodia and Cofilin in NK-92 Cells in a Dose- and Time-Dependent Manner. Eur. J. Histochem. 2018, 62, 2848. [Google Scholar] [CrossRef] [PubMed]
- Rigiracciolo, D.C.; Santolla, M.F.; Lappano, R.; Vivacqua, A.; Cirillo, F.; Galli, G.R.; Talia, M.; Muglia, L.; Pellegrino, M.; Nohata, N.; et al. Focal Adhesion Kinase (FAK) Activation by Estrogens Involves GPER in Triple-Negative Breast Cancer Cells. J. Exp. Clin. Cancer Res. 2019, 38, 58. [Google Scholar] [CrossRef]
- Nalluri, S.M.; O’Connor, J.W.; Virgi, G.A.; Stewart, S.E.; Ye, D.; Gomez, E.W. TGFβ1-Induced Expression of Caldesmon Mediates Epithelial–Mesenchymal Transition. Cytoskeleton 2018, 75, 201–212. [Google Scholar] [CrossRef]
- Johansson, M.W.; Khanna, M.; Bortnov, V.; Annis, D.S.; Nguyen, C.L.; Mosher, D.F. IL-5-Stimulated Eosinophils Adherent to Periostin Undergo Stereotypic Morphological Changes and ADAM8-Dependent Migration. Clin. Exp. Allergy 2017, 47, 1263–1274. [Google Scholar] [CrossRef]
- Daubon, T.; Spuul, P.; Alonso, F.; Fremaux, I.; Génot, E. VEGF-A Stimulates Podosome-Mediated Collagen-IV Proteolysis in Microvascular Endothelial Cells. J. Cell Sci. 2016, 129, 2586–2598. [Google Scholar] [CrossRef]
- Williams, K.S.; Killebrew, D.A.; Clary, G.P.; Seawell, J.A.; Meeker, R.B. Differential Regulation of Macrophage Phenotype by Mature and Pro-Nerve Growth Factor. J. Neuroimmunol. 2015, 285, 76–93. [Google Scholar] [CrossRef] [PubMed]
- Serafino, A.; Andreola, F.; Pittaluga, E.; Krasnowska, E.K.; Nicotera, G.; Sferrazza, G.; Sinibaldi Vallebona, P.; Pierimarchi, P.; Garaci, E. Thymosin A1 Modifies Podosome Architecture and Promptly Stimulates the Expression of Podosomal Markers in Mature Macrophages. Expert Opin. Biol. Ther. 2015, 15, 101–116. [Google Scholar] [CrossRef] [PubMed]
- Kung, M.-L.; Tsai, H.-E.; Hu, T.-H.; Kuo, H.-M.; Liu, L.-F.; Chen, S.-C.; Lin, P.-R.; Ma, Y.-L.; Wang, E.-M.; Liu, G.-S.; et al. Hepatoma-Derived Growth Factor Stimulates Podosome Rosettes Formation in NIH/3T3 Cells through the Activation of Phosphatidylinositol 3-Kinase/Akt Pathway. Biochem. Biophys. Res. Commun. 2012, 425, 169–176. [Google Scholar] [CrossRef] [PubMed]
- Le Roux-Goglin, E.; Varon, C.; Spuul, P.; Asencio, C.; Mégraud, F.; Génot, E. Helicobacter Infection Induces Podosome Assembly in Primary Hepatocytes In Vitro. Eur. J. Cell Biol. 2012, 91, 161–170. [Google Scholar] [CrossRef]
- Tatin, F.; Grise, F.; Reuzeau, E.; Genot, E.; Moreau, V. Sodium Fluoride Induces Podosome Formation in Endothelial Cells. Biol. Cell 2010, 102, 489–498. [Google Scholar] [CrossRef] [PubMed]
- Sa, G.; Liu, Z.; Ren, J.; Wan, Q.; Xiong, X.; Yu, Z.; Chen, H.; Zhao, Y.; He, S. Keratinocyte Growth Factor (KGF) Induces Podosome Formation via Integrin-Erk1/2 Signaling in Human Immortalized Oral Epithelial Cells. Cell. Signal. 2019, 61, 39–47. [Google Scholar] [CrossRef]
- Griera, M.; Martin-Villar, E.; Banon-Rodríguez, I.; Blundell, M.P.; Jones, G.E.; Anton, I.M.; Thrasher, A.J.; Rodriguez-Puyol, M.; Calle, Y. Integrin Linked Kinase (ILK) Regulates Podosome Maturation and Stability in Dendritic Cells. Int. J. Biochem. Cell Biol. 2014, 50, 47–54. [Google Scholar] [CrossRef]
- Mu, X.; Wang, X.; Huang, W.; Wang, R.-T.; Essandoh, K.; Li, Y.; Pugh, A.M.; Peng, J.; Deng, S.; Wang, Y.; et al. Circulating Exosomes Isolated from Septic Mice Induce Cardiovascular Hyperpermeability Through Promoting Podosome Cluster Formation. Shock 2018, 49, 429–441. [Google Scholar] [CrossRef]
- Singh, A.; Gill, G.; Kaur, H.; Amhmed, M.; Jakhu, H. Role of Osteopontin in Bone Remodeling and Orthodontic Tooth Movement: A Review. Prog. Orthod. 2018, 19, 18. [Google Scholar] [CrossRef]
- Mader, C.C.; Oser, M.; Magalhaes, M.A.O.; Bravo-Cordero, J.J.; Condeelis, J.; Koleske, A.J.; Gil-Henn, H. An EGFR–Src–Arg–Cortactin Pathway Mediates Functional Maturation of Invadopodia and Breast Cancer Cell Invasion. Cancer Res. 2011, 71, 1730–1741. [Google Scholar] [CrossRef] [PubMed]
- Eckert, M.A.; Lwin, T.M.; Chang, A.T.; Kim, J.; Danis, E.; Ohno-Machado, L.; Yang, J. Twist1-Induced Invadopodia Formation Promotes Tumor Metastasis. Cancer Cell 2011, 19, 372–386. [Google Scholar] [CrossRef]
- Pignatelli, J.; Tumbarello, D.A.; Schmidt, R.P.; Turner, C.E. Hic-5 Promotes Invadopodia Formation and Invasion during TGF-β–Induced Epithelial–Mesenchymal Transition. J. Cell Biol. 2012, 197, 421–437. [Google Scholar] [CrossRef] [PubMed]
- Hoshino, D.; Branch, K.M.; Weaver, A.M. Signaling Inputs to Invadopodia and Podosomes. J. Cell Sci. 2013, 126, 2979–2989. [Google Scholar] [CrossRef]
- Rajadurai, C.V.; Havrylov, S.; Zaoui, K.; Vaillancourt, R.; Stuible, M.; Naujokas, M.; Zuo, D.; Tremblay, M.L.; Park, M. Met Receptor Tyrosine Kinase Signals through a Cortactin–Gab1 Scaffold Complex, to Mediate Invadopodia. J. Cell Sci. 2012, 125, 2940–2953. [Google Scholar] [CrossRef]
- Makowiecka, A.; Simiczyjew, A.; Nowak, D.; Mazur, A.J. Varying Effects of EGF, HGF and TGFβ on Formation of Invadopodia and Invasiveness of Melanoma Cell Lines of Different Origin. Eur. J. Histochem. 2016, 60, 2728. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.; Zhu, M.; Yu, S.; Hai, L.; Zhang, L.; Zhang, C.; Zhao, P.; Zhou, H.; Wang, S.; Yang, X. Arg Kinase Mediates CXCL12/CXCR4-Induced Invadopodia Formation and Invasion of Glioma Cells. Exp. Cell Res. 2020, 389, 216–229. [Google Scholar] [CrossRef]
- Love, A.M.; Prince, D.J.; Jessen, J.R. Vangl2-Dependent Regulation of Membrane Protrusions and Directed Migration Requires a Fibronectin Extracellular Matrix. Development 2018, 145, dev165472. [Google Scholar] [CrossRef] [PubMed]
- Singh, R. Central Role of PI3K-SYK Interaction in Fibrinogen-Induced Lamellipodia and Filopodia Formation in Platelets. FEBS Open Bio. 2016, 6, 1285–1296. [Google Scholar] [CrossRef]
- Wang, W.Y.; Pearson, A.T.; Kutys, M.L.; Choi, C.K.; Wozniak, M.A.; Baker, B.M.; Chen, C.S. Extracellular Matrix Alignment Dictates the Organization of Focal Adhesions and Directs Uniaxial Cell Migration. APL Bioeng. 2018, 2, 046107. [Google Scholar] [CrossRef] [PubMed]
- Diaz, C.; Neubauer, S.; Rechenmacher, F.; Kessler, H.; Missirlis, D. Recruitment of ανβ3 Integrin to α5β1 Integrin-Induced Clusters Enables Focal Adhesion Maturation and Cell Spreading. J. Cell Sci. 2020, 133, jcs232702. [Google Scholar] [CrossRef]
- Stritt, S.; Thielmann, I.; Dütting, S.; Stegner, D.; Nieswandt, B. Phospholipase D Is a Central Regulator of Collagen I-Induced Cytoskeletal Rearrangement and Podosome Formation in Megakaryocytes. J. Thromb. Haemost. 2014, 12, 1364–1371. [Google Scholar] [CrossRef]
- Schachtner, H.; Calaminus, S.D.J.; Sinclair, A.; Monypenny, J.; Blundell, M.P.; Leon, C.; Holyoake, T.L.; Thrasher, A.J.; Michie, A.M.; Vukovic, M.; et al. Megakaryocytes Assemble Podosomes That Degrade Matrix and Protrude through Basement Membrane. Blood 2013, 121, 2542–2552. [Google Scholar] [CrossRef] [PubMed]
- Suzuki, T.; Shoji, S.; Yamamoto, K.; Nada, S.; Okada, M.; Yamamoto, T.; Honda, Z. Essential Roles of Lyn in Fibronectin-Mediated Filamentous Actin Assembly and Cell Motility in Mast Cells. J. Immunol. 1998, 161, 3694–3701. [Google Scholar] [PubMed]
- Beaty, B.T.; Sharma, V.P.; Bravo-Cordero, J.J.; Simpson, M.A.; Eddy, R.J.; Koleske, A.J.; Condeelis, J. Β1 Integrin Regulates Arg to Promote Invadopodial Maturation and Matrix Degradation. MBoC 2013, 24, 1661–1675. [Google Scholar] [CrossRef]
- Artym, V.V.; Swatkoski, S.; Matsumoto, K.; Campbell, C.B.; Petrie, R.J.; Dimitriadis, E.K.; Li, X.; Mueller, S.C.; Bugge, T.H.; Gucek, M.; et al. Dense Fibrillar Collagen Is a Potent Inducer of Invadopodia via a Specific Signaling Network. J. Cell Biol. 2015, 208, 331–350. [Google Scholar] [CrossRef]
- Di Martino, J.; Moreau, V.; Saltel, F. Type I Collagen Fibrils: An Inducer of Invadosomes. Oncotarget 2015, 6, 28519–28520. [Google Scholar] [CrossRef] [PubMed]
- Zhao, P.; Xu, Y.; Wei, Y.; Qiu, Q.; Chew, T.-L.; Kang, Y.; Cheng, C. The CD44s Splice Isoform Is a Central Mediator for Invadopodia Activity. J. Cell Sci. 2016, 129, 1355–1365. [Google Scholar] [CrossRef] [PubMed]
- Burridge, K. Focal Adhesions: A Personal Perspective on a Half Century of Progress. FEBS J. 2017, 284, 3355–3361. [Google Scholar] [CrossRef] [PubMed]
- Tai, Y.-L.; Chen, L.-C.; Shen, T.-L. Emerging Roles of Focal Adhesion Kinase in Cancer. Biomed. Res. Int. 2015, 2015, 690690. [Google Scholar] [CrossRef]
- Tomakidi, P.; Schulz, S.; Proksch, S.; Weber, W.; Steinberg, T. Focal Adhesion Kinase (FAK) Perspectives in Mechanobiology: Implications for Cell Behaviour. Cell Tissue Res. 2014, 357, 515–526. [Google Scholar] [CrossRef]
- Schlaepfer, D.D.; Mitra, S.K.; Ilic, D. Control of Motile and Invasive Cell Phenotypes by Focal Adhesion Kinase. Biochim. Biophys. Acta Mol. Cell Res. 2004, 1692, 77–102. [Google Scholar] [CrossRef]
- Sarangi, B.R.; Gupta, M.; Doss, B.L.; Tissot, N.; Lam, F.; Mège, R.-M.; Borghi, N.; Ladoux, B. Coordination between Intra- and Extracellular Forces Regulates Focal Adhesion Dynamics. Nano Lett. 2017, 17, 399–406. [Google Scholar] [CrossRef] [PubMed]
- Larsen, M.; Artym, V.V.; Green, J.A.; Yamada, K.M. The Matrix Reorganized: Extracellular Matrix Remodeling and Integrin Signaling. Curr. Opin. Cell Biol. 2006, 18, 463–471. [Google Scholar] [CrossRef]
- Hu, Y.-L.; Lu, S.; Szeto, K.W.; Sun, J.; Wang, Y.; Lasheras, J.C.; Chien, S. FAK and Paxillin Dynamics at Focal Adhesions in the Protrusions of Migrating Cells. Sci. Rep. 2014, 4, 6024. [Google Scholar] [CrossRef] [PubMed]
- Ciobanasu, C.; Faivre, B.; Le Clainche, C. Integrating Actin Dynamics, Mechanotransduction and Integrin Activation: The Multiple Functions of Actin Binding Proteins in Focal Adhesions. Eur. J. Cell Biol. 2013, 92, 339–348. [Google Scholar] [CrossRef]
- Stutchbury, B.; Atherton, P.; Tsang, R.; Wang, D.-Y.; Ballestrem, C. Distinct Focal Adhesion Protein Modules Control Different Aspects of Mechanotransduction. J. Cell Sci. 2017, 130, 1612–1624. [Google Scholar] [CrossRef] [PubMed]
- Zhou, D.W.; Lee, T.T.; Weng, S.; Fu, J.; García, A.J. Effects of Substrate Stiffness and Actomyosin Contractility on Coupling between Force Transmission and Vinculin-Paxillin Recruitment at Single Focal Adhesions. Mol. Biol. Cell 2017, 28, 1901–1911. [Google Scholar] [CrossRef]
- Grashoff, C.; Hoffman, B.D.; Brenner, M.D.; Zhou, R.; Parsons, M.; Yang, M.T.; McLean, M.A.; Sligar, S.G.; Chen, C.S.; Ha, T.; et al. Measuring Mechanical Tension across Vinculin Reveals Regulation of Focal Adhesion Dynamics. Nature 2010, 466, 263–266. [Google Scholar] [CrossRef] [PubMed]
- Atherton, P.; Stutchbury, B.; Wang, D.-Y.; Jethwa, D.; Tsang, R.; Meiler-Rodriguez, E.; Wang, P.; Bate, N.; Zent, R.; Barsukov, I.L.; et al. Vinculin Controls Talin Engagement with the Actomyosin Machinery. Nat. Commun. 2015, 6, 10038. [Google Scholar] [CrossRef] [PubMed]
- Omachi, T.; Ichikawa, T.; Kimura, Y.; Ueda, K.; Kioka, N. Vinculin Association with Actin Cytoskeleton Is Necessary for Stiffness-Dependent Regulation of Vinculin Behavior. PLoS ONE 2017, 12, e0175324. [Google Scholar] [CrossRef]
- Calderwood, D.A.; Campbell, I.D.; Critchley, D.R. Talins and Kindlins: Partners in Integrin-Mediated Adhesion. Nat. Rev. Mol. Cell Biol. 2013, 14, 503–517. [Google Scholar] [CrossRef] [PubMed]
- Chan, Z.C.-K.; Kwan, H.-L.R.; Wong, Y.S.; Jiang, Z.; Zhou, Z.; Tam, K.W.; Chan, Y.-S.; Chan, C.B.; Lee, C.W. Site-Directed MT1-MMP Trafficking and Surface Insertion Regulate AChR Clustering and Remodeling at Developing NMJs. eLife 2020, 9, e54379. [Google Scholar] [CrossRef]
- El Azzouzi, K.; Wiesner, C.; Linder, S. Metalloproteinase MT1-MMP Islets Act as Memory Devices for Podosome Reemergence. J. Cell Biol. 2016, 213, 109–125. [Google Scholar] [CrossRef] [PubMed]
- Infante, E.; Castagnino, A.; Ferrari, R.; Monteiro, P.; Agüera-González, S.; Paul-Gilloteaux, P.; Domingues, M.J.; Maiuri, P.; Raab, M.; Shanahan, C.M.; et al. LINC Complex-Lis1 Interplay Controls MT1-MMP Matrix Digest-on-Demand Response for Confined Tumor Cell Migration. Nat. Commun. 2018, 9, 2443. [Google Scholar] [CrossRef]
- Kuo, S.-L.; Chen, C.-L.; Pan, Y.-R.; Chiu, W.-T.; Chen, H.-C. Biogenesis of Podosome Rosettes through Fission. Sci. Rep. 2018, 8, 524. [Google Scholar] [CrossRef] [PubMed]
- Van den Dries, K.; Schwartz, S.L.; Byars, J.; Meddens, M.B.M.; Bolomini-Vittori, M.; Lidke, D.S.; Figdor, C.G.; Lidke, K.A.; Cambi, A. Dual-Color Superresolution Microscopy Reveals Nanoscale Organization of Mechanosensory Podosomes. Mol. Biol. Cell 2013, 24, 2112–2123. [Google Scholar] [CrossRef]
- Shattil, S.J.; O’Toole, T.; Eigenthaler, M.; Thon, V.; Williams, M.; Babior, B.M.; Ginsberg, M.H. Beta 3-Endonexin, a Novel Polypeptide That Interacts Specifically with the Cytoplasmic Tail of the Integrin Beta 3 Subunit. J. Cell Biol. 1995, 131, 807–816. [Google Scholar] [CrossRef] [PubMed]
- Ray, R.M.; Li, C.; Bhattacharya, S.; Naren, A.P.; Johnson, L.R. Spermine, a Molecular Switch Regulating EGFR, Integrin Β3, Src, and FAK Scaffolding. Cell. Signal. 2012, 24, 931–942. [Google Scholar] [CrossRef]
- Proag, A.; Bouissou, A.; Mangeat, T.; Voituriez, R.; Delobelle, P.; Thibault, C.; Vieu, C.; Maridonneau-Parini, I.; Poincloux, R. Working Together: Spatial Synchrony in the Force and Actin Dynamics of Podosome First Neighbors. ACS Nano 2015, 9, 3800–3813. [Google Scholar] [CrossRef]
- Bhuwania, R.; Cornfine, S.; Fang, Z.; Krüger, M.; Luna, E.J.; Linder, S. Supervillin Couples Myosin-Dependent Contractility to Podosomes and Enables Their Turnover. J. Cell Sci. 2012, 125, 2300–2314. [Google Scholar] [CrossRef] [PubMed]
- Joosten, B.; Willemse, M.; Fransen, J.; Cambi, A.; van den Dries, K. Super-Resolution Correlative Light and Electron Microscopy (SR-CLEM) Reveals Novel Ultrastructural Insights Into Dendritic Cell Podosomes. Front. Immunol. 2018, 9, 1908. [Google Scholar] [CrossRef]
- Cervero, P.; Wiesner, C.; Bouissou, A.; Poincloux, R.; Linder, S. Lymphocyte-Specific Protein 1 Regulates Mechanosensory Oscillation of Podosomes and Actin Isoform-Based Actomyosin Symmetry Breaking. Nat. Commun. 2018, 9, 515. [Google Scholar] [CrossRef] [PubMed]
- Petropoulos, C.; Oddou, C.; Emadali, A.; Hiriart-Bryant, E.; Boyault, C.; Faurobert, E.; Vande Pol, S.; Kim-Kaneyama, J.-R.; Kraut, A.; Coute, Y.; et al. Roles of Paxillin Family Members in Adhesion and ECM Degradation Coupling at Invadosomes. J. Cell Biol. 2016, 213, 585–599. [Google Scholar] [CrossRef] [PubMed]
- Walde, M.; Monypenny, J.; Heintzmann, R.; Jones, G.E.; Cox, S. Vinculin Binding Angle in Podosomes Revealed by High Resolution Microscopy. PLoS ONE 2014, 9, e88251. [Google Scholar] [CrossRef]
- Praekelt, U.; Kopp, P.M.; Rehm, K.; Linder, S.; Bate, N.; Patel, B.; Debrand, E.; Manso, A.M.; Ross, R.S.; Conti, F.; et al. New Isoform-Specific Monoclonal Antibodies Reveal Different Sub-Cellular Localisations for Talin1 and Talin2. Eur. J. Cell Biol. 2012, 91, 180–191. [Google Scholar] [CrossRef]
- Labernadie, A.; Thibault, C.; Vieu, C.; Maridonneau-Parini, I.; Charrière, G.M. Dynamics of Podosome Stiffness Revealed by Atomic Force Microscopy. Proc. Natl. Acad. Sci. USA 2010, 107, 21016–21021. [Google Scholar] [CrossRef]
- Horton, M.A.; Nesbit, M.A.; Helfrich, M.H. Interaction of Osteopontin with Osteoclast Integrinsa. Ann. N. Y. Acad. Sci. 1995, 760, 190–200. [Google Scholar] [CrossRef]
- Juin, A.; Billottet, C.; Moreau, V.; Destaing, O.; Albiges-Rizo, C.; Rosenbaum, J.; Génot, E.; Saltel, F. Physiological Type I Collagen Organization Induces the Formation of a Novel Class of Linear Invadosomes. MBoC 2011, 23, 297–309. [Google Scholar] [CrossRef]
- Ferrari, R.; Infante, E.; Chavrier, P. Nucleus–Invadopodia Duo During Cancer Invasion. Trends Cell Biol. 2019, 29, 93–96. [Google Scholar] [CrossRef] [PubMed]
- Castro-Castro, A.; Marchesin, V.; Monteiro, P.; Lodillinsky, C.; Rossé, C.; Chavrier, P. Cellular and Molecular Mechanisms of MT1-MMP-Dependent Cancer Cell Invasion. Annu. Rev. Cell Dev. Biol 2016, 3220, 555–576. [Google Scholar] [CrossRef]
- Hastie, E.L.; Sherwood, D.R. A New Front in Cell Invasion: The Invadopodial Membrane. Eur. J. Cell Biol. 2016, 95, 441–448. [Google Scholar] [CrossRef] [PubMed]
- Meirson, T.; Gil-Henn, H. Targeting Invadopodia for Blocking Breast Cancer Metastasis. Drug Resist. Updates 2018, 39, 1–17. [Google Scholar] [CrossRef] [PubMed]
- Evans, J.G.; Correia, I.; Krasavina, O.; Watson, N.; Matsudaira, P. Macrophage Podosomes Assemble at the Leading Lamella by Growth and Fragmentation. J. Cell Biol. 2003, 161, 697–705. [Google Scholar] [CrossRef] [PubMed]
- Calle, Y.; Chou, H.; Thrasher, A.J.; Jones, G.E. Wiskott–Aldrich Syndrome Protein and the Cytoskeletal Dynamics of Dendritic Cells. J. Pathol. 2004, 204, 460–469. [Google Scholar] [CrossRef]
- Levy-Apter, E.; Finkelshtein, E.; Vemulapalli, V.; Li, S.S.-C.; Bedford, M.T.; Elson, A. Adaptor Protein GRB2 Promotes Src Tyrosine Kinase Activation and Podosomal Organization by Protein-Tyrosine Phosphatase ϵ in Osteoclasts. J. Biol. Chem. 2014, 289, 36048–36058. [Google Scholar] [CrossRef]
- Tsuboi, S. Requirement for a Complex of Wiskott-Aldrich Syndrome Protein (WASP) with WASP Interacting Protein in Podosome Formation in Macrophages. J. Immunol. 2007, 178, 2987–2995. [Google Scholar] [CrossRef] [PubMed]
- Pignatelli, J.; Bravo-Cordero, J.J.; Roh-Johnson, M.; Gandhi, S.J.; Wang, Y.; Chen, X.; Eddy, R.J.; Xue, A.; Singer, R.H.; Hodgson, L.; et al. Macrophage-Dependent Tumor Cell Transendothelial Migration Is Mediated by Notch1/MenaINV-Initiated Invadopodium Formation. Sci. Rep. 2016, 6, 37874. [Google Scholar] [CrossRef]
- Oser, M.; Dovas, A.; Cox, D.; Condeelis, J. Nck1 and Grb2 Localization Patterns Can Distinguish Invadopodia from Podosomes. Eur. J. Cell Biol. 2011, 90, 181–188. [Google Scholar] [CrossRef]
- Weidmann, M.D.; Surve, C.R.; Eddy, R.J.; Chen, X.; Gertler, F.B.; Sharma, V.P.; Condeelis, J.S. Mena(INV) Dysregulates Cortactin Phosphorylation to Promote Invadopodium Maturation. Sci. Rep. 2016, 6, 36142. [Google Scholar] [CrossRef] [PubMed]
- Hughes, S.K.; Oudin, M.J.; Tadros, J.; Neil, J.; Del Rosario, A.; Joughin, B.A.; Ritsma, L.; Wyckoff, J.; Vasile, E.; Eddy, R.; et al. PTP1B-Dependent Regulation of Receptor Tyrosine Kinase Signaling by the Actin-Binding Protein Mena. MBoC 2015, 26, 3867–3878. [Google Scholar] [CrossRef]
- Rajadurai, C.V.; Havrylov, S.; Coelho, P.P.; Ratcliffe, C.D.H.; Zaoui, K.; Huang, B.H.; Monast, A.; Chughtai, N.; Sangwan, V.; Gertler, F.B.; et al. 5′-Inositol Phosphatase SHIP2 Recruits Mena to Stabilize Invadopodia for Cancer Cell Invasion. J. Cell Biol. 2016, 214, 719–734. [Google Scholar] [CrossRef]
- Gorai, S.; Paul, D.; Haloi, N.; Borah, R.; Santra, M.K.; Manna, D. Mechanistic Insights into the Phosphatidylinositol Binding Properties of the Pleckstrin Homology Domain of Lamellipodin. Mol. BioSyst. 2016, 12, 747–757. [Google Scholar] [CrossRef] [PubMed]
- Bravo-Cordero, J.J.; Magalhaes, M.A.O.; Eddy, R.J.; Hodgson, L.; Condeelis, J. Functions of Cofilin in Cell Locomotion and Invasion. Nat. Rev. Mol. Cell Biol. 2013, 14, 405–415. [Google Scholar] [CrossRef]
- Beaty, B.T.; Condeelis, J. Digging a Little Deeper: The Stages of Invadopodium Formation and Maturation. Eur. J. Cell Biol. 2014, 93, 438–444. [Google Scholar] [CrossRef]
- Watson, J.R.; Owen, D.; Mott, H.R. Cdc42 in Actin Dynamics: An Ordered Pathway Governed by Complex Equilibria and Directional Effector Handover. Small GTPases 2017, 8, 237–244. [Google Scholar] [CrossRef] [PubMed]
- Murphy, D.A.; Courtneidge, S.A. The “ins” and “Outs” of Podosomes and Invadopodia: Characteristics, Formation and Function. Nat. Rev. Mol. Cell Biol. 2011, 12, 413–426. [Google Scholar] [CrossRef]
- Jacob, A.; Linklater, E.; Bayless, B.A.; Lyons, T.; Prekeris, R. The Role and Regulation of Rab40b-Tks5 Complex during Invadopodia Formation and Cancer Cell Invasion. J. Cell Sci. 2016, 129, 4341–4353. [Google Scholar] [CrossRef]
- Williams, K.C.; McNeilly, R.E.; Coppolino, M.G. SNAP23, Syntaxin4, and Vesicle-Associated Membrane Protein 7 (VAMP7) Mediate Trafficking of Membrane Type 1-Matrix Metalloproteinase (MT1-MMP) during Invadopodium Formation and Tumor Cell Invasion. Mol. Biol Cell. 2014, 25, 2061–2070. [Google Scholar] [CrossRef]
- Steffen, A.; Le Dez, G.; Poincloux, R.; Recchi, C.; Nassoy, P.; Rottner, K.; Galli, T.; Chavrier, P. MT1-MMP-Dependent Invasion Is Regulated by TI-VAMP/VAMP7. Curr. Biol. 2008, 18, 926–931. [Google Scholar] [CrossRef]
- Vieira, L.F.D.A.; Lins, M.P.; Viana, I.M.M.N.; Dos Santos, J.E.; Smaniotto, S.; Reis, M.D.D.S. Metallic Nanoparticles Reduce the Migration of Human Fibroblasts In Vitro. Nanoscale Res. Lett. 2017, 12, 200. [Google Scholar] [CrossRef]
- Lo, H.-M.; Ma, M.-C.; Shieh, J.; Chen, H.-L.; Wu, W. Naked Physically Synthesized Gold Nanoparticles Affect Migration, Mitochondrial Activity, and Proliferation of Vascular Smooth Muscle Cells. Int. J. Nanomed. 2018, 13, 3163–3176. [Google Scholar] [CrossRef] [PubMed]
- Královec, K.; Havelek, R.; Koutová, D.; Veverka, P.; Kubíčková, L.; Brázda, P.; Kohout, J.; Herynek, V.; Vosmanská, M.; Kaman, O. Magnetic Nanoparticles of Ga-Substituted ε-Fe2O3 for Biomedical Applications: Magnetic Properties, Transverse Relaxivity, and Effects of Silica-Coated Particles on Cytoskeletal Networks. J. Biomed. Mater. Res. Part A 2020, 108, 1563–1578. [Google Scholar] [CrossRef] [PubMed]
- Sheykhzadeh, S.; Luo, M.; Peng, B.; White, J.; Abdalla, Y.; Tang, T.; Mäkilä, E.; Voelcker, N.H.; Tong, W.Y. Transferrin-Targeted Porous Silicon Nanoparticles Reduce Glioblastoma Cell Migration across Tight Extracellular Space. Sci. Rep. 2020, 10, 2320. [Google Scholar] [CrossRef]
- Popara, J.; Accomasso, L.; Vitale, E.; Gallina, C.; Roggio, D.; Iannuzzi, A.; Raimondo, S.; Rastaldo, R.; Alberto, G.; Catalano, F.; et al. Silica Nanoparticles Actively Engage with Mesenchymal Stem Cells in Improving Acute Functional Cardiac Integration. Nanomedicine 2018, 13, 1121–1138. [Google Scholar] [CrossRef] [PubMed]
- Kenific, C.M.; Wittmann, T.; Debnath, J. Autophagy in Adhesion and Migration. J. Cell Sci. 2016, 129, 3685–3693. [Google Scholar] [CrossRef] [PubMed]
- Sharifi, M.N.; Mowers, E.E.; Drake, L.E.; Collier, C.; Chen, H.; Zamora, M.; Mui, S.; Macleod, K.F. Autophagy Promotes Focal Adhesion Disassembly and Cell Motility of Metastatic Tumor Cells through the Direct Interaction of Paxillin with LC3. Cell Rep. 2016, 15, 1660–1672. [Google Scholar] [CrossRef] [PubMed]
- Kenific, C.M.; Stehbens, S.J.; Goldsmith, J.; Leidal, A.M.; Faure, N.; Ye, J.; Wittmann, T.; Debnath, J. NBR1 Enables Autophagy-Dependent Focal Adhesion Turnover. J. Cell Biol. 2016, 212, 577–590. [Google Scholar] [CrossRef]
- Dower, C.M.; Wills, C.A.; Frisch, S.M.; Wang, H.-G. Mechanisms and Context Underlying the Role of Autophagy in Cancer Metastasis. Autophagy 2018, 14, 1110–1128. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Yin, S.; Zhang, L.; Shi, K.; Tang, J.; Zhang, Z.; He, Q. A Tumor-Activatable Particle with Antimetastatic Potential in Breast Cancer via Inhibiting the Autophagy-Dependent Disassembly of Focal Adhesion. Biomaterials 2018, 168, 1–9. [Google Scholar] [CrossRef]
- Shin, T.H.; Lee, D.Y.; Ketebo, A.A.; Lee, S.; Manavalan, B.; Basith, S.; Ahn, C.; Kang, S.H.; Park, S.; Lee, G. Silica-Coated Magnetic Nanoparticles Decrease Human Bone Marrow-Derived Mesenchymal Stem Cell Migratory Activity by Reducing Membrane Fluidity and Impairing Focal Adhesion. Nanomaterials 2019, 9, 1475. [Google Scholar] [CrossRef]
- Mulens-Arias, V.; Rojas, J.M.; Sanz-Ortega, L.; Portilla, Y.; Pérez-Yagüe, S.; Barber, D.F. Polyethylenimine-Coated Superparamagnetic Iron Oxide Nanoparticles Impair in Vitro and in Vivo Angiogenesis. Nanomed. Nanotechnol. Biol. Med. 2019, 21, 102063. [Google Scholar] [CrossRef] [PubMed]
- Ma, X.; Hartmann, R.; De Aberasturi, D.J.; Yang, F.; Soenen, S.J.H.; Manshian, B.B.; Franz, J.; Valdeperez, D.; Pelaz, B.; Feliu, N.; et al. Colloidal Gold Nanoparticles Induce Changes in Cellular and Subcellular Morphology. ACS Nano 2017, 11, 7807–7820. [Google Scholar] [CrossRef]
- Královec, K.; Melounková, L.; Slováková, M.; Mannová, N.; Sedlák, M.; Bartáček, J.; Havelek, R. Disruption of Cell Adhesion and Cytoskeletal Networks by Thiol-Functionalized Silica-Coated Iron Oxide Nanoparticles. Int. J. Mol. Sci. 2020, 21, 9350. [Google Scholar] [CrossRef] [PubMed]
- Ketebo, A.A.; Shin, T.H.; Jun, M.; Lee, G.; Park, S. Effect of Silica-Coated Magnetic Nanoparticles on Rigidity Sensing of Human Embryonic Kidney Cells. J. Nanobiotechnology 2020, 18, 170. [Google Scholar] [CrossRef] [PubMed]
- Marcus, J.; Bejerano-Sagie, M.; Patterson, N.; Bagchi, S.; Verkhusha, V.V.; Connolly, D.; Goldberg, G.L.; Golden, A.; Sharma, V.P.; Condeelis, J.; et al. Septin 9 Isoforms Promote Tumorigenesis in Mammary Epithelial Cells by Increasing Migration and ECM Degradation through Metalloproteinase Secretion at Focal Adhesions. Oncogene 2019, 38, 5839. [Google Scholar] [CrossRef]
- Mulens-Arias, V.; Balfourier, A.; Nicolás-Boluda, A.; Carn, F.; Gazeau, F. Disturbance of Adhesomes by Gold Nanoparticles Reveals a Size- and Cell Type-Bias. Biomater. Sci. 2019, 7, 389–408. [Google Scholar] [CrossRef]
- Soenen, S.J.H.; Nuytten, N.; De Meyer, S.F.; De Smedt, S.C.; De Cuyper, M. High Intracellular Iron Oxide Nanoparticle Concentrations Affect Cellular Cytoskeleton and Focal Adhesion Kinase-Mediated Signaling. Small 2010, 6, 832–842. [Google Scholar] [CrossRef] [PubMed]
- Tay, C.Y.; Cai, P.; Setyawati, M.I.; Fang, W.; Tan, L.P.; Hong, C.H.L.; Chen, X.; Leong, D.T. Nanoparticles Strengthen Intracellular Tension and Retard Cellular Migration. Nano Lett. 2014, 14, 83–88. [Google Scholar] [CrossRef]
- Discher, D.E.; Janmey, P.; Wang, Y. Tissue Cells Feel and Respond to the Stiffness of Their Substrate. Science 2005, 310, 1139–1143. [Google Scholar] [CrossRef]
- Ingber, D.E. Tensegrity: The Architectural Basis of Cellular Mechanotransduction. Annu. Rev. Physiol. 1997, 59, 575–599. [Google Scholar] [CrossRef] [PubMed]
- Humphrey, J.D.; Dufresne, E.R.; Schwartz, M.A. Mechanotransduction and Extracellular Matrix Homeostasis. Nat. Rev. Mol. Cell Biol. 2014, 15, 802–812. [Google Scholar] [CrossRef]
- Fouchard, J.; Bimbard, C.; Bufi, N.; Durand-Smet, P.; Proag, A.; Richert, A.; Cardoso, O.; Asnacios, A. Three-Dimensional Cell Body Shape Dictates the Onset of Traction Force Generation and Growth of Focal Adhesions. Proc. Natl. Acad. Sci. USA 2014, 111, 13075–13080. [Google Scholar] [CrossRef] [PubMed]
- Wu, Z.; Plotnikov, S.V.; Moalim, A.Y.; Waterman, C.M.; Liu, J. Two Distinct Actin Networks Mediate Traction Oscillations to Confer Focal Adhesion Mechanosensing. Biophys. J. 2017, 112, 780–794. [Google Scholar] [CrossRef]
- Ibrahim, M.; Schoelermann, J.; Mustafa, K.; Cimpan, M.R. TiO2 Nanoparticles Disrupt Cell Adhesion and the Architecture of Cytoskeletal Networks of Human Osteoblast-like Cells in a Size Dependent Manner. J. Biomed. Mater. Res. Part A 2018, 106, 2582–2593. [Google Scholar] [CrossRef]
- Miyauchi, T.; Yamada, M.; Yamamoto, A.; Iwasa, F.; Suzawa, T.; Kamijo, R.; Baba, K.; Ogawa, T. The Enhanced Characteristics of Osteoblast Adhesion to Photofunctionalized Nanoscale TiO2 Layers on Biomaterials Surfaces. Biomaterials 2010, 31, 3827–3839. [Google Scholar] [CrossRef]
- Lipski, A.M.; Pino, C.J.; Haselton, F.R.; Chen, I.-W.; Shastri, V.P. The Effect of Silica Nanoparticle-Modified Surfaces on Cell Morphology, Cytoskeletal Organization and Function. Biomaterials 2008, 29, 3836–3846. [Google Scholar] [CrossRef]
- Perez, J.E.; Fage, F.; Pereira, D.; Abou-Hassan, A.; Asnacios, S.; Asnacios, A.; Wilhelm, C. Transient Cell Stiffening Triggered by Magnetic Nanoparticle Exposure. J. Nanobiotechnol. 2021, 19, 117. [Google Scholar] [CrossRef] [PubMed]
- Lyu, Z.; Wang, H.; Wang, Y.; Ding, K.; Liu, H.; Yuan, L.; Shi, X.; Wang, M.; Wang, Y.; Chen, H. Maintaining the Pluripotency of Mouse Embryonic Stem Cells on Gold Nanoparticle Layers with Nanoscale but Not Microscale Surface Roughness. Nanoscale 2014, 6, 6959. [Google Scholar] [CrossRef]
- Chen, W.; Villa-Diaz, L.G.; Sun, Y.; Weng, S.; Kim, J.K.; Lam, R.H.W.; Han, L.; Fan, R.; Krebsbach, P.H.; Fu, J. Nanotopography Influences Adhesion, Spreading, and Self-Renewal of Human Embryonic Stem Cells. ACS Nano 2012, 6, 4094–4103. [Google Scholar] [CrossRef]
- Azatov, M.; Sun, X.; Suberi, A.; Fourkas, J.T.; Upadhyaya, A. Topography on a Subcellular Scale Modulates Cellular Adhesions and Actin Stress Fiber Dynamics in Tumor Associated Fibroblasts. Phys. Biol. 2017, 14, 065003. [Google Scholar] [CrossRef]
- Bello, D.G.; Fouillen, A.; Badia, A.; Nanci, A. A Nanoporous Titanium Surface Promotes the Maturation of Focal Adhesions and Formation of Filopodia with Distinctive Nanoscale Protrusions by Osteogenic Cells. Acta Biomater. 2017, 60, 339–349. [Google Scholar] [CrossRef] [PubMed]
- Arnold, M.; Schwieder, M.; Blümmel, J.; Cavalcanti-Adam, E.A.; López-Garcia, M.; Kessler, H.; Geiger, B.; Spatz, J.P. Cell Interactions with Hierarchically Structured Nano-Patterned Adhesive Surfaces. Soft Matter 2009, 5, 72–77. [Google Scholar] [CrossRef] [PubMed]
- Abagnale, G.; Steger, M.; Nguyen, V.H.; Hersch, N.; Sechi, A.; Joussen, S.; Denecke, B.; Merkel, R.; Hoffmann, B.; Dreser, A.; et al. Surface Topography Enhances Differentiation of Mesenchymal Stem Cells towards Osteogenic and Adipogenic Lineages. Biomaterials 2015, 61, 316–326. [Google Scholar] [CrossRef]
- Posa, F.; Baha-Schwab, E.H.; Wei, Q.; Di Benedetto, A.; Neubauer, S.; Reichart, F.; Kessler, H.; Spatz, J.P.; Albiges-Rizo, C.; Mori, G.; et al. Surface Co-Presentation of BMP-2 and Integrin Selective Ligands at the Nanoscale Favors A5β1 Integrin-Mediated Adhesion. Biomaterials 2021, 267, 120484. [Google Scholar] [CrossRef]
- Kang, H.; Wong, D.S.H.; Yan, X.; Jung, H.J.; Kim, S.; Lin, S.; Wei, K.; Li, G.; Dravid, V.P.; Bian, L. Remote Control of Multimodal Nanoscale Ligand Oscillations Regulates Stem Cell Adhesion and Differentiation. ACS Nano 2017, 11, 9636–9649. [Google Scholar] [CrossRef]
- Kang, H.; Jung, H.J.; Wong, D.S.H.; Kim, S.K.; Lin, S.; Chan, K.F.; Zhang, L.; Li, G.; Dravid, V.P.; Bian, L. Remote Control of Heterodimeric Magnetic Nanoswitch Regulates the Adhesion and Differentiation of Stem Cells. J. Am. Chem. Soc. 2018, 140, 5909–5913. [Google Scholar] [CrossRef]
- Okada, T.; Ogura, T. Nanoscale Imaging of the Adhesion Core Including Integrin Β1 on Intact Living Cells Using Scanning Electron-Assisted Dielectric-Impedance Microscopy. PLoS ONE 2018, 13, e0204133. [Google Scholar] [CrossRef]
- Rojas, J.M.; Sanz-Ortega, L.; Mulens-Arias, V.; Gutiérrez, L.; Pérez-Yagüe, S.; Barber, D.F. Superparamagnetic Iron Oxide Nanoparticle Uptake Alters M2 Macrophage Phenotype, Iron Metabolism, Migration and Invasion. Nanomed. Nanotechnol. Biol. Med. 2016, 12, 1127–1138. [Google Scholar] [CrossRef] [PubMed]
- Mulens-Arias, V.; Rojas, J.M.; Pérez-Yagüe, S.; Morales, M.P.; Barber, D.F. Polyethylenimine-Coated SPIONs Trigger Macrophage Activation through TLR-4 Signaling and ROS Production and Modulate Podosome Dynamics. Biomaterials 2015, 52, 494–506. [Google Scholar] [CrossRef]
- Akatsuka, S.; Yamashita, Y.; Ohara, H.; Liu, Y.-T.; Izumiya, M.; Abe, K.; Ochiai, M.; Jiang, L.; Nagai, H.; Okazaki, Y.; et al. Fenton Reaction Induced Cancer in Wild Type Rats Recapitulates Genomic Alterations Observed in Human Cancer. PLoS ONE 2012, 7, e43403. [Google Scholar] [CrossRef] [PubMed]
- Brillo, V.; Chieregato, L.; Leanza, L.; Muccioli, S.; Costa, R. Mitochondrial Dynamics, ROS, and Cell Signaling: A Blended Overview. Life 2021, 11, 332. [Google Scholar] [CrossRef]
- Xu, J.; Yang, J.; Nyga, A.; Ehteramyan, M.; Moraga, A.; Wu, Y.; Zeng, L.; Knight, M.M.; Shelton, J.C. Cobalt (II) Ions and Nanoparticles Induce Macrophage Retention by ROS-Mediated down-Regulation of RhoA Expression. Acta Biomater. 2018, 72, 434–446. [Google Scholar] [CrossRef] [PubMed]
- Linder, S.; Aepfelbacher, M. Podosomes: Adhesion Hot-Spots of Invasive Cells. Trends Cell Biol. 2003, 13, 376–385. [Google Scholar] [CrossRef]
- Mulens-Arias, V.; Rojas, J.M.; Pérez-Yagüe, S.; Morales, M.d.P.; Barber, D.F. Polyethylenimine-Coated SPION Exhibits Potential Intrinsic Anti-Metastatic Properties Inhibiting Migration and Invasion of Pancreatic Tumor Cells. J. Control. Release 2015, 216, 78–92. [Google Scholar] [CrossRef]
- Simone, T.M.; Higgins, C.E.; Czekay, R.-P.; Law, B.K.; Higgins, S.P.; Archambeault, J.; Kutz, S.M.; Higgins, P.J. SERPINE1: A Molecular Switch in the Proliferation-Migration Dichotomy in Wound-”Activated” Keratinocytes. Adv. Wound Care 2014, 3, 281–290. [Google Scholar] [CrossRef]
- Shih, Y.-P.; Takada, Y.; Lo, S.H. Silencing of DLC1 Upregulates PAI-1 Expression and Reduces Migration in Normal Prostate Cells. Mol. Cancer Res. 2012, 10, 34–39. [Google Scholar] [CrossRef]
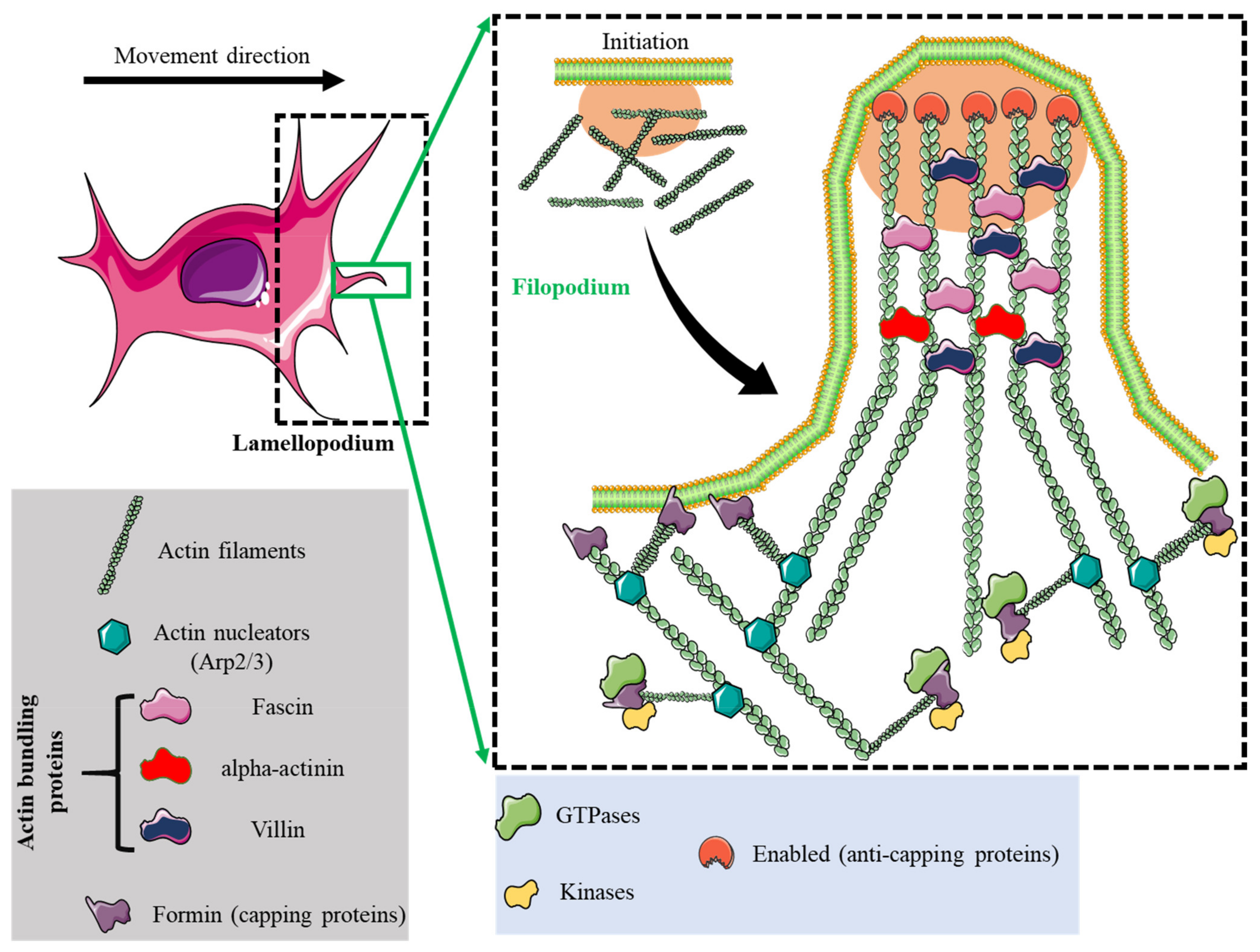
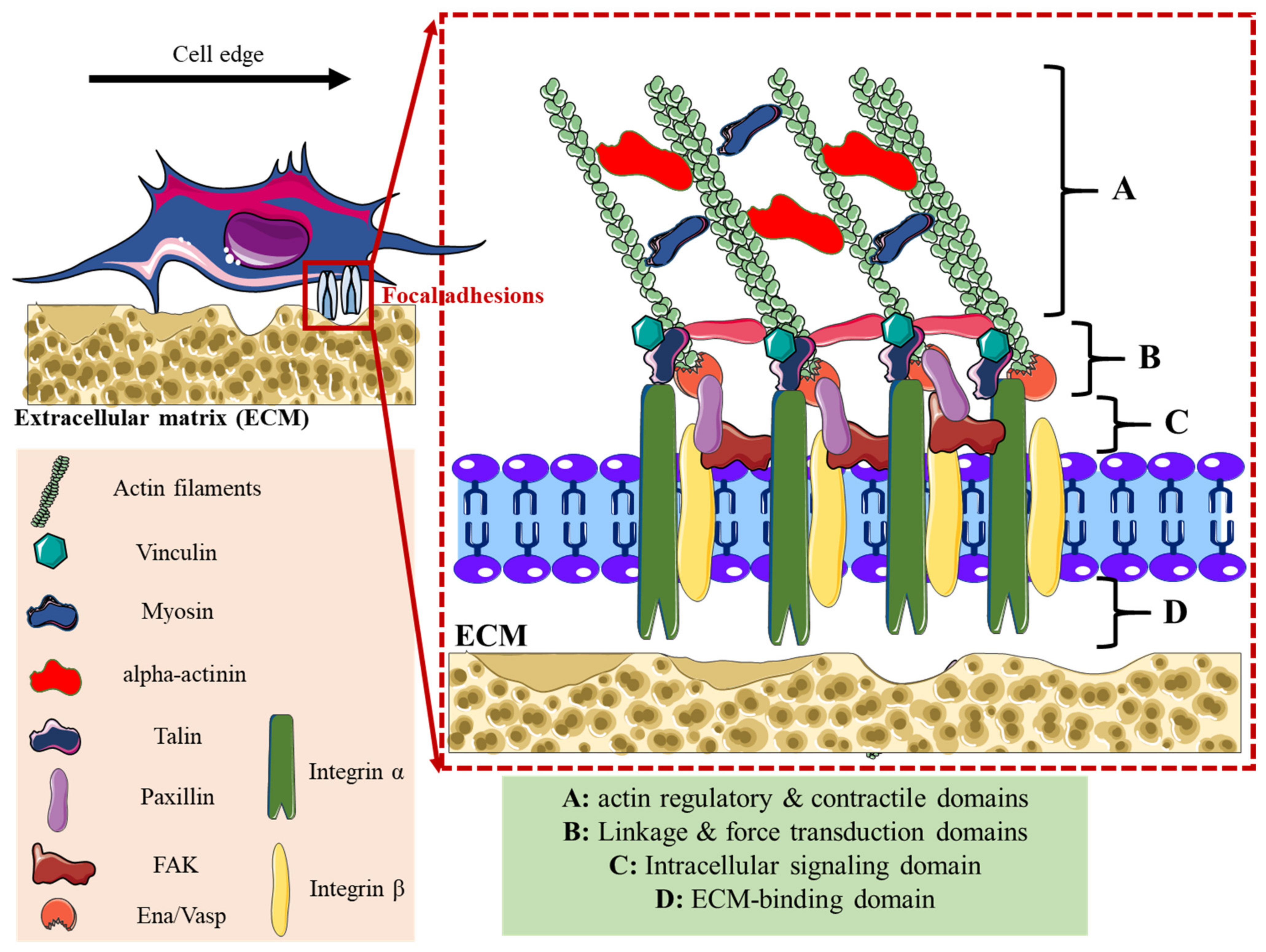
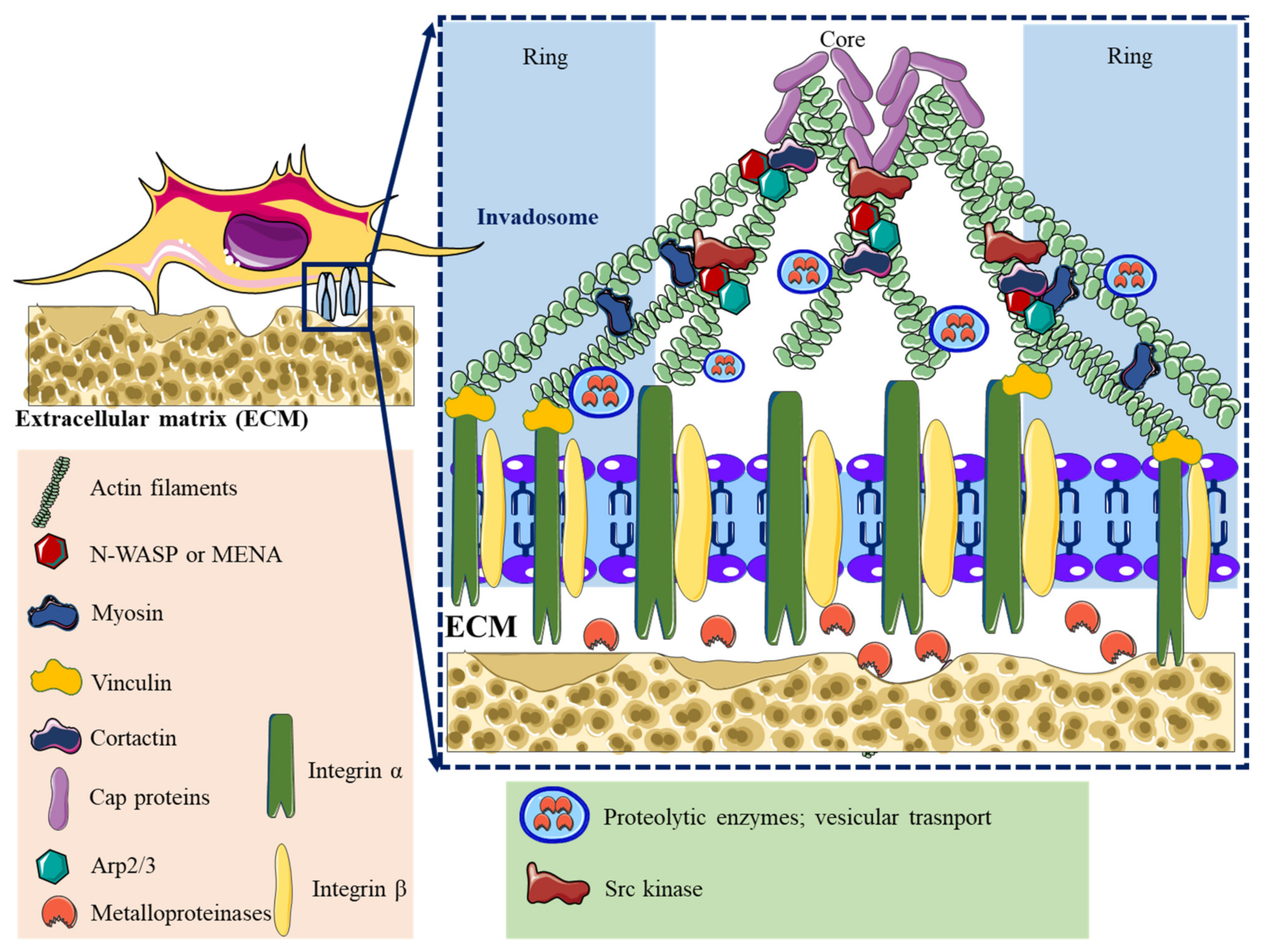
| Lamellipodia | Filopodia | Focal Adhesions | Podosomes | Invadopodia | |
|---|---|---|---|---|---|
| Structure | Sheet-like protrusions that attach to ECM driven by branched actin arrangements | Often originating at lamellipodium as finger-like extensions driven by linear actin polymerization | Clusters of transmembrane receptors, integrins, and cytosolic proteins driven by parallel actin filaments branched at the end | A discrete actin-rich core surrounded by a ring of actin-associated and signaling proteins, driven by branched and unbranched actin filaments | A discrete actin-rich core is surrounded by a ring of actin-associated and signaling proteins. Often linked to tumor cells driven by parallel actin filaments within the tip and branched at the base |
| Cellular location | Leading-edge | Embedded within lamellipodia | Leading-edge of the cell | Ventral cell surface, often clustered behind the leading edge of the cell | Ventral cell surface, often situated under the nucleus |
| Dimesions | Width: 0.1–0.2 μm | Width: 0.1–0.3 μm; length: 3–10 μm | Width: 2–6 μm | Width: 0.5–2 μm; length: 0.5–1 μm [45] | Width: 0.5–2 μm; length: >2 μm |
| Pericellular proteolysis | Minimal | No | Minimal [46] | Yes, through MT1MMP and UPAR | Yes, through MMP2, MMP9, MT1MMP, seprase, UPAR, ADAM12, ADAM15, and ADAM19 |
| Duration of structure | Minutes | Minutes | Hours, it depends on the rate of cell migration | Minutes [47] | Hours [48] |
| Soluble stimuli | HGF [49,50], TNFα and TNFβ [51], endothelin-3 [52], CLCF-1 [53] | VEGF-A [54], GDF-5 [55], EGF [56], HGF [57], TNFα [58] and TNFβ [51], leptin [59] | Estrogen [60], TGFβ1 [61], endothelin-3 [52] | IL-5 [62], VEGF-A [63], pro-NGF [64], thymosin α1 [65], hepatoma-derived growth factor (HDGF) [66], TGFβ1 [67], NaF [68], KGF [69], SDF-1α [70], exosomes [71], osteopontin [72] | EGF [73], PDGF [74], TGFβ [75], VEGF [76], HGF [77,78], SDF-1 [79] |
| ECM stimuli | Fibronectin [80], fibrinogen [81] | Fibronectin [80], fibrinogen [81] | Fibronectin [82,83], | Collagen I [84], fibronectin [85,86], fibrinogen [85], | Fibrinogen [87], Collagen I [88,89], hyaluronan [90] |
| Nanoparticle | Cell | FA Markers | Probable Signaling Direction | Time (h) | Effect |
|---|---|---|---|---|---|
| SiO2 | Human mesenchymal stem cells | Vinculin | Inside–out | 16 | Increased FA size and maturation [147] |
| SiO2 (50, 100, 300 nm) | Bovine aortic endothelial cells (BAEC) | F-actin/vinculin | Outside–in | 24 | Decreased FA size [169] |
| SiO2 (50, 100, 300 nm) | Mouse calvarial preosteoblasts (MC3T3-E1) | F-actin/vinculin | Outside–in | 24 | Increased FA size [169] |
| SiO2@IONP | Human bone marrow-derived mesenchymal stem cells (hBM-MSCs) | F-actin/vinculin | Inside–out | 12 | Decreased FA size, decreased phospho-Src, reduced phospho-FAK, reduced traction forces and cell migration [153] |
| Ga-substituted ε-Fe2O3 | A549 | F-actin/vinculin | Inside–out | 24 | Decreased FA area and reduced cell adhesion [145] |
| Resovist, Endorem, and magnetoliposomes | Primary human blood outgrowth endothelial cells (hBOECs) | F-actin/vinculin | Inside–out | 24 | Decreased FA formation and maturation Reduced phospho-FAK Reduced cell migration [160] |
| AuNP (16, 30, 40 nm) | Mouse endothelial cell(SVEC4-10) | F-actin/vinculin and F-actin/zyxin | Outside–in | 2 and 24 | Decreased total FA size, reduced MT1-MMP recruitment Reduced MMP2 and ECM degradation [159] |
| AuNP (16, 30, 40 nm) | Mouse mesenchymal stem cells | F-actin/vinculin and F-actin/zyxin | Outside–in | 2 and 24 | Decreased total and mature FA, reduced ECM degradation [159] |
| AuNP | Human umbilical vein endothelial cells (HUVECs) | F-actin/vinculin | Inside–out | 24 | Reduced FA size and impaired F-actin and tubulin polymerization [155] |
| SiO2 | Oral mucosa cells TR156 | F-actin/vinculin | Inside–out | 12 | Increased F-actin filament traction, increased FA formation Decreased cell migration [161] |
| TiO2 | Oral mucosa cells TR156 | F-actin/vinculin | Inside–out | 12 | Increased F-actin filament traction, increased FA formation Decreased cell migration [161] |
| Hydroxyapatite | Oral mucosa cells TR156 | F-actin/vinculin | Inside–out | 12 | Increased F-actin filament traction, increased FA formation Decreased cell migration [161] |
| TiO2 | Human osteoblast-like cells SaOS-2 | F-actin/p-FAK | Inside–out | 24 | Decreased FA area, reduced cell migration Reduced vinculin [167] |
| Polyethylenimine@IONP | HUVEC | F-actin/vinculin p-cortactin | Inside–out | 6 | Lower phospho-cortactin+ FA, reduced cell migrationReduced phospho-Src and phospho-cortactin ROS involvement [154] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mulens-Arias, V. Dissecting the Inorganic Nanoparticle-Driven Interferences on Adhesome Dynamics. J. Nanotheranostics 2021, 2, 174-195. https://doi.org/10.3390/jnt2030011
Mulens-Arias V. Dissecting the Inorganic Nanoparticle-Driven Interferences on Adhesome Dynamics. Journal of Nanotheranostics. 2021; 2(3):174-195. https://doi.org/10.3390/jnt2030011
Chicago/Turabian StyleMulens-Arias, Vladimir. 2021. "Dissecting the Inorganic Nanoparticle-Driven Interferences on Adhesome Dynamics" Journal of Nanotheranostics 2, no. 3: 174-195. https://doi.org/10.3390/jnt2030011
APA StyleMulens-Arias, V. (2021). Dissecting the Inorganic Nanoparticle-Driven Interferences on Adhesome Dynamics. Journal of Nanotheranostics, 2(3), 174-195. https://doi.org/10.3390/jnt2030011





