Abstract
Vision transformers (ViTs) have recently gained traction in plant disease classification due to their strong performance in visual recognition tasks. However, their application to tomato leaf disease recognition remains challenged by two factors, namely the need for models that can generalise across diverse disease conditions and the absence of a unified framework for systematic comparison. Existing ViT-based approaches often yield inconsistent results across datasets and disease types, limiting their reliability and practical deployment. To address these limitations, this study proposes the ViT-Based Robust Framework (ViT-RoT), a novel benchmarking framework designed to systematically evaluate the performance of various ViT architectures in tomato leaf disease classification. The framework facilitates the systematic classification of state-of-the-art ViT variants into high-, moderate-, and low-performing groups for tomato leaf disease recognition. A thorough empirical analysis is conducted using one publicly available benchmark dataset, assessed through standard evaluation metrics. Results demonstrate that the ConvNeXt-Small and Swin-Small models consistently achieve superior accuracy and robustness across all datasets. Beyond identifying the most effective ViT variant, the study highlights critical considerations for designing ViT-based models that are not only accurate but also efficient and adaptable to real-world agricultural applications. This study contributes a structured foundation for future research and development in vision-based plant disease diagnosis.
1. Introduction
The agricultural sector is critical to global food security and contributes significantly to national economies. In this context, tomato cultivation is a major contributor to local and international markets [1]. The demand for tomatoes has increased in recent years, mainly driven by the expanding fast-food industry and the growing consumption of processed tomato-based products. The rising demand has led to more efforts to improve tomato production. Tomato farming also plays a vital role in supporting the livelihoods of smallholder farmers, particularly in developing countries. In many regions, it serves as a cash crop that enhances household income and rural employment opportunities. However, tomato crops are highly vulnerable to a variety of leaf diseases, such as early blight, late blight, and leaf mold, which can adversely affect the plant and significantly reduce yield quality and quantity, consequently leading to economic losses [2]. Therefore, timely and accurate identification of these diseases is essential for effective application and sustainable crop management.
Traditionally, disease detection in plant leaves has relied on manual inspection by agricultural experts and farmers, which is labour-intensive, time-consuming and prone to human error [3]. To overcome these limitations, in the past, researchers have utilised artificial intelligence (AI) and computer vision-based solutions [4]. Convolutional neural networks (CNNs), in particular, have demonstrated significant effectiveness in image-based plant disease recognition tasks [5]. To improve performance and better adapt to the characteristics of specific plant disease datasets, custom-designed CNNs are commonly employed in this domain [6,7]. More recently, transfer learning has gained prominence as an effective alternative, leveraging pre-trained CNNs to enhance classification accuracy while minimising reliance on large labelled datasets. Building on this approach, various pre-trained CNN models have been employed for plant leaf disease classification, each offering distinct advantages and strengths [8]. VGG16 and VGG19 are widely used baselines due to their computational efficiency, while ResNet50 and ResNet101 leverage skip connections to achieve greater depth and generalisability in distinguishing visually similar leaf diseases [9,10,11]. MobileNet’s lightweight architecture makes it particularly well-suited for deploying leaf disease classification models on mobile and embedded agricultural devices [12]. EfficientNet balances accuracy and efficiency using compound scaling, while DenseNet enhances leaf disease detection through feature reuse across layers [13,14,15]. However, the inherent limitations of convolutional operations often hinder CNNs from capturing subtle and infrequent features in leaf images, which are crucial for distinguishing visually similar disease patterns [16].
In recent years, vision transformers (ViTs) have emerged as a robust alternative to CNNs for a range of computer vision tasks. Originally introduced for natural language processing (NLP), ViTs gained popularity after outperforming CNNs on various benchmark datasets, particularly in image classification tasks with large amounts of labelled data [17,18,19]. Recent studies have shown that ViTs effectively capture intricate features, enhancing their ability to distinguish between subtle disease patterns, such as those found in plants, where differences are difficult to detect [20,21]. Additionally, unlike traditional deep learning methods that often require extensive preprocessing and manual feature extraction, ViTs minimise the need for specialised knowledge in feature engineering [22]. This is particularly valuable in agriculture, where effective disease detection enhances trust in AI and informs expert decision-making. As plant disease detection advances, ViTs are expected to provide more accurate and efficient solutions for sustainable crop management. Recently, researchers have developed ViT variants to enhance the efficiency of image-based classification tasks, which leverage the transformer model’s ability to accurately capture contextual relationships and features within images. Notable variants that are used in plant disease recognition include compact convolutional transformers (CCTs) [23], Swin transformers [24], MobileViT [25], MaxViT [17], and EfficientViT [26].
Despite the growing adoption of ViTs in plant disease recognition, their practical application in agriculture remains limited due to several unresolved challenges. Specifically, these include (1) the absence of high-performing ViT models that can accurately classify diverse and visually similar leaf diseases under real-world agricultural conditions and (2) the lack of a systematic comparative framework to evaluate and benchmark the effectiveness of various ViT variants using leaf images. While several ViT-based approaches have demonstrated encouraging results, their performance often varies across datasets and disease types, making generalisation difficult in uncontrolled environments. Furthermore, practical deployment in agricultural settings requires models that are not only accurate but also computationally efficient and adaptable to resource-constrained environments. To address these limitations, this study introduces ViT-RoT, a benchmarking framework designed to systematically evaluate ViT architectures for tomato leaf disease recognition. By establishing a structured comparative analysis of ViT variants, ViT-RoT provides insights into the trade-offs between accuracy, efficiency, and generalisability, ensuring that selected models are optimised for real-world agricultural applications. This research conducts a comprehensive empirical study to identify the most robust ViT models for accurately classifying tomato leaf diseases under diverse conditions. The study aims to establish a well-defined evaluation framework that enhances disease detection in real-world agricultural settings by incorporating systematic benchmarking and performance analysis. The key contributions of this study are outlined below.
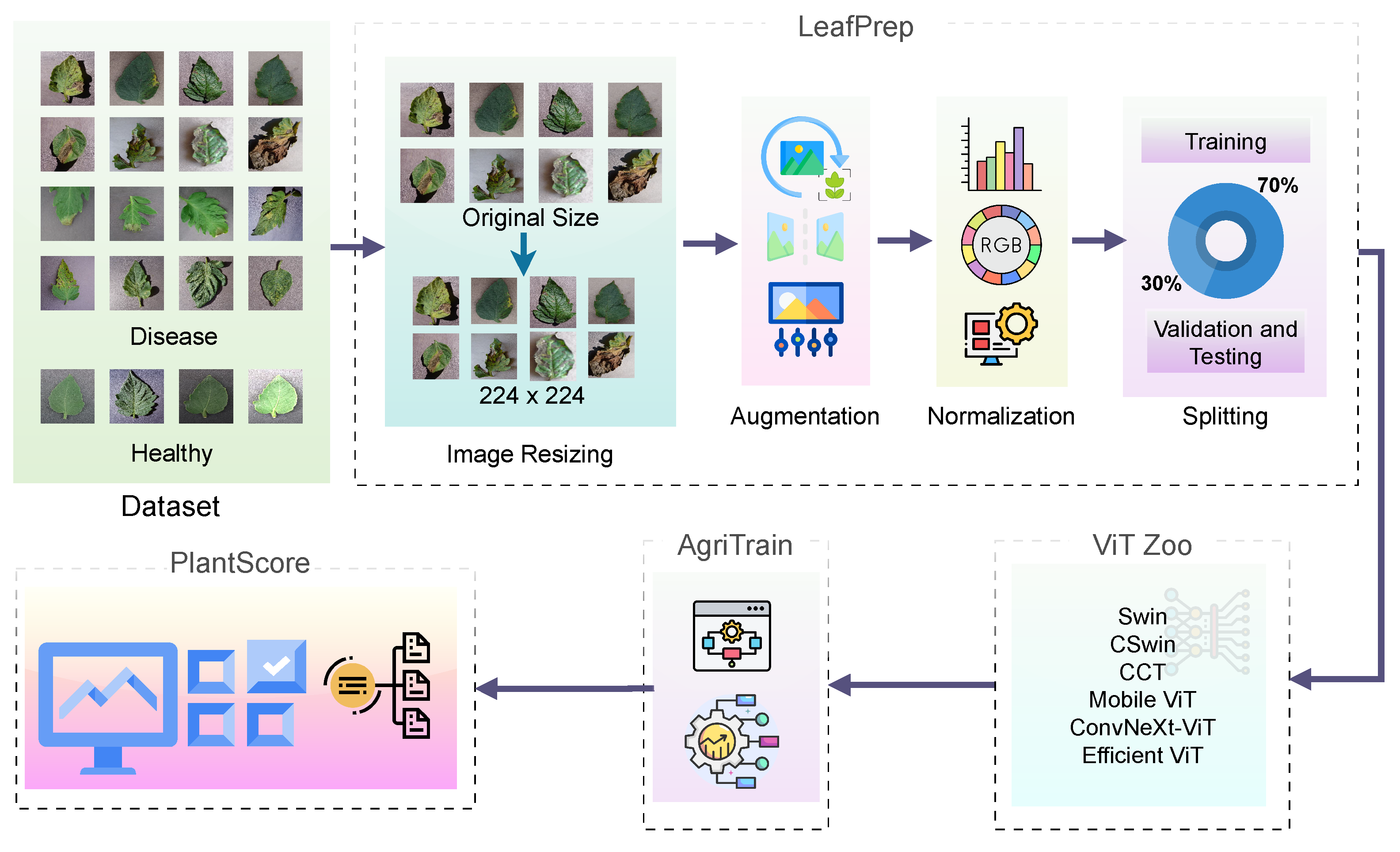
- ViT-RoT, as shown in Figure 1, a novel benchmarking framework, is introduced to systematically evaluate the performance of ViT architectures in tomato leaf disease recognition.
- A comprehensive comparative and empirical analysis of multiple state-of-the-art ViT variants is conducted under consistent experimental settings. This enables an objective evaluation of each model’s capability in recognising complex disease patterns in tomato leaf images.
- Extensive performance benchmarking is conducted on three benchmark datasets using standard evaluation metrics to comprehensively assess the classification effectiveness of each ViT variant to classify images into high-, moderate-, and low-performing ViT variants. The results demonstrate that ConvNeXt-Small and Swin-Small consistently outperform all other ViT variants in tomato disease recognition.
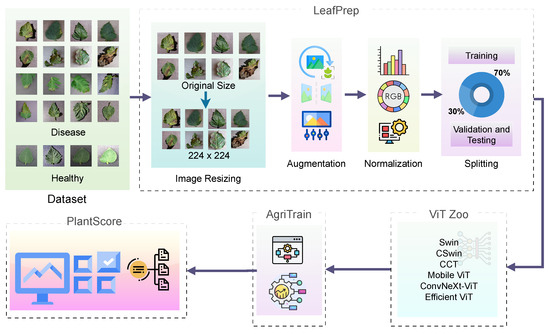
Figure 1.
Overall flow of the proposed ViT-RoT framework for tomato leaf disease recognition.
Figure 1.
Overall flow of the proposed ViT-RoT framework for tomato leaf disease recognition.
The remainder of this paper is organized as follows: Section 2 discusses related works in the field of transformer-based tomato leaf disease recognition; Section 3 describes the proposed methodology and the experimental setup; Section 4 presents the results; and Section 5 concludes the paper with future research directions.
2. Related Work
Over the years, numerous studies have investigated the application of deep learning techniques for automated leaf-based plant disease detection, with a particular emphasis on the use of CNN-based pre-trained models [27,28] and, more recently, ViTs [29,30]. These models have been applied across various crops, including tomato plants, to identify and classify diseases from leaf images. This section categorises and reviews a selection of closely related studies on tomato leaf disease recognition into two main approaches, namely (1) CNN-based methods and (2) ViT-based methods. For a more comprehensive overview, readers are encouraged to refer to recent survey papers [5,31].
2.1. CNN-Based Approaches
CNN-based approaches have consistently outperformed traditional machine learning methods in plant disease recognition from leaf images, with notable success in classifying tomato diseases. Maeda et al. [32] evaluated five CNN models, such as AlexNet, GoogleNet, Inception V3, ResNet-18, and ResNet-50, for classifying nine distinct tomato diseases and healthy leaves, and they reported that GoogleNet achieved the highest performance with an AUC score of 99.72%. However, the study concluded that GoogleNet’s architectural complexity may lead to longer training times and higher computational demands compared to more lightweight models. In [33], a comparative analysis of several CNN-based architectures for the classification of tomato diseases was performed. The findings indicated that the ResNet-50 architecture achieved superior performance, with an accuracy of 96.51% compared to 95.83% for AlexNet and 95.66% for GoogleNet. These results were obtained using the stochastic gradient descent (SGD) optimiser. However, despite its high accuracy, the ResNet-50-based model demonstrated relatively slow inference times, which may have hindered its practical applicability in real-time disease detection scenarios. In [34], the authors demonstrated that the pre-trained convolutional neural network models EfficientNet-B4 and EfficientNet-B5 outperformed several other state-of-the-art architectures previously regarded as highly accurate. However, EfficientNet-B4 had 19 million parameters and EfficientNet-B5 had 30 million parameters. As a result, more computational time was required to accommodate the increased resource demands. Yulita et al. [35] proposed a tomato disease detection method that used DenseNet as the backbone. The model was trained on 18,160 images from the PlantVillage repository and achieved a classification accuracy of 95.40% after 30 training iterations, successfully distinguishing between nine disease categories and healthy leaves.
2.2. ViT-Based Approaches
Self-attention is the fundamental mechanism enabling ViTs to capture global dependencies in an image. Unlike CNNs, which rely on local convolutional filters, self-attention dynamically assigns weights to different parts of the image based on their importance. Formally, given input patch embeddings , the self-attention mechanism computes attention scores using query (), key (), and value () matrices:
where is the scaling factor. This allows ViTs to weigh different patches based on relevance, which enables robust feature extraction for complex leaf disease patterns [36].
The recent popularity in the adoption of ViTs for plant leaf disease classification is attributed to their self-attention mechanisms, which facilitate improved classification accuracy while reducing the need for extensive pre-processing. For instance, Reedha et al. [37] demonstrated the effectiveness of the convolutional-free ViT model, which leverages the self-attention mechanism to transform an image into a sequence of patches for processing by a standard transformer encoder. Despite the limited size of the dataset, the authors reported high classification performance, which they attributed to the application of data augmentation, transfer learning, and the relatively small number of target classes. Thai et al. [29] applied a ViT-based approach to cassava leaf disease identification, achieving at least a 1% higher accuracy than popular CNN models and a 90% F1 score. However, the model is large, with 85.79 million parameters, which limits its applicability in resource-constrained Internet of Things (IoT) environments. To address this, they employed quantisation, reducing the model’s size by three times before deploying it on a Raspberry Pi 4 module.
Researchers have proposed various ViT variants to address specific challenges in image-based leaf disease classification, each offering distinct advantages regarding computational efficiency, classification accuracy, and adaptability to diverse application contexts. In [38], the authors proposed a model called TLMViT, which integrates transfer learning with ViT architectures for plant disease classification. This approach enhances feature extraction by first utilising pre-trained CNN models, such as VGG19 or ResNet50, followed by a ViT at a deeper layer. The model was evaluated on the PlantVillage and wheat datasets, achieving validation accuracies of 98.81% and 99.86%, respectively. Recent advancements in the Swin transformer have made it highly efficient in processing images, making it well-suited for real-time detection systems that need fast responses. For instance, Sun et al. [24] employed the PlantVillage and Tomato-Village datasets to evaluate a Swin transformer-based approach for tomato disease classification. The results demonstrated that the proposed system exhibited high effectiveness in disease detection, achieving an accuracy of 99.7%. Additionally, the integration of the CNN-attention module improved the model’s performance.
In a similar vein, an extended study by Thakur et al. [39] presented PlantXViT, a lightweight model that integrates CNNs and ViTs for efficient plant disease identification across various crops, specifically designed for deployment in IoT-based smart agriculture systems. Evaluations on five datasets show PlantXViT outperforms state-of-the-art models, achieving accuracies of 93.5%, 92.5%, and 98.3% on apple, maize, and rice datasets, respectively. Similarly, MobileViT is a lightweight hybrid model combining the strengths of CNNs and ViTs for efficient and accurate image classification [25]. Building upon this, Han et al. [40] proposed an enhanced MobileViT network, incorporating a Squeeze-and-Excitation module for improved feature fusion and a global attention mechanism to enhance feature representation, thereby increasing classification accuracy. Experimental results show the improved model achieves an 88.86% recognition accuracy, which is 4.28% points higher than the original MobileViT, outperforming other CNN models. In a related MobileViT advancement, Tonmoy et al. [41] introduced MobilePlantViT, a hybrid ViT model for efficient plant disease classification. With only 0.69 million parameters, it outperformed larger models like MobileViTv1 and MobileViTv2, achieving test accuracies from 80% to over 99% across various datasets. This highlights its potential for lightweight, resource-efficient AI in smart agriculture. Hossain et al. [17] evaluated different transformer-based models for classifying tomato leaf diseases. They found that MaxViT outperformed the others, achieving an accuracy of 97%. In another study by Emmanuel and Hidayaturrahman [26], pre-trained ViT models, including EfficientViT b-series, EfficientViT m-series, and MobileViT, were used for plant disease classification. The research compared those ViT models and concluded that EfficientViT b2 is the best model for plant disease classification, provided that training time and parameter size are not a concern.
In summary, the literature shows a growing interest in applying ViTs for plant disease detection, with several studies highlighting their effectiveness in classifying plant diseases from leaf images. While these models show great novelty, challenges still exist, especially in adapting them to handle the variability of agricultural conditions and their potential in tomato leaf disease recognition. Issues such as the need for more robust models and the lack of a standard framework to evaluate different ViT variants for this task remain unexplored.
3. Proposed Method
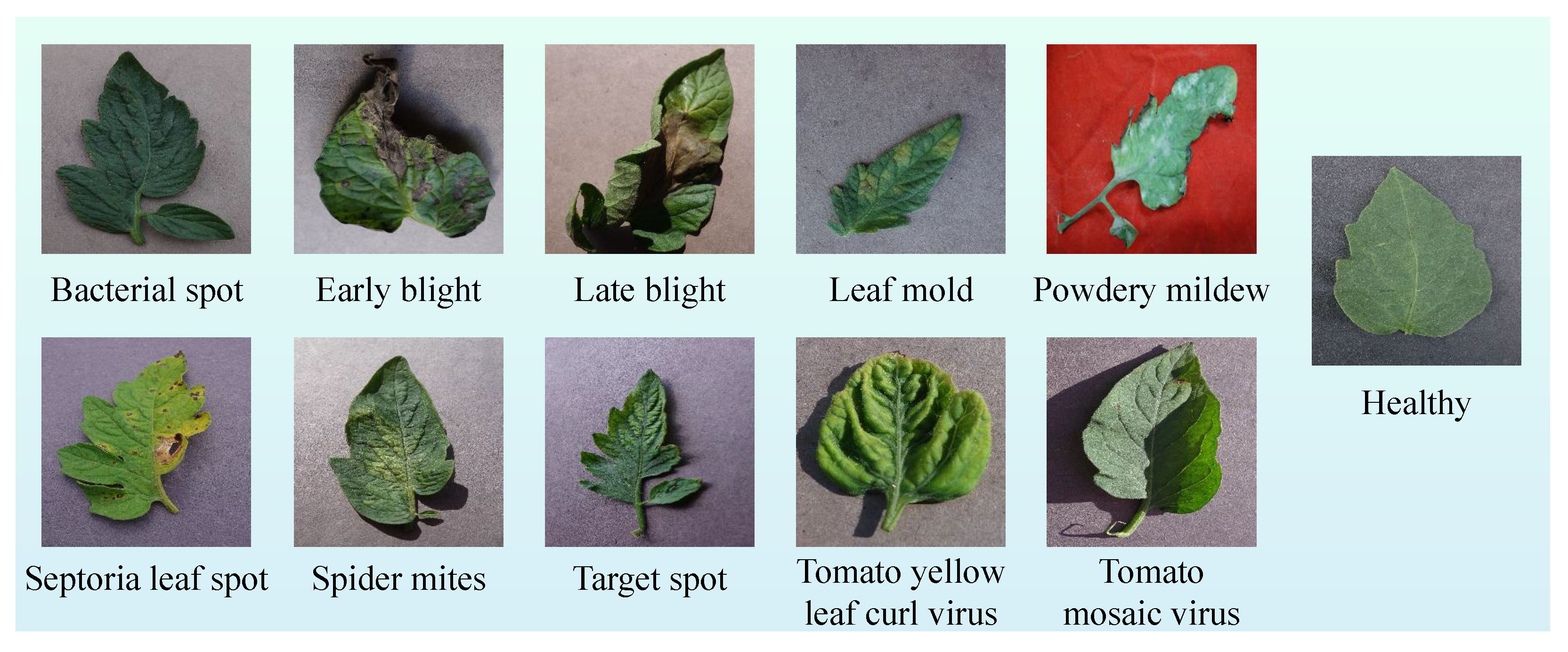
In this section, the ViT-Based Robust Framework for tomato leaf disease recognition (ViT-RoT) is introduced as a unified framework for the detection and classification of tomato plant diseases using ViT architectures. The ViT-RoT is designed to address the need for accurate and efficient plant disease recognition by leveraging the powerful feature extraction capabilities of ViTs. The framework systematically evaluates the performance of various ViT-based models under a carefully designed and standardised experimental setup, ensuring consistency and fairness in comparisons. All experiments were conducted using the tomato leaf data from the Tomato Leaves Dataset, which provides a comprehensive collection of images depicting different disease types, including common and rare infections. Figure 2 illustrates sample images from the dataset, showcasing both diseased and healthy tomato leaves to highlight the visual differences used for classification.
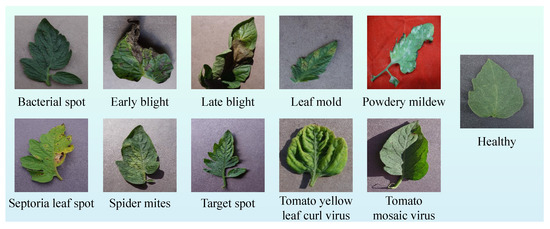
Figure 2.
Sample images from the Tomato Leaves Dataset, including examples of leaves affected by various diseases and a healthy leaf.
The proposed ViT-RoT framework integrates LeafPrep, a standardised preprocessing pipeline; ViT Zoo, a module for managing model variants; AgriTrain, a specialised training strategy; and PlantScore, a uniform evaluation protocol to facilitate reliable benchmarking across multiple state-of-the-art transformer models. The overall pipeline of the proposed ViT-RoT framework is shown in Figure 1.
3.1. LeafPrep—Preprocessing Pipeline
The first module of the ViT-RoT framework is the preprocessing stage, namely LeafPrep. This module plays a critical role in ensuring data consistency and proper preparation before model training and evaluation. LeafPrep is designed to standardise the input images and enhance the model’s ability to generalise across varying conditions. It consists of four essential steps, namely image resizing, augmentation, normalisation, and train–validation–test splitting.
First, each raw input image is resized to a fixed resolution of pixels to match the input size requirements of ViT architectures:
where represents the resizing operation.
Following resizing, a series of data augmentation techniques are applied to increase dataset diversity and improve model robustness. The main augmentations used in LeafPrep are
- RandomHorizontalFlip: Flips the image horizontally with a probability of 0.5.
- RandomVerticalFlip: Flips the image vertically with a probability of 0.5.
- RandomRotation: Applies random rotations within a specified angle range.
- RandomResizedCrop: Randomly crops and resizes the image to enhance spatial variability.
- ColorJitter: Adjusts brightness and contrast to introduce colour variations.
- Normalise: Standardises pixel values to zero mean and unit variance.
These operations can be formally represented as
where may include the above transformations.
After augmentation, the images are normalised to have zero mean and unit variance, which is essential for stabilising and accelerating the training process. The normalisation step can be mathematically expressed as
where denotes the pixel value at spatial location and channel c, while and are the mean and standard deviation of channel c computed across the training dataset.
Finally, the preprocessed dataset is split into two disjoint subsets, which are the training and validation sets. If the entire dataset is denoted by , the split can be expressed as
This commonly uses a ratio such as for training and for validation. Through these carefully designed preprocessing steps, LeafPrep ensures that the input to the ViT-based models is standardised, diverse, and representative of real-world variability.
3.2. ViT Zoo—Model Variant Module
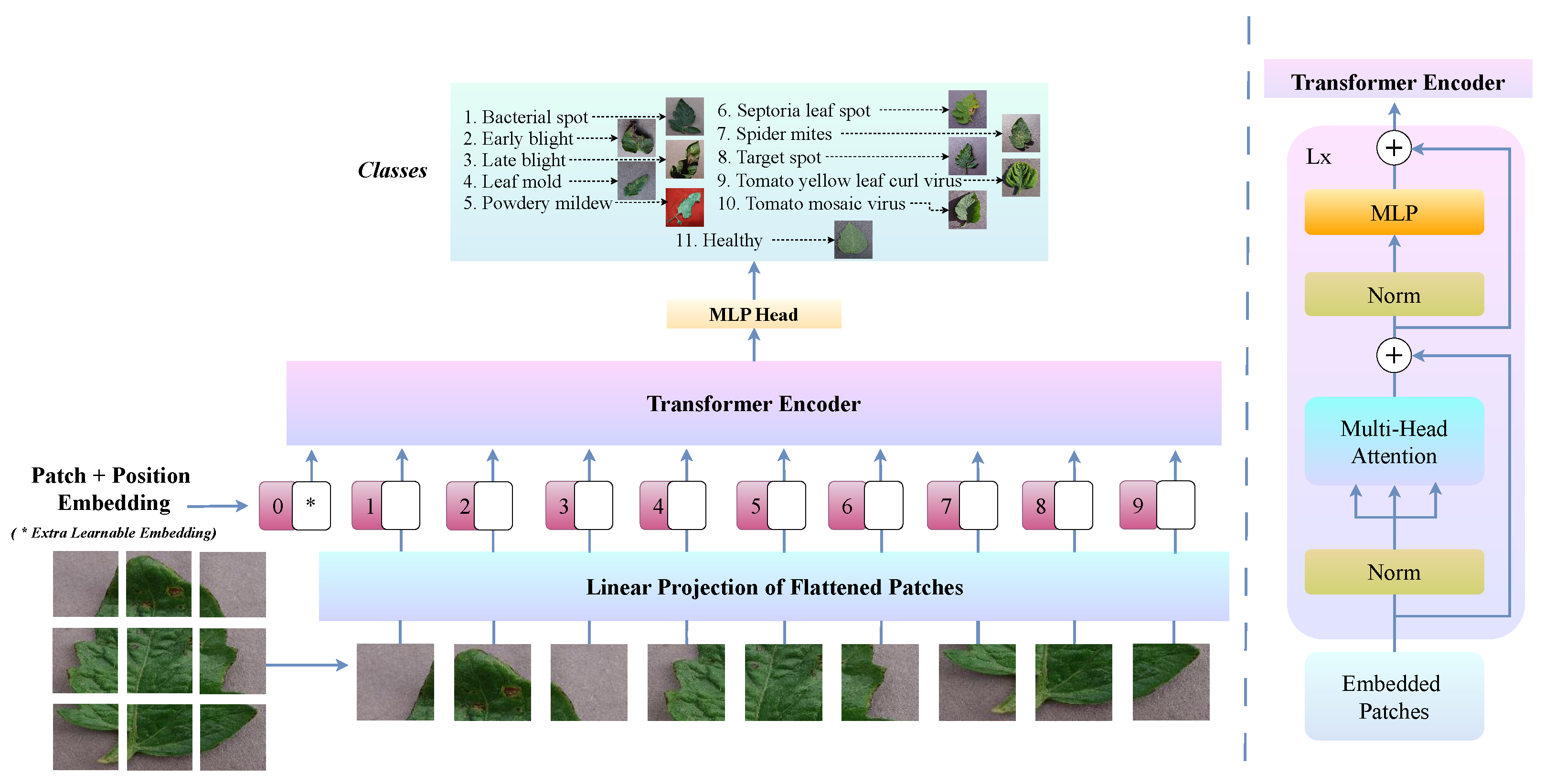
After the preprocessing stage, the subsequent module in the proposed ViT-RoT framework, namely ViT Zoo, is dedicated to classifying tomato leaf diseases utilising ViTs. ViTs have recently emerged as a powerful alternative to traditional CNNs for image classification tasks. Unlike CNNs, which rely on localised receptive fields and convolutional operations, ViTs treat an image as a sequence of patches and apply a self-attention mechanism to capture global contextual information.
Formally, given a preprocessed input image , it is divided into N non-overlapping patches , where each patch is for patch size . The sequence of flattened patches is then linearly embedded using a learnable projection matrix , resulting in
where denotes the positional embedding added to retain positional information.
The embedded patches are subsequently processed through L transformer encoder layers. Each layer consists of a multi-head self-attention (MSA) mechanism and a feed-forward network (FFN), defined as
for . The final output is then passed through a classification head, typically a linear layer, to predict the disease class.
Figure 3 illustrates the ViT architecture, highlighting its key components and structure specifically adapted for tomato leaf disease classification. This design allows ViTs to effectively capture complex disease patterns and subtle variations that may be overlooked by CNNs.
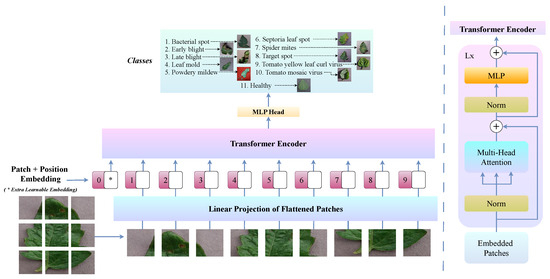
Figure 3.
Overview of the ViT architecture, showcasing its core components and structure for tomato leaf disease classification.
The overall ViT-based classification process is summarised in Algorithm 1. The tomato leaf disease classification algorithm using ViT begins by dividing a normalised image into small patches, projecting them into a latent space with positional embeddings and processing them through multiple transformer layers. After successive applications of MSA and FFN with residual connections, a classification head predicts the disease label.
The ViT Zoo module leverages several state-of-the-art ViT-based models, each offering unique architectural innovations tailored to enhance performance in plant disease recognition. Apart from the primary ViT, the models employed, such as CCT, Swin transformer, MobileViT, ConvNeXt-ViT, and EfficientViT, are described in detail in the next subsections.
| Algorithm 1 Tomato Leaf Disease Classification using ViT |
|
3.2.1. CCT
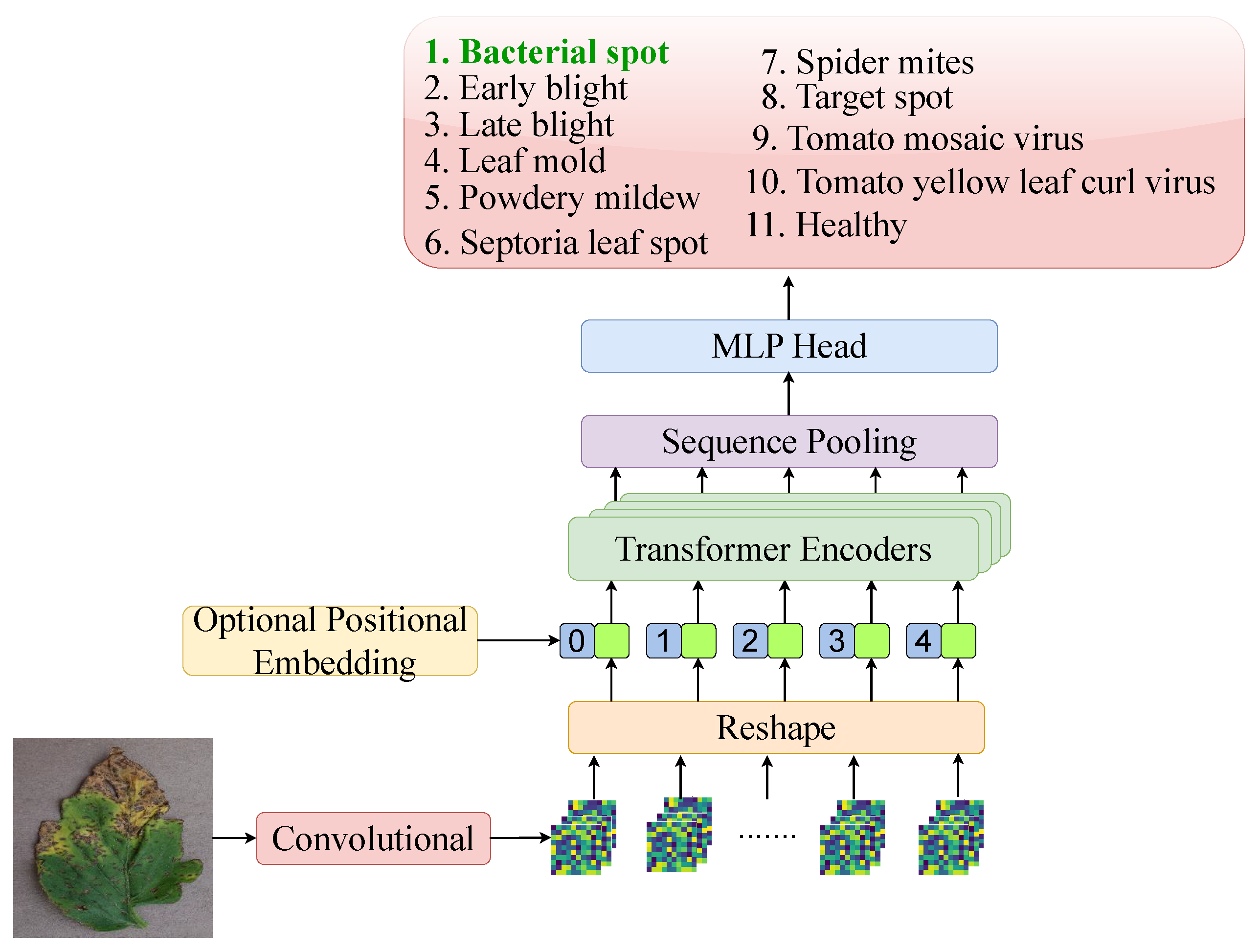
The CCT architecture, as shown in Figure 4, merges convolutional layers with transformer architectures by replacing patch embeddings with convolutional tokenisers. This design adds inductive biases like locality and translation equivariance, missing in vanilla ViTs. CCT also replaces class tokens and positional embeddings with sequence pooling, enhancing flexibility and reducing input structure dependence [42]. It outperforms lightweight transformers like ViT-Lite and ConViT, especially on tasks with limited data and high spatial variance, making it ideal for plant disease recognition in leaf imagery.
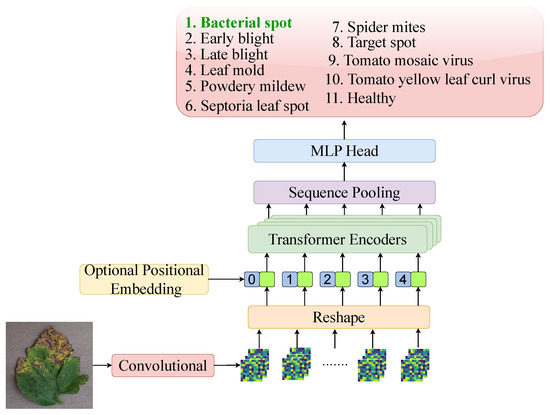
Figure 4.
Overview of the CCT architecture, showcasing its core components and structure for tomato leaf disease classification.
3.2.2. Swin Transformer
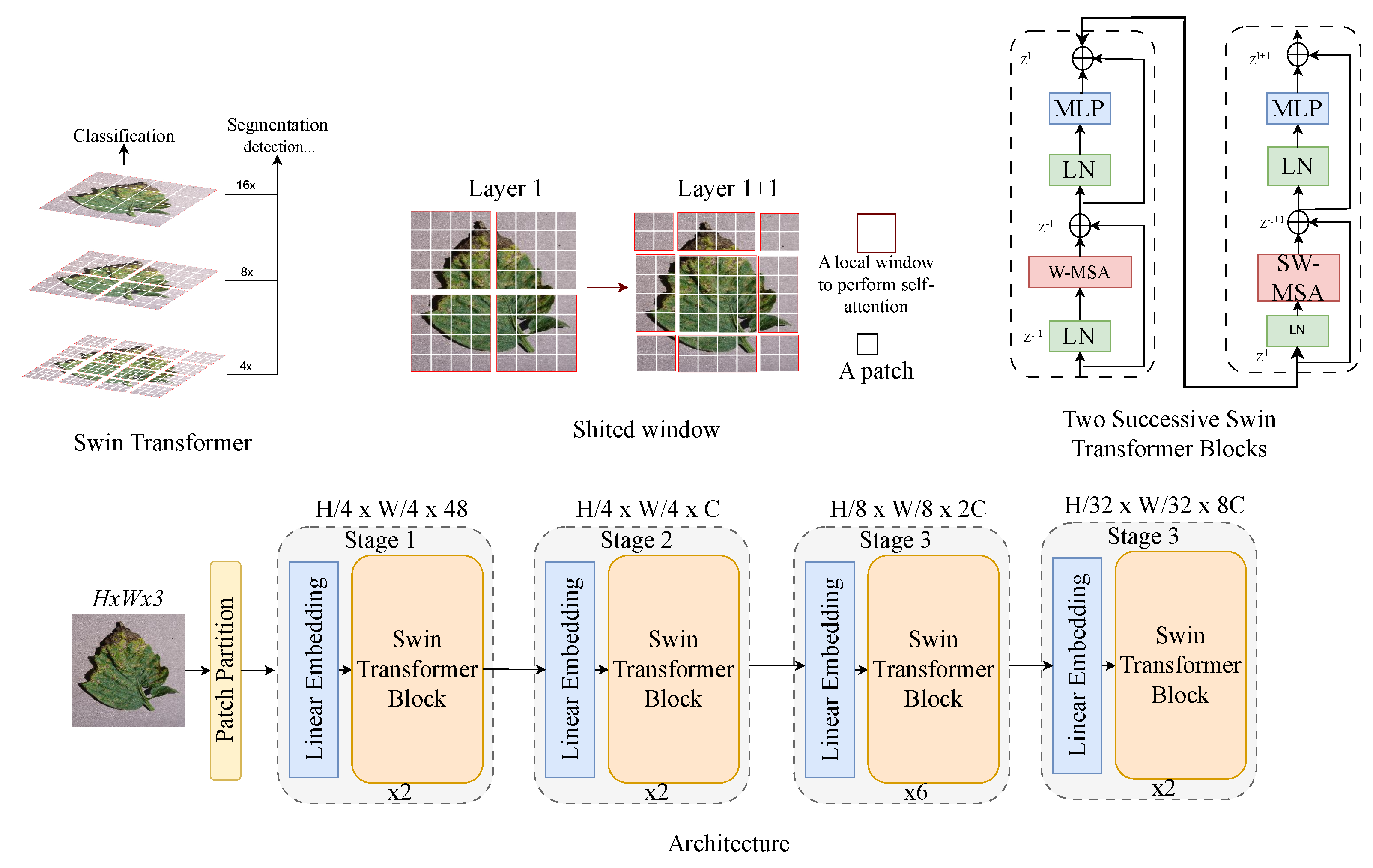
Swin transformer (i.e., shifted window transformer), as shown in Figure 5, is a hierarchical ViT designed for various computer vision tasks. Unlike standard ViTs that use fixed-size patches, Swin transformer computes self-attention within local non-overlapping windows, reducing computational complexity [43]. A shifted windowing mechanism between layers enables cross-window interaction, enhancing model expressiveness. This design improves efficiency while maintaining long-range dependency modelling, which makes it effective for tasks like image classification and object detection and segmentation, where both local and global patterns are crucial.
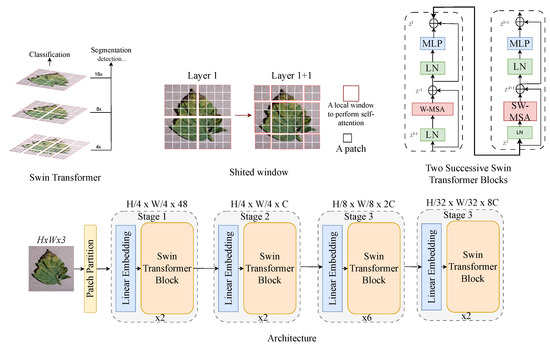
Figure 5.
Swin ViT architecture: a modular framework for tomato leaf disease recognition.
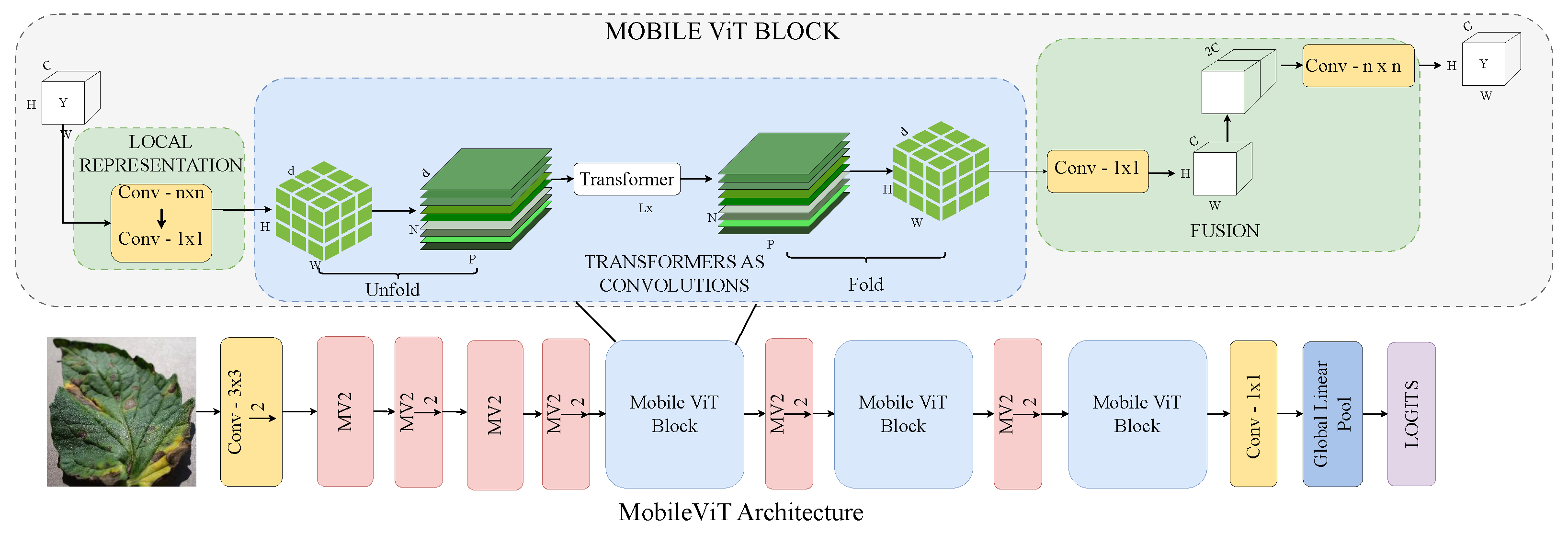
3.2.3. MobileViT
MobileViT, as shown in Figure 6, is a lightweight, mobile-friendly ViT designed to combine the strengths of CNNs and transformers, making it suitable for resource-constrained devices [44]. It replaces traditional local convolutions with global context modelling through transformer layers, capturing both local and long-range dependencies while maintaining efficiency. Its compact design and generalisation ability make it ideal for mobile-based agricultural diagnosis, such as real-time tomato leaf disease detection.
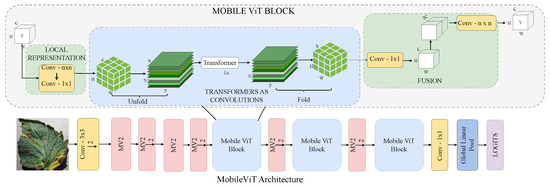
Figure 6.
MobileViT architecture: lightweight and efficient transformer design for tomato leaf disease recognition.
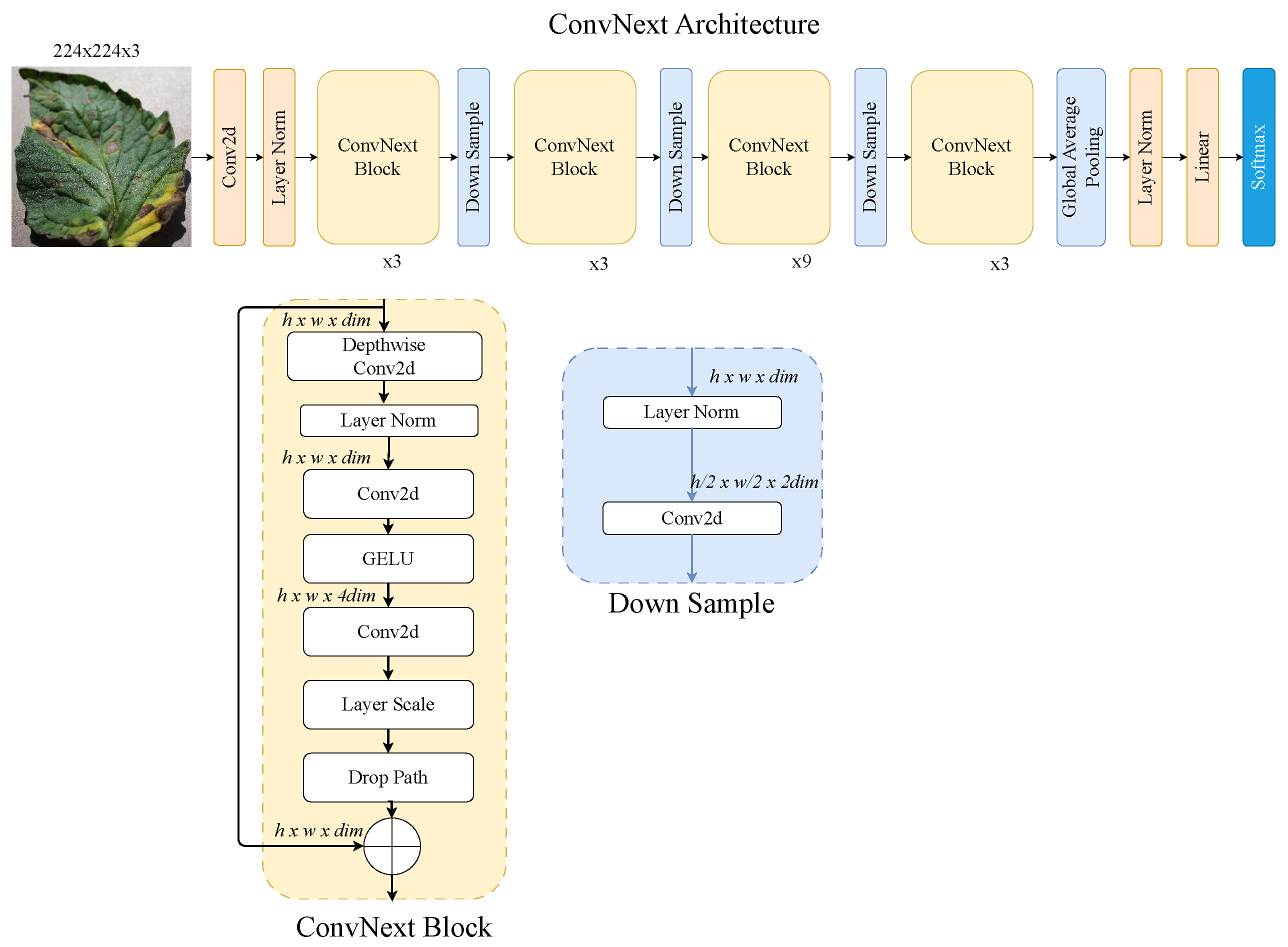
3.2.4. ConvNeXt–ViT
The ConvNeXt–ViT hybrid architecture, as shown in Figure 7, merges the strengths of convolutional networks and ViTs for efficient image understanding. ConvNeXt, a modernised CNN inspired by transformer principles, provides a strong convolutional backbone with improved spatial hierarchies and stability [45]. Combined with a ViT module, the hybrid model captures both localised features and global context. This design boosts feature richness and performance, making it ideal for complex tasks in image analysis.
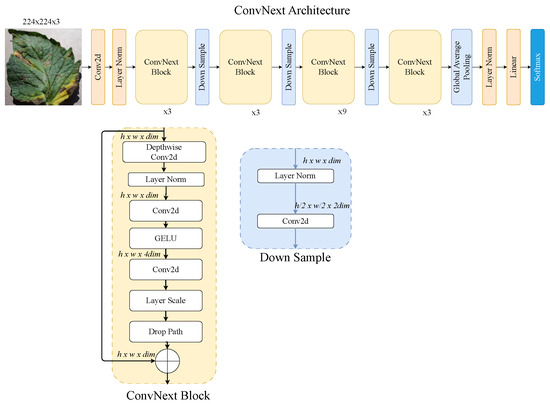
Figure 7.
ConvNeXt-ViT architecture: hybrid convolutional transformer model for tomato leaf disease recognition.
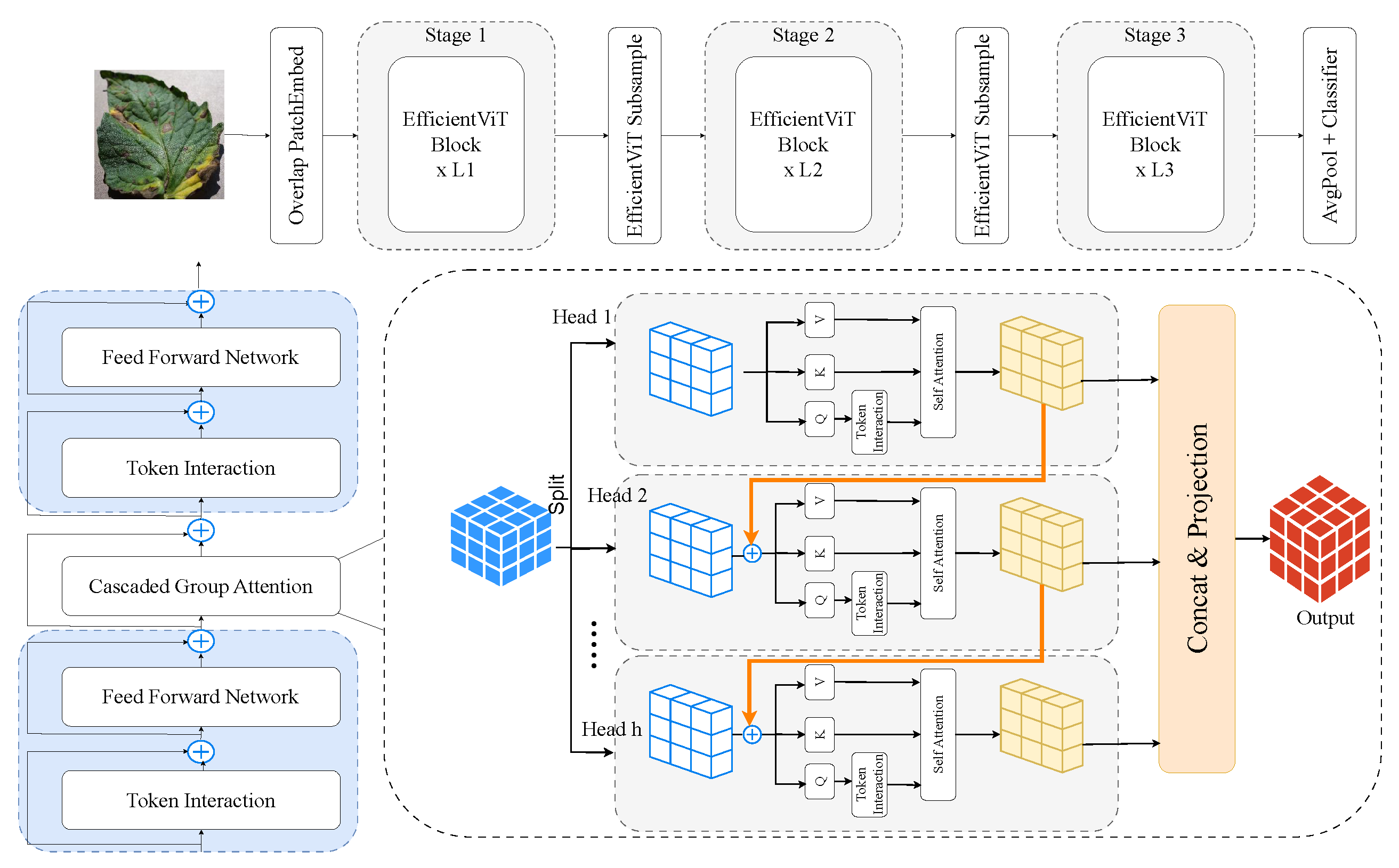
3.2.5. EfficientViT
EfficientViT, as shown in Figure 8, is a family of resource-efficient ViTs designed to balance accuracy and latency. It uses depthwise convolutions, efficient attention modules, and multi-resolution processing to reduce complexity while preserving performance [46]. Well-suited for edge deployment, EfficientViT offers fast inference on low-power devices and handles high-resolution agricultural images effectively, making it ideal for real-time tomato leaf disease detection in precision farming.
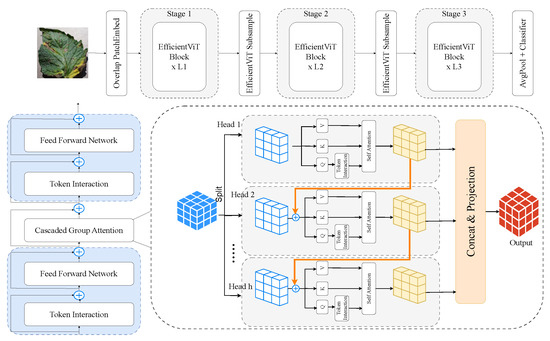
Figure 8.
EfficientViT architecture: optimised transformer framework for tomato leaf disease recognition.
3.3. AgriTrain—Training Strategy
The AgriTrain module of the ViT-RoT framework defines the training strategy employed to optimise the ViT models for robust tomato leaf disease recognition. A supervised learning approach was utilised, where the objective is to minimise the discrepancy between the predicted class probabilities and the ground-truth labels.
Given a training set consisting of N normalized images and their corresponding labels for C classes, the model with parameters is optimized by minimizing the cross-entropy loss , defined as
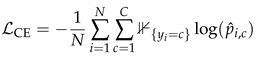 where denotes the predicted probability of sample i belonging to class c and
where denotes the predicted probability of sample i belonging to class c and 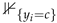 is an indicator function that equals 1 if and 0 otherwise.
is an indicator function that equals 1 if and 0 otherwise.
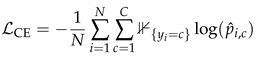
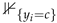 is an indicator function that equals 1 if and 0 otherwise.
is an indicator function that equals 1 if and 0 otherwise.Model training was conducted using the AdamW optimiser, an improved variant of the Adam optimiser that incorporates decoupled weight decay to prevent overfitting. The parameter update rule for AdamW is given by
where is the learning rate, and are biased estimates of the first and second moments of the gradients, is a small constant for numerical stability, and is the weight decay coefficient.
During training, multiple evaluation metrics were continuously monitored. To mitigate the risk of overfitting, an early stopping mechanism was employed. Training was halted if the validation loss did not improve over a specified patience period p. The AgriTrain strategy is summarised in Algorithm 2.
| Algorithm 2 AgriTrain—Supervised Training Strategy for ViT-RoT |
|
The progression of the model’s performance was visualized using training and validation loss curves to monitor learning behaviour and identify potential overfitting or underfitting. Following training, the best model checkpoint, selected based on the lowest validation loss, was used for further qualitative analysis. Inference was carried out on randomly selected validation samples to assess the real-world effectiveness of the model. The models were trained using the default hyperparameters provided by their respective official implementations, as summarized in Table 1. These configurations include parameters such as learning rate, batch size, optimiser, and loss function, ensuring consistency and fair comparison between models.

Table 1.
AgriTrain—hyperparameter comparison for ViT-RoT. CE denotes cross-entropy loss; LS-CE denotes label smoothing cross-entropy.
3.4. PlantScore—Evaluation Metrics
To evaluate the performance of the trained models, four widely used evaluation metrics, such as accuracy, precision, recall, and F1-score, were employed in the PlantScore module. These metrics provide a comprehensive view of classification effectiveness, particularly in scenarios with class imbalance. Let TP denote true Positive, TN denote true negatives, FP denote false positives, and FN denote false negatives. The metrics are subsequently defined as follows:
Accuracy measures the proportion of total correct predictions out of all predictions.
Precision quantifies the proportion of correct positive predictions among all predicted positives.
Recall measures the proportion of actual positive cases that were correctly predicted.
The F1-score is the harmonic mean of precision and recall, providing a single score that balances both concerns.
For multi-class classification, these metrics were computed for each class and aggregated using macro averaging, ensuring equal weight was given to all classes regardless of their frequency in the dataset.
4. Experiments
4.1. Datasets
The Tomato Leaf Disease Dataset, utilized in the ViT-RoT framework experiments, comprises 23,531 RGB images across 11 distinct classes, encompassing a variety of tomato leaf diseases along with healthy leaves. These classes include Bacterial Spot, Early Blight, Healthy, Late Blight, Leaf Mold, Powdery Mildew, Septoria Leaf Spot, Spider Mites (Two-spotted Spider Mite), Target Spot, Tomato Mosaic Virus, and Tomato Yellow Leaf Curl Virus.
The dataset comprises images resized to 224 × 224 pixels to align with the input requirements of modern convolutional and transformer-based neural network architectures. Primarily captured using mobile phone cameras in controlled laboratory conditions, the dataset offers high-resolution imagery. A smaller subset includes images from semi-natural agricultural environments, introducing limited variability in lighting and background to enhance representational diversity.
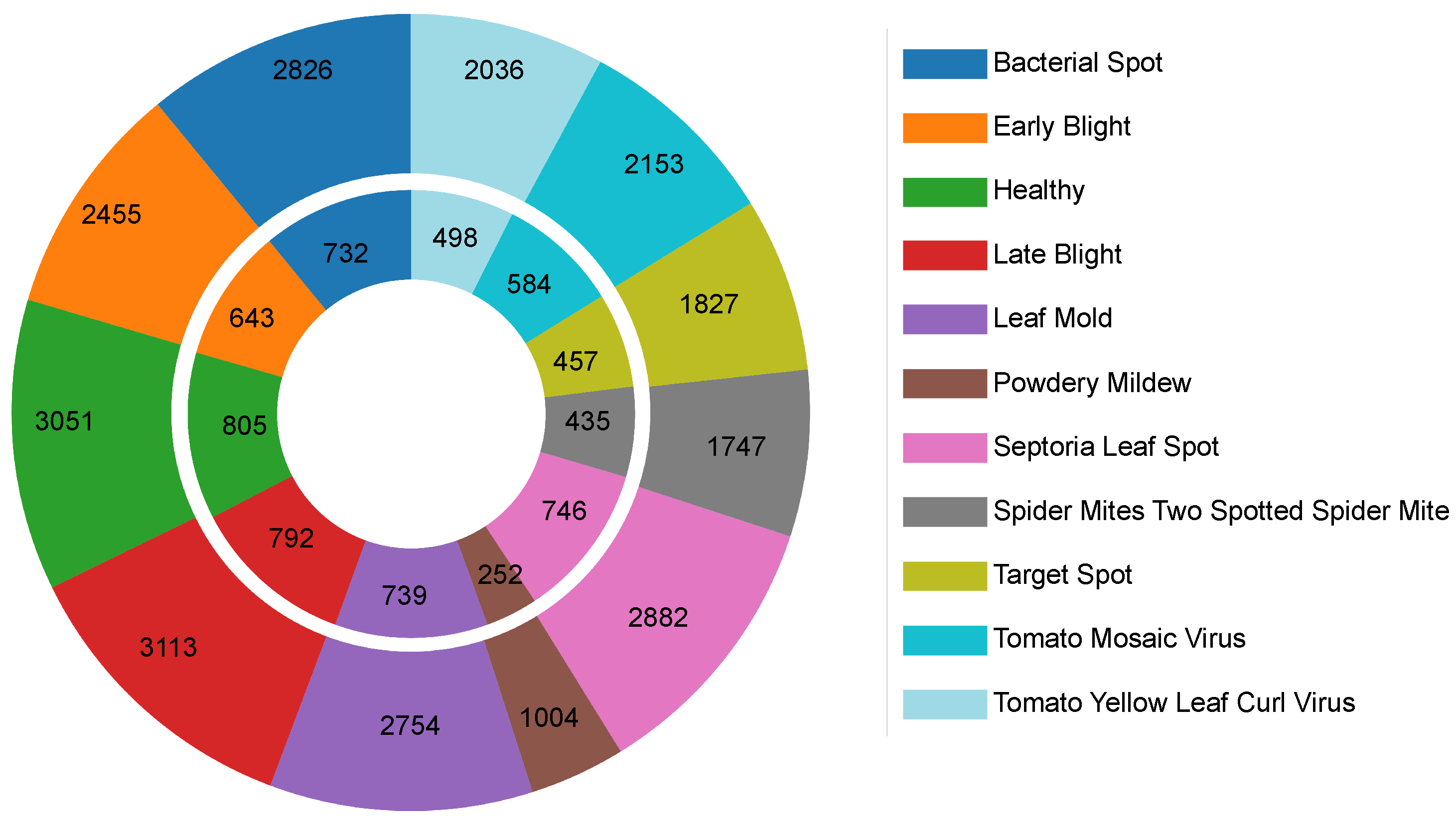
The dataset was split into 18,848 training images and 4683 validation images, as shown in Figure 9. This partitioning supports effective model training and evaluation while partially addressing class imbalance. Class representation ranges from 1256 images (Powdery Mildew) to 3905 images (Late Blight), ensuring sufficient samples for robust classification. The mix of controlled and semi-controlled settings makes the dataset suitable for evaluating data-efficient vision transformers in plant disease recognition. However, the limited inclusion of uncontrolled, real-world agricultural scenarios may hinder direct generalization to field conditions without further domain adaptation.
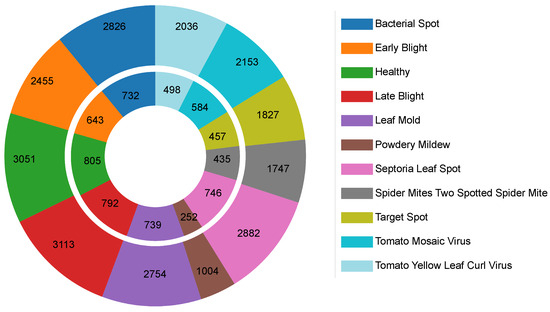
Figure 9.
The disease and healthy class distribution in the tomato leaf dataset: (exterior) training set and (interior) validation sets.
The dataset is publicly available on Kaggle (https://www.kaggle.com/datasets/ashishmotwani/tomato/data, accessed on 10 June 2025), promoting reproducibility and facilitating further research in agricultural AI applications.
4.2. Research Setup
The ViT-RoT framework experiments were conducted on a laptop equipped with an NVIDIA RTX 4070 GPU (8GB VRAM), an Intel Core i9 CPU (5.40 GHz clock speed), and 32GB RAM. A batch size of 16 was optimised for the GPU’s memory constraints. Experiments were run on Windows 11, with a fixed random seed ensuring reproducibility of data shuffling, augmentation, and model initialisation.
4.3. Results and Discussions
Several recent studies have explored the effectiveness of various CNN- and ViT-based architectures for leaf disease classification. Table 2 provides a comparative summary, highlighting the models used and performance metrics reported in the literature.

Table 2.
Performance of CNN- and ViT-based models for tomato plant disease classification.
Table 3 presents a detailed comparative evaluation of the ViT variants incorporated in the ViT-RoT framework across multiple metrics, including loss, accuracy, precision, recall, F1-score, and the number of training epochs. Each model’s performance is accompanied by its respective ranking (as a superscript) across all evaluation metrics. This detailed comparison highlights the nuances of model trade-offs between the number of training epochs, model accuracy, and other important evaluation criteria, which provides a comprehensive overview of the strengths and weaknesses of each variant.

Table 3.
Results obtained for all the ViT variants integrated in the ViT-RoT framework with ranks given in superscripts.
EfficientViT-B0 demonstrates the best performance in terms of loss (0.0427), reflecting superior convergence, while it also ranks fourth in accuracy and precision and fifth in recall and F1-score. Although EfficientViT-B0 achieved the lowest loss, Swin-Small and ConvNeXt-Small jointly achieved the highest accuracy (0.9904), positioning them as the leading models in terms of classification correctness. The ViT models, including ViT-Tiny, ViT-Small, and ViT-Base, exhibit high accuracy values, with ViT-Tiny achieving 0.9867, ranking it in the top 10 across all metrics. These models tend to perform well in terms of accuracy, precision, and recall, although they show higher loss values compared to other models. MobileViT models, such as MobileViT-XXSmall and MobileViT-XSmall, generally show slightly lower performance metrics, with accuracy values around 0.9855, and higher loss values, indicating a trade-off between model size and performance.
The EfficientViT models display competitive performance, with EfficientViT-B0 achieving the highest accuracy among all models (0.9900) and ranking consistently well across all metrics. In contrast, EfficientViT-B2 shows lower performance, particularly in accuracy (0.9779) and other metrics, which places it at the bottom of the rankings. The Swin transformer models, including Swin-Tiny, Swin-Small, and Swin-Base, exhibit robust performance, particularly Swin-Small, which ranks first in accuracy (0.9904). Similarly, the ConvNeXt models demonstrate high performance, with ConvNeXt-Small ranking highly in terms of accuracy and F1-score. The CCT models, on the other hand, lag behind in comparison, with CCT-7×7×2×224 showing the lowest accuracy and loss, indicating less efficient performance overall.
According to the results obtained, the ViT variants integrated in the ViT-RoT framework are classified into three categories, namely top performers, moderate performers, and low performers.
4.3.1. Top Performers
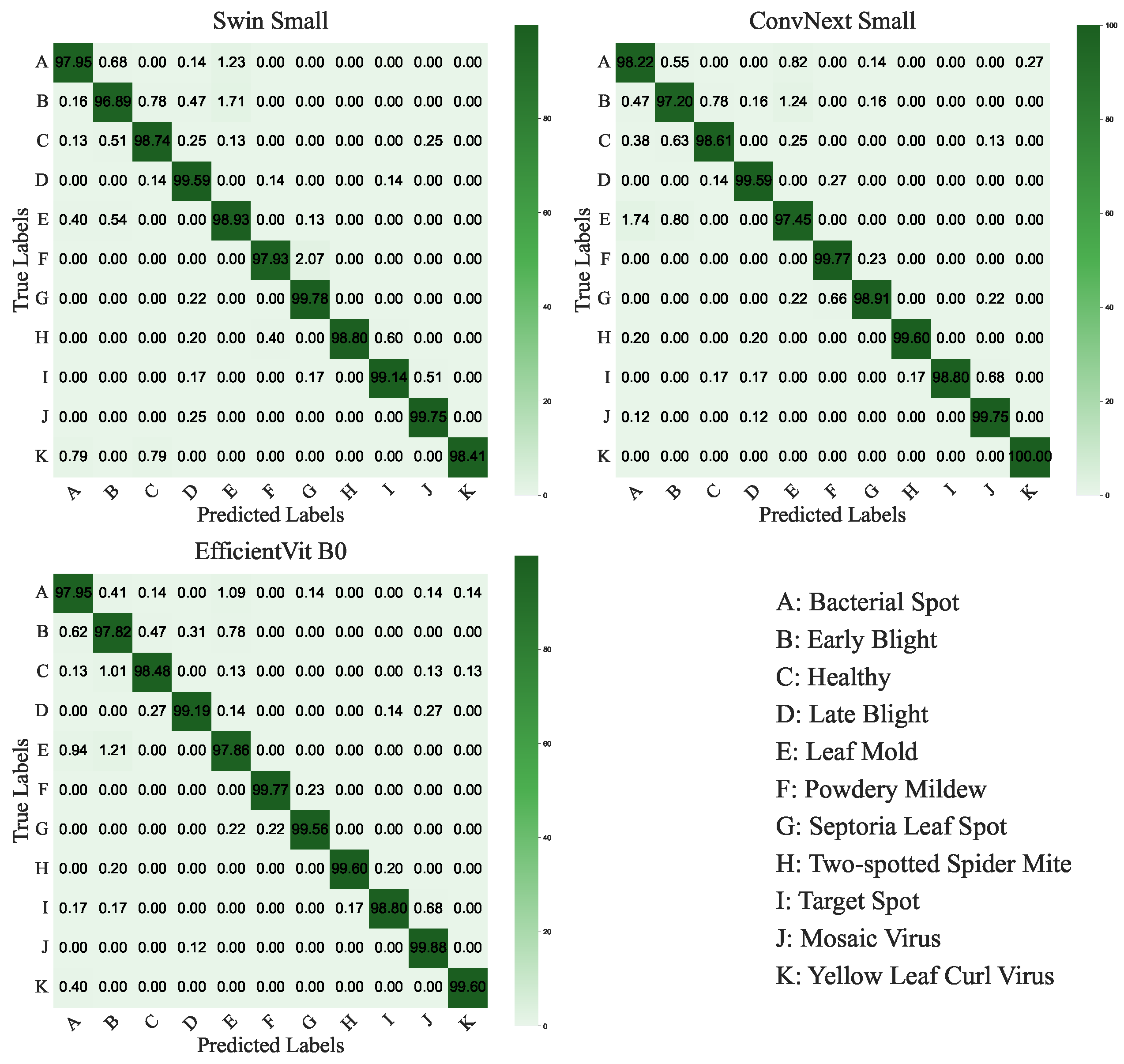
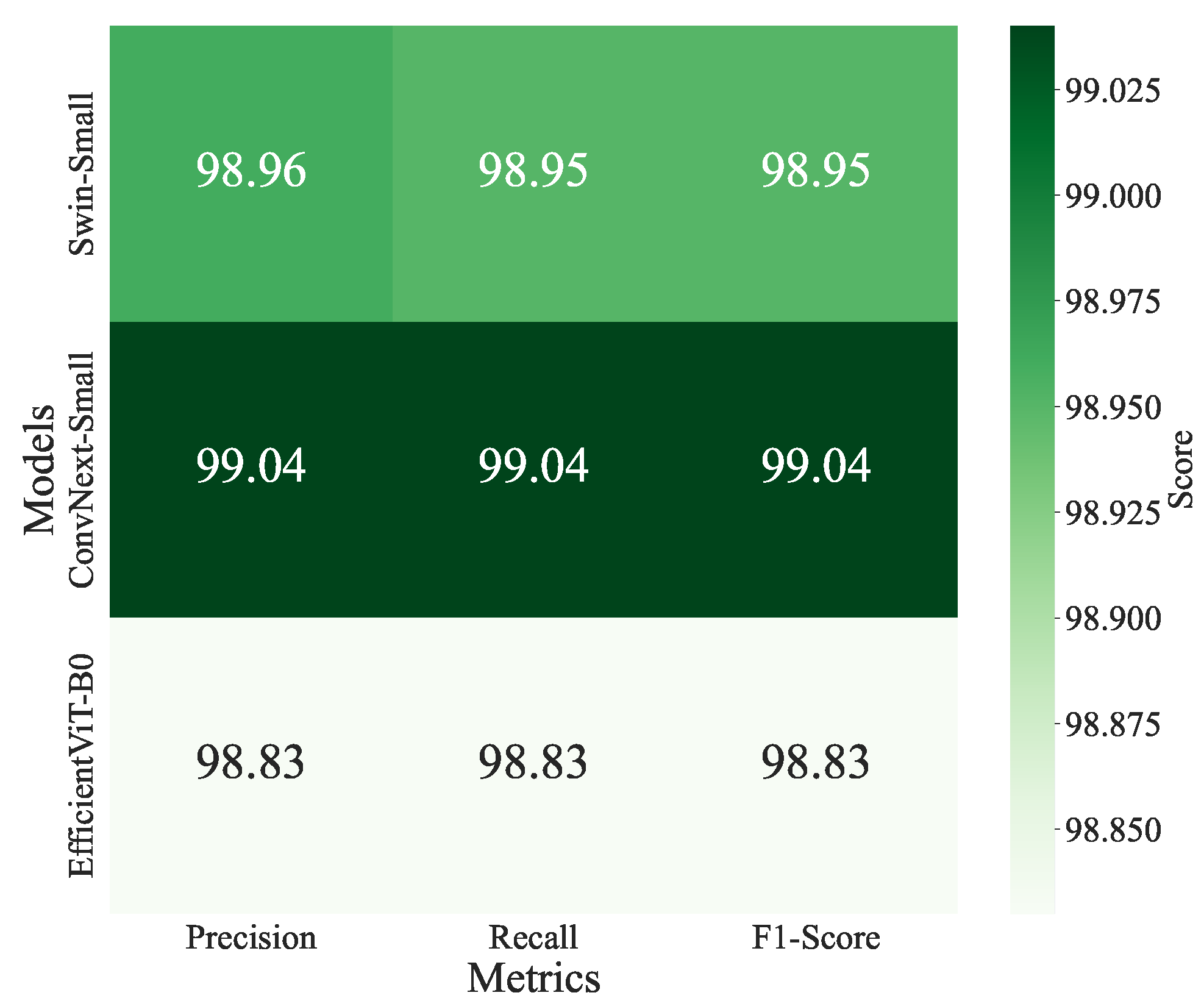
ConvNeXt-Small and Swin-Small models achieved the highest accuracy of 0.9904, with losses of 0.0542 and 0.0697, respectively, and precision, recall, and F1-scores of 0.9905 and 0.9904. EfficientViT-B0 follows closely with an accuracy of 0.9900 and the lowest loss of 0.0427. Swin-Base (0.9903) and ConvNeXt-Tiny (0.9899) also perform strongly, balancing accuracy and computational demands. The confusion matrices for ConvNeXt-Small, Swin-Small, and EfficientViT-B0 are shown in Figure 10 and illustrate high per-class accuracy across all disease categories. Notably, ConvNeXt-Small demonstrates excellent consistency across both common and less frequent classes, with minimal confusion observed among visually similar diseases such as Powdery Mildew and Septoria Leaf Spot. Swin-Small also maintains strong overall performance, though it exhibits slightly more confusion in classes with overlapping visual symptoms. EfficientViT-B0, despite being more lightweight, exhibits competitive class-level precision and recall, effectively handling challenging classes like Powdery Mildew and Spider Mites. These observations highlight the robustness of ViT-based models in capturing fine-grained visual patterns and their ability to handle class imbalance effectively. Additionally, the heatmap of precision, recall, and F1-scores for the top-performing models (Figure 11) reinforces this consistency, with ConvNeXt-Small exhibiting a slight advantage in precision, recording a value of 0.9905.
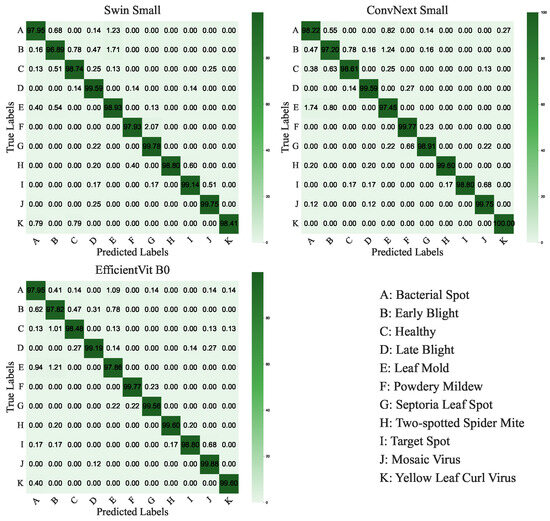
Figure 10.
Confusion matrices for the top-performing ViT-RoT models (Swin-Small, ConvNeXt-Small, EfficientViT-B0) on the Tomato Leaf Disease Dataset, illustrating per-class classification accuracy across 11 classes.
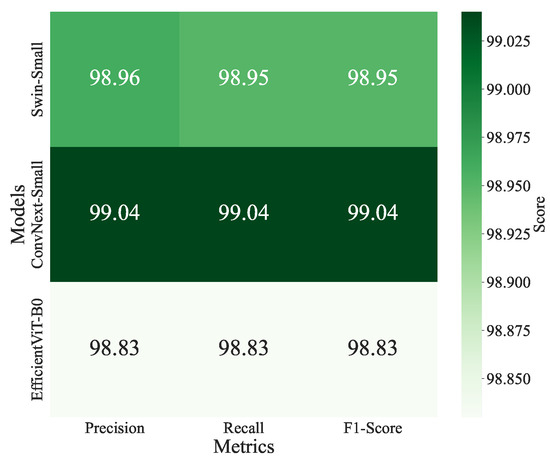
Figure 11.
Heatmap of validation precision, recall, and F1-scores for the top-performing models, Swin-Small, ConvNeXt-Small, and EfficientViT-B0, on a tomato disease classification task.
4.3.2. Moderate Performers
ViT-Small (0.9898), Swin-Tiny (0.9885), ViT-Base (0.9883), and EfficientViT-M5 (0.9883) achieve high accuracies with losses between 0.0490 and 0.0582, offering robust performance. MobileViT-XXSmall and XSmall (0.9855) provide lightweight alternatives, suitable for resource-constrained environments, with losses of 0.0597 and 0.0665, respectively.
4.3.3. Low Performers
EfficientViT-B2 (0.9779) and CCT-7×7×2×224 (0.9791) exhibit the lowest accuracies, with higher losses of 0.0834 and 0.1590, respectively, suggesting training instability. MobileViT-Small (0.9813) and CCT-14×7×2×224 (0.9835) also trail, limited by capacity or optimisation challenges.
4.3.4. Performance Trends and Architectural Insights
This section analyses the performance of various ViT variants and hybrid architectures, focusing on their efficiency, accuracy, and training behaviour. Key observations are summarised below.
- Efficiency vs. Accuracy: Optimised models like ConvNeXt-Small, Swin-Small, and EfficientViT-B0 outperform larger models (e.g., ViT-Base), suggesting that architectural efficiency is critical for the dataset’s moderate size.
- Hierarchical Attention: Hierarchical models (ConvNeXt, Swin) consistently outperform global attention models (ViT), leveraging localised feature extraction to handle noise and class imbalance effectively. This is evident in ConvNeXt-Small and Swin-Small’s top accuracies (0.9904).
- Lightweight Models: EfficientViT-B0 (0.9900) and MobileViT-XXSmall/XSmall (0.9855) achieve high accuracies with reduced computational demands, ideal for edge deployment in agricultural diagnostics.
- Training Stability: Lower losses correlate with higher accuracies (e.g., EfficientViT-B0: 0.0427, 0.9900; ConvNeXt-Small: 0.0542, 0.9904), except for EfficientViT-B2 (0.0834, 0.9779), indicating optimisation challenges.
Architectural designs drive the following differences:
- ConvNeXt: ConvNeXt-Small’s top performance (0.9904) stems from its modernised convolutional design with transformer-inspired elements, excelling in noisy and imbalanced data. ConvNeXt-Tiny (98.99%) reinforces this robustness.
- Swin Transformers: Swin-Small and Swin-Base leverage hierarchical shifted window-based attention, ideal for multi-scale feature extraction in mixed image conditions. Swin-Tiny (0.9885) highlights scalability.
- EfficientViT: EfficientViT-B0’s low loss and high accuracy reflect optimised multi-scale attention. EfficientViT-B2’s lower performance suggests scaling limitations.
- ViT: ViT-Small (0.9898) outperforms ViT-Base (98.83%) due to better generalisation, as larger ViTs risk overfitting on moderate-sized datasets.
- MobileViT and CCT: MobileViT-XXSmall/XSmall offer efficiency, but MobileViT-Small (0.9813) and CCT models (0.9791–0.9835) underperform due to limited capacity or high losses.
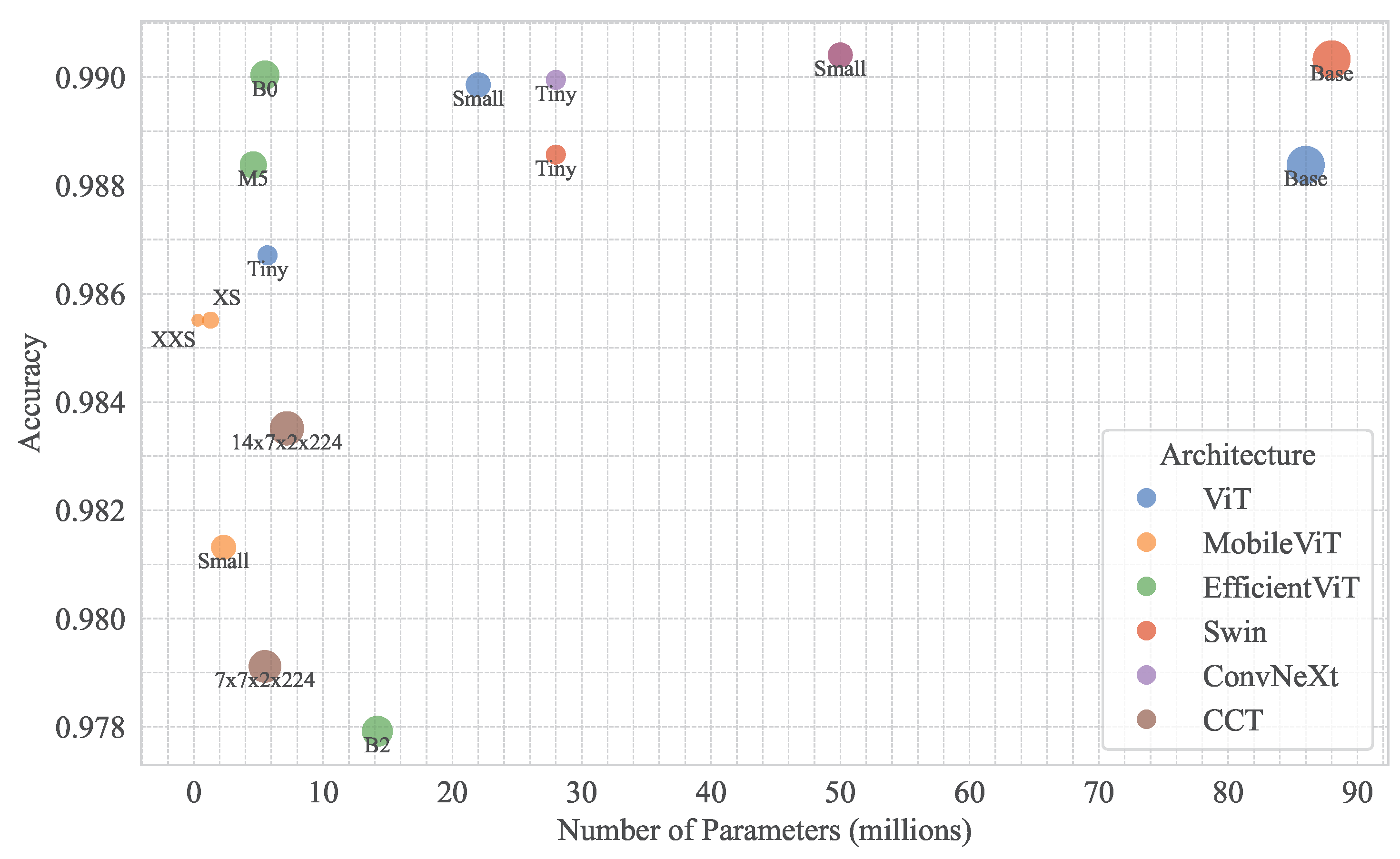
4.3.5. Efficiency and Practical Implications
Model efficiency is critical for agricultural diagnostics, particularly for edge deployment. Figure 12 illustrates parameter counts vs. model accuracy across models, highlighting lightweight models like EfficientViT-B0 (0.9900) and MobileViT-XXSmall/XSmall (0.9855) with fewer parameters, suitable for resource-constrained devices. ConvNeXt-Small and Swin-Small, with marginal accuracy gains (0.9904), are ideal for high-stakes applications requiring maximum precision.
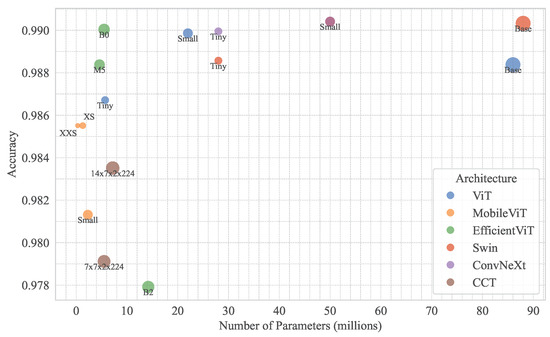
Figure 12.
Relationship between model accuracy and parameter count across different vision transformer architecture variants.
The ViT-RoT framework’s high accuracies and robust handling of class imbalance make it a promising tool for automated tomato disease diagnosis. ConvNeXt-Small and Swin-Small are recommended for production environments due to their superior performance, while EfficientViT-B0 offers a compelling trade-off for edge devices.
Table 4 highlights the trade-offs between model size, computational complexity, and inference speed. MobileViT-XXSmall is the most compact (1.27 M parameters) and fastest (3.965 ms), while EfficientViT-B0 offers the lowest FLOPs (0.10B), making both highly suitable for edge deployment. In contrast, models like ViT-Base and Swin-Base have significantly higher parameter counts (86.57 M and 87.77 M, respectively) and FLOPs, resulting in slower inference times, which makes them less ideal for low-resource settings. ConvNeXt-Small and Swin-Small offer a strong balance, with higher accuracy and moderate resource demands, making them suitable for production environments.

Table 4.
Comparison of deep learning models by parameter count, computational complexity, and inference efficiency.
Nonetheless, several limitations must be acknowledged. Although the dataset includes both controlled and outdoor images with occlusion and natural backgrounds, it does not fully capture the variability of real-world field conditions such as dynamic lighting, motion blur, and diverse sensor characteristics. The use of a single dataset also limits generalizability to other cultivars, regions, or unseen diseases. Additionally, contextual factors like environmental conditions and disease progression over time are not considered.
ViTs also require larger training datasets than CNNs due to their reduced inductive bias. This can be a challenge in agriculture, where labeled data is often limited, impacting training efficiency and generalization [51]. Additionally, ViTs exhibit higher computational complexity, as their self-attention mechanism scales quadratically with the number of image patches, whereas CNNs scale linearly with the number of pixels processed. This computational overhead makes ViTs less efficient for resource-constrained environments compared to traditional convolutional architectures [51].
5. Conclusions
This study presents the ViT-RoT framework, a systematic approach for evaluating and benchmarking ViT models for tomato leaf disease classification. Through comprehensive empirical analysis, the framework categorises ViT variants into high-, moderate-, and low-performing groups based on their effectiveness in tomato disease recognition. Notably, ConvNeXt-Small and Swin-Small emerged as top performers, demonstrating superior accuracy and robustness across multiple datasets. The findings emphasise the importance of selecting models that balance high performance, generalisability, and efficiency, making them well-suited for real-world agricultural applications. By providing a structured evaluation framework, ViT-RoT offers valuable insights for advancing AI-driven plant disease detection and lays the groundwork for sustainable crop management solutions.
The ViT-RoT framework highlights the critical role of model robustness in addressing real-world disease detection challenges, but opportunities for further refinement remain. Future work can focus on enhancing dataset diversity by incorporating images from varied agricultural regions and field conditions to improve model generalisation. Additionally, optimising ViT architectures for deployment on mobile and edge devices will enable real-time disease detection in resource-constrained environments. Integrating temporal data and environmental factors, such as humidity and temperature, could further enhance predictive accuracy and adaptability, enabling earlier and more precise interventions. Furthermore, exploring class balancing strategies such as oversampling, class reweighting, or focal loss will be essential to address dataset imbalance and improve detection accuracy for under-represented disease categories. These advancements will strengthen the framework’s applicability, paving the way for more effective AI-based solutions in precision agriculture.
Author Contributions
Conceptualization, S.N. (Sathiyamohan Nishankar); methodology, S.N. (Sathiyamohan Nishankar), V.P., T.M., S.N. (Sivaraj Nimishan), S.T. and Y.S.; software, S.N. (Sathiyamohan Nishankar), V.P. and T.M.; validation, S.N. (Sathiyamohan Nishankar), S.T. and Y.S.; formal analysis, S.T.; investigation, S.T. and Y.S.; resources, S.N. (Sathiyamohan Nishankar), V.P. and T.M.; data curation, S.N. (Sathiyamohan Nishankar) and T.M.; writing—original draft preparation, S.N. (Sathiyamohan Nishankar) and S.N. (Sivaraj Nimishan); writing—review and editing, S.T. and Y.S.; visualization, S.N. (Sathiyamohan Nishankar), V.P. and T.M.; supervision, S.T. and Y.S.; project administration, S.T. All authors have read and agreed to the published version of the manuscript.
Funding
This research received no external funding.
Data Availability Statement
The authors used a publicly available dataset downloaded from https://www.kaggle.com/datasets/ashishmotwani/tomato/data, accessed on 10 June 2025.
Conflicts of Interest
The authors declare no conflicts of interest.
References
- Sundararaman, B.; Jagdev, S.; Khatri, N. Transformative role of artificial intelligence in advancing sustainable tomato (Solanum lycopersicum) disease management for global food security: A comprehensive review. Sustainability 2023, 15, 11681. [Google Scholar] [CrossRef]
- Trivedi, N.K.; Gautam, V.; Anand, A.; Aljahdali, H.M.; Villar, S.G.; Anand, D.; Goyal, N.; Kadry, S. Early detection and classification of tomato leaf disease using high-performance deep neural network. Sensors 2021, 21, 7987. [Google Scholar] [CrossRef] [PubMed]
- Thuseethan, S.; Vigneshwaran, P.; Charles, J.; Wimalasooriya, C. Siamese network-based lightweight framework for tomato leaf disease recognition. Computers 2024, 13, 323. [Google Scholar] [CrossRef]
- Jafar, A.; Bibi, N.; Naqvi, R.A.; Sadeghi-Niaraki, A.; Jeong, D. Revolutionizing agriculture with artificial intelligence: Plant disease detection methods, applications, and their limitations. Front. Plant Sci. 2024, 15, 1356260. [Google Scholar] [CrossRef]
- George, R.; Thuseethan, S.; Ragel, R.G.; Mahendrakumaran, K.; Nimishan, S.; Wimalasooriya, C.; Alazab, M. Past, present and future of deep plant leaf disease recognition: A survey. Comput. Electron. Agric. 2025, 234, 110128. [Google Scholar] [CrossRef]
- Paul, S.G.; Biswas, A.A.; Saha, A.; Zulfiker, M.S.; Ritu, N.A.; Zahan, I.; Rahman, M.; Islam, M.A. A real-time application-based convolutional neural network approach for tomato leaf disease classification. Array 2023, 19, 100313. [Google Scholar] [CrossRef]
- Islam, S.U.; Zaib, S.; Ferraioli, G.; Pascazio, V.; Schirinzi, G.; Husnain, G. Enhanced deep learning architecture for rapid and accurate tomato plant disease diagnosis. AgriEngineering 2024, 6, 375–395. [Google Scholar] [CrossRef]
- Ahmad, I.; Hamid, M.; Yousaf, S.; Shah, S.T.; Ahmad, M.O. Optimizing pretrained convolutional neural networks for tomato leaf disease detection. Complexity 2020, 2020, 8812019. [Google Scholar] [CrossRef]
- Too, E.C.; Yujian, L.; Njuki, S.; Yingchun, L. A comparative study of fine-tuning deep learning models for plant disease identification. Comput. Electron. Agric. 2019, 161, 272–279. [Google Scholar] [CrossRef]
- Tan, L.; Lu, J.; Jiang, H. Tomato leaf diseases classification based on leaf images: A comparison between classical machine learning and deep learning methods. AgriEngineering 2021, 3, 542–558. [Google Scholar] [CrossRef]
- Karande, S.; Garg, B. Performance evaluation and optimization of convolutional neural network architectures for Tomato plant disease eleven classes based on augmented leaf images dataset. Neural Comput. Appl. 2024, 36, 11919–11943. [Google Scholar] [CrossRef]
- Ahmed, A.A.; Reddy, G.H. A mobile-based system for detecting plant leaf diseases using deep learning. AgriEngineering 2021, 3, 478–493. [Google Scholar] [CrossRef]
- Nazir, T.; Iqbal, M.M.; Jabbar, S.; Hussain, A.; Albathan, M. EfficientPNet—an optimized and efficient deep learning approach for classifying disease of potato plant leaves. Agriculture 2023, 13, 841. [Google Scholar] [CrossRef]
- Ali, A.H.; Youssef, A.; Abdelal, M.; Raja, M.A. An ensemble of deep learning architectures for accurate plant disease classification. Ecol. Inform. 2024, 81, 102618. [Google Scholar] [CrossRef]
- Liu, W.; Yu, L.; Luo, J. A hybrid attention-enhanced DenseNet neural network model based on improved U-Net for rice leaf disease identification. Front. Plant Sci. 2022, 13, 922809. [Google Scholar] [CrossRef]
- Karimanzira, D. Context-Aware Tomato Leaf Disease Detection Using Deep Learning in an Operational Framework. Electronics 2025, 14, 661. [Google Scholar] [CrossRef]
- Hossain, S.; Tanzim Reza, M.; Chakrabarty, A.; Jung, Y.J. Aggregating different scales of attention on feature variants for tomato leaf disease diagnosis from image data: A transformer driven study. Sensors 2023, 23, 3751. [Google Scholar] [CrossRef]
- Jamil, S.; Jalil Piran, M.; Kwon, O.J. A comprehensive survey of transformers for computer vision. Drones 2023, 7, 287. [Google Scholar] [CrossRef]
- Maurício, J.; Domingues, I.; Bernardino, J. Comparing vision transformers and convolutional neural networks for image classification: A literature review. Appl. Sci. 2023, 13, 5521. [Google Scholar] [CrossRef]
- Parez, S.; Dilshad, N.; Alghamdi, N.S.; Alanazi, T.M.; Lee, J.W. Visual intelligence in precision agriculture: Exploring plant disease detection via efficient vision transformers. Sensors 2023, 23, 6949. [Google Scholar] [CrossRef]
- Al Mamun, A.; Ahmedt-Aristizabal, D.; Zhang, M.; Hossen, M.I.; Hayder, Z.; Awrangjeb, M. Plant Disease Detection Using Self-supervised Learning: A Systematic Review. IEEE Access 2024, 12, 171926–171943. [Google Scholar] [CrossRef]
- Alzahrani, M.S.; Alsaade, F.W. Transform and deep learning algorithms for the early detection and recognition of tomato leaf disease. Agronomy 2023, 13, 1184. [Google Scholar] [CrossRef]
- Jajja, A.I.; Abbas, A.; Khattak, H.A.; Niedbała, G.; Khalid, A.; Rauf, H.T.; Kujawa, S. Compact convolutional transformer (CCT)-Based approach for whitefly attack detection in cotton crops. Agriculture 2022, 12, 1529. [Google Scholar] [CrossRef]
- Sun, Y.; Ning, L.; Zhao, B.; Yan, J. Tomato Leaf Disease Classification by Combining EfficientNetv2 and a Swin Transformer. Appl. Sci. 2024, 14, 7472. [Google Scholar] [CrossRef]
- Zhang, M.; Lin, Z.; Tang, S.; Lin, C.; Zhang, L.; Dong, W.; Zhong, N. Dual-Attention-Enhanced MobileViT Network: A Lightweight Model for Rice Disease Identification in Field-Captured Images. Agriculture 2025, 15, 571. [Google Scholar] [CrossRef]
- Hamdi, E.B.; Hidayaturrahmana. Ensemble of pre-trained vision transformer models in plant disease classification, an efficient approach. Procedia Comput. Sci. 2024, 245, 565–573. [Google Scholar] [CrossRef]
- Tugrul, B.; Elfatimi, E.; Eryigit, R. Convolutional neural networks in detection of plant leaf diseases: A review. Agriculture 2022, 12, 1192. [Google Scholar] [CrossRef]
- Alam, T.S.; Jowthi, C.B.; Pathak, A. Comparing pre-trained models for efficient leaf disease detection: A study on custom CNN. J. Electr. Syst. Inf. Technol. 2024, 11, 12. [Google Scholar] [CrossRef]
- Thai, H.T.; Tran-Van, N.Y.; Le, K.H. Artificial cognition for early leaf disease detection using vision transformers. In Proceedings of the 2021 International Conference on Advanced Technologies for Communications (ATC), Ho Chi Minh City, Vietnam, 14–16 October 2021; IEEE: Piscataway, NJ, USA, 2021; pp. 33–38. [Google Scholar]
- Barman, U.; Sarma, P.; Rahman, M.; Deka, V.; Lahkar, S.; Sharma, V.; Saikia, M.J. Vit-SmartAgri: Vision transformer and smartphone-based plant disease detection for smart agriculture. Agronomy 2024, 14, 327. [Google Scholar] [CrossRef]
- Sharma, S.; Sharma, G.; Menghani, E. Tomato plant disease detection with pretrained CNNs: Review of performance assessment and visual presentation. In Artificial Intelligence in Medicine and Healthcare; CRC Press: Boca Raton, FL, USA, 2025; pp. 67–85. [Google Scholar]
- Maeda-Gutiérrez, V.; Galvan-Tejada, C.E.; Zanella-Calzada, L.A.; Celaya-Padilla, J.M.; Galván-Tejada, J.I.; Gamboa-Rosales, H.; Luna-Garcia, H.; Magallanes-Quintanar, R.; Guerrero Mendez, C.A.; Olvera-Olvera, C.A. Comparison of convolutional neural network architectures for classification of tomato plant diseases. Appl. Sci. 2020, 10, 1245. [Google Scholar] [CrossRef]
- Zhang, K.; Wu, Q.; Liu, A.; Meng, X. Can deep learning identify tomato leaf disease? Adv. Multimed. 2018, 2018, 6710865. [Google Scholar] [CrossRef]
- Atila, Ü.; Uçar, M.; Akyol, K.; Uçar, E. Plant leaf disease classification using EfficientNet deep learning model. Ecol. Inform. 2021, 61, 101182. [Google Scholar] [CrossRef]
- Yulita, I.N.; Amri, N.A.; Hidayat, A. Mobile application for tomato plant leaf disease detection using a dense convolutional network architecture. Computation 2023, 11, 20. [Google Scholar] [CrossRef]
- Dosovitskiy, A.; Beyer, L.; Kolesnikov, A.; Weissenborn, D.; Zhai, X.; Unterthiner, T.; Dehghani, M.; Minderer, M.; Heigold, G.; Gelly, S.; et al. An Image is Worth 16x16 Words: Transformers for Image Recognition at Scale. arXiv 2021, arXiv:2010.11929. [Google Scholar]
- Reedha, R.; Dericquebourg, E.; Canals, R.; Hafiane, A. Transformer neural network for weed and crop classification of high resolution UAV images. Remote Sens. 2022, 14, 592. [Google Scholar] [CrossRef]
- Tabbakh, A.; Barpanda, S.S. A deep features extraction model based on the transfer learning model and vision transformer “tlmvit” for plant disease classification. IEEE Access 2023, 11, 45377–45392. [Google Scholar] [CrossRef]
- Thakur, P.S.; Khanna, P.; Sheorey, T.; Ojha, A. Explainable vision transformer enabled convolutional neural network for plant disease identification: PlantXViT. arXiv 2022, arXiv:2207.07919. [Google Scholar]
- Han, Z.; Sun, J. Tomato leaf diseases recognition model based on improved MobileVit. In Proceedings of the 2024 IEEE 4th International Conference on Information Technology, Big Data and Artificial Intelligence (ICIBA), Chongqing, China, 6–8 December 2024; IEEE: Piscataway, NJ, USA, 2024; Volume 4, pp. 1205–1209. [Google Scholar]
- Tonmoy, M.R.; Hossain, M.M.; Dey, N.; Mridha, M. MobilePlantViT: A Mobile-friendly Hybrid ViT for Generalized Plant Disease Image Classification. arXiv 2025, arXiv:2503.16628. [Google Scholar]
- Mahabub, S.; Jahan, I.; Hasan, M.N.; Islam, M.S.; Akter, L.; Musfiqur, M.; Foysal, R.; Onik, M.K.R. Efficient detection of tomato leaf diseases using optimized Compact Convolutional Transformers (CCT) Model. Magna Sci. Adv. Res. Rev. 2024, 12, 39–53. [Google Scholar] [CrossRef]
- Liu, Z.; Lin, Y.; Cao, Y.; Hu, H.; Wei, Y.; Zhang, Z.; Lin, S.; Guo, B. Swin transformer: Hierarchical vision transformer using shifted windows. In Proceedings of the IEEE/CVF International Conference on Computer Vision, Montreal, QC, Canada, 10–17 October 2021; pp. 10012–10022. [Google Scholar]
- Singh, A.K.; Rao, A.; Chattopadhyay, P.; Maurya, R.; Singh, L. Effective plant disease diagnosis using Vision Transformer trained with leafy-generative adversarial network-generated images. Expert Syst. Appl. 2024, 254, 124387. [Google Scholar] [CrossRef]
- Huang, X.; Xu, D.; Chen, Y.; Zhang, Q.; Feng, P.; Ma, Y.; Dong, Q.; Yu, F. EConv-ViT: A strongly generalized apple leaf disease classification model based on the fusion of ConvNeXt and Transformer. Inf. Process. Agric. 2025, in press. [CrossRef]
- Liu, X.; Peng, H.; Zheng, N.; Yang, Y.; Hu, H.; Yuan, Y. Efficientvit: Memory efficient vision transformer with cascaded group attention. In Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition, Vancouver, BC, Canada, 17–24 June 2023; pp. 14420–14430. [Google Scholar]
- Saleem, M.H.; Potgieter, J.; Arif, K.M. Tomato plant disease detection using transfer learning with C-GAN synthetic images. Comput. Electron. Agric. 2021, 187, 106279. [Google Scholar] [CrossRef]
- Haider, F.A.; Abbas, A.; Mehmood, Z. Tomato Leaf Diseases Detection Using Deep Learning Technique; IntechOpen: London, UK, 2021. [Google Scholar] [CrossRef]
- Ashwinkumar, S.; Rajagopal, S.; Aarathy, V.B.S.; Magudeeswaran, P. Detecting tomato leaf diseases by image processing through deep convolutional neural networks. Artif. Intell. Agric. 2023, 9, 100301. [Google Scholar] [CrossRef]
- Rahim, M.M.A.; Islam, M.T.; Islam, M.S. A robust deep learning approach for tomato plant leaf disease localization and classification. Sci. Rep. 2022, 12, 21498. [Google Scholar] [CrossRef]
- Ruan, B.K.; Shuai, H.H.; Cheng, W.H. Vision Transformers: State of the Art and Research Challenges. arXiv 2022. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).