Recognition of Strawberry Powdery Mildew in Complex Backgrounds: A Comparative Study of Deep Learning Models
Abstract
1. Introduction
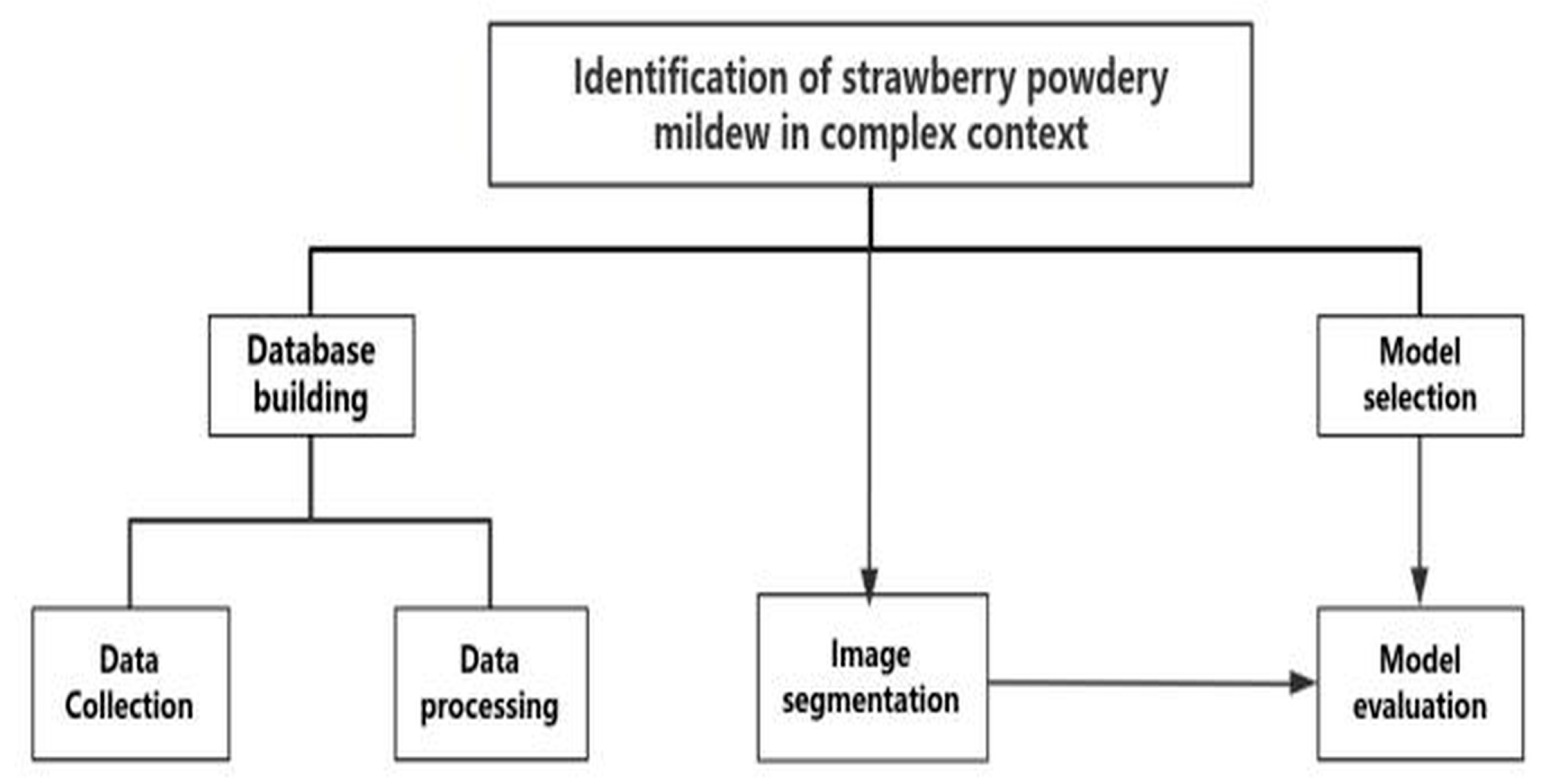
2. Materials and Methods
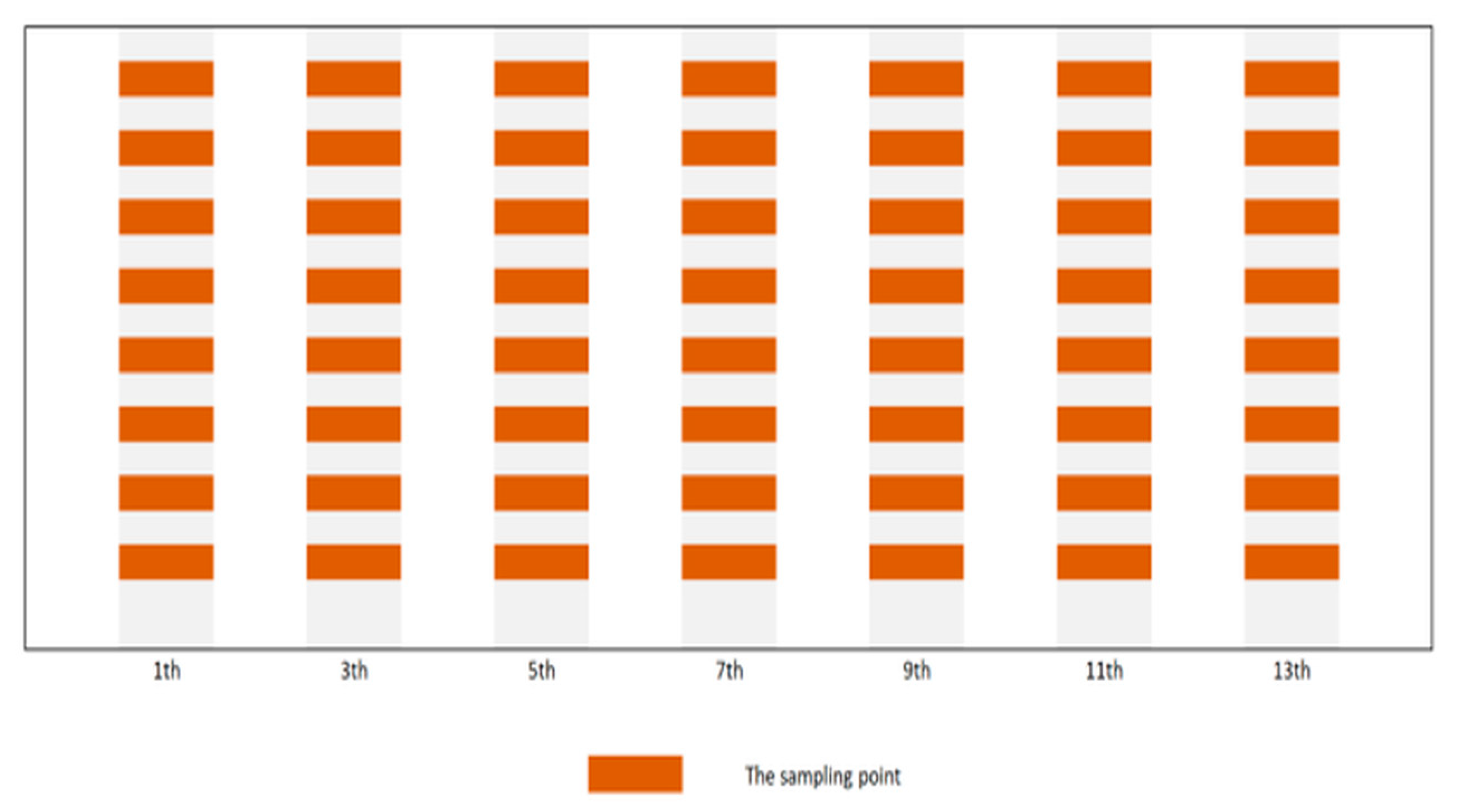
2.1. Dataset Construction and Preprocessing
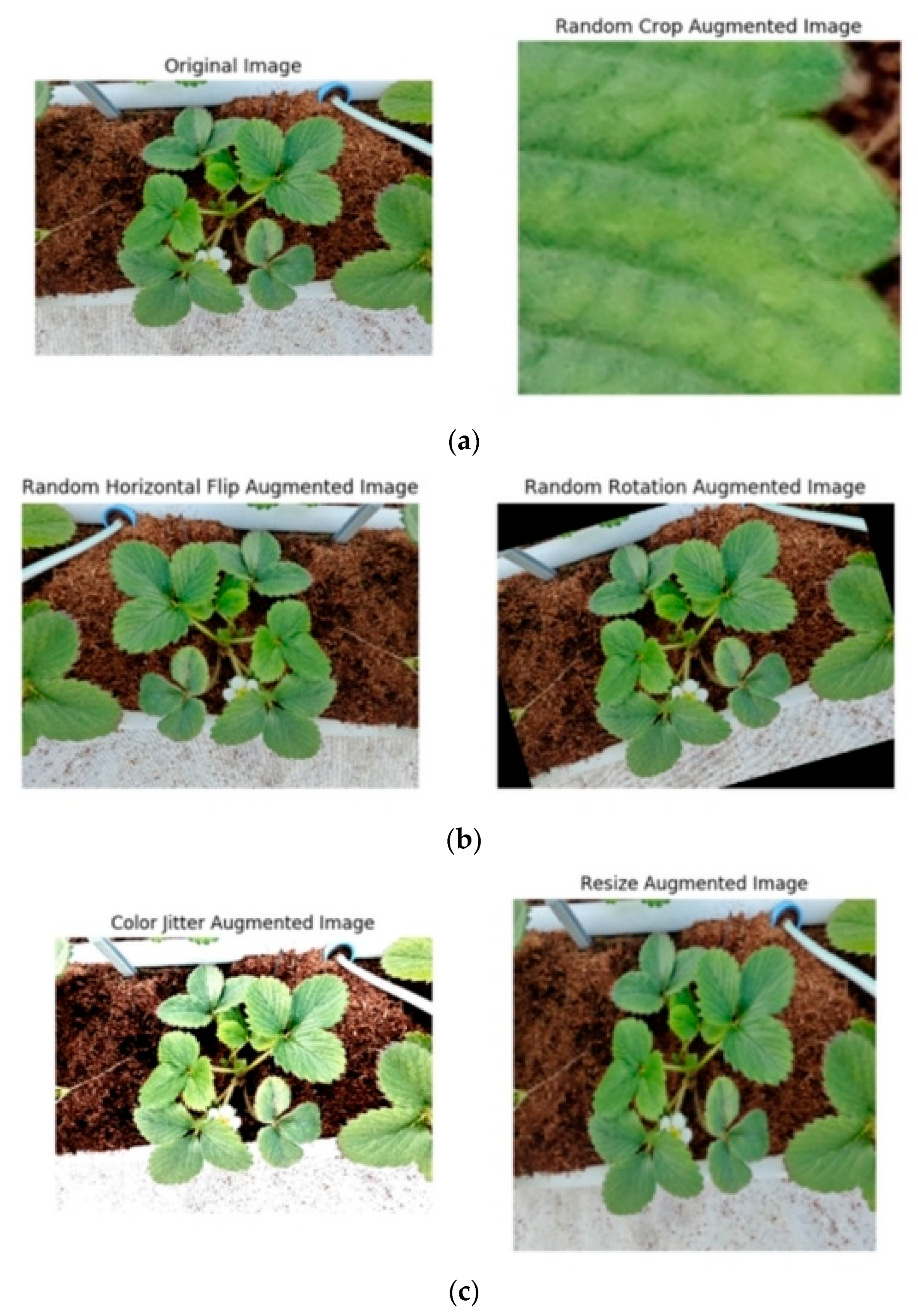
2.2. Data Augmentation
- Random rotation within the range [−10°, 10°]
- Horizontal and vertical flipping
- Random cropping with a scale range of 0.8–1.0
- Addition of Gaussian noise (mean = 0, variance = 0.01)
- Color jittering to simulate natural light variations
2.3. Image Segmentation
- Lower bound: (35, 43, 46)
- Upper bound: (77, 255, 255)
2.4. Model Architecture and Training
2.5. Training Settings
- GPU: NVIDIA Tesla V100
- CPU: 2 Cores
- RAM: 16 GB
- Video Memory: 16 GB
- Disk: 100 GB
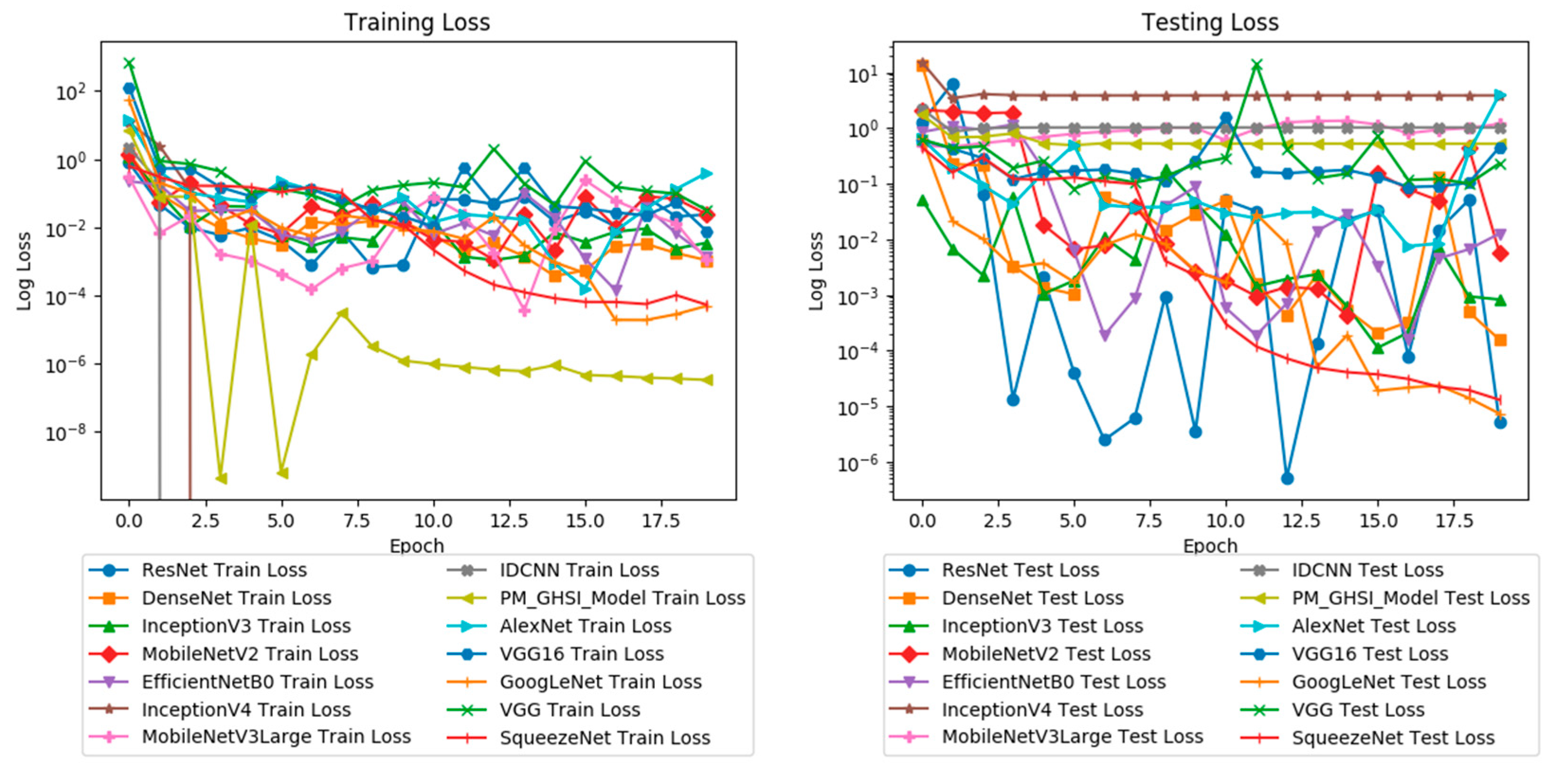
3. Results
3.1. Model Performance Evaluation
- where:
- TP = True Positives
- TN = True Negatives
- FP = False Positives
- FN = False Negatives
- M = Total number of samples
- Incorporating channel attention mechanisms, residual connections, or multi-branch feature aggregation into SqueezeNet to enhance its expressive power.
- Extracting knowledge from high-performance models such as DenseNet-121 or InceptionV4 to make lightweight variants of SqueezeNet approach the performance of deeper models while maintaining computational efficiency.
- Using SqueezeNet in a multi-stage framework as a preliminary filter to identify candidate regions for further inspection by more accurate models.
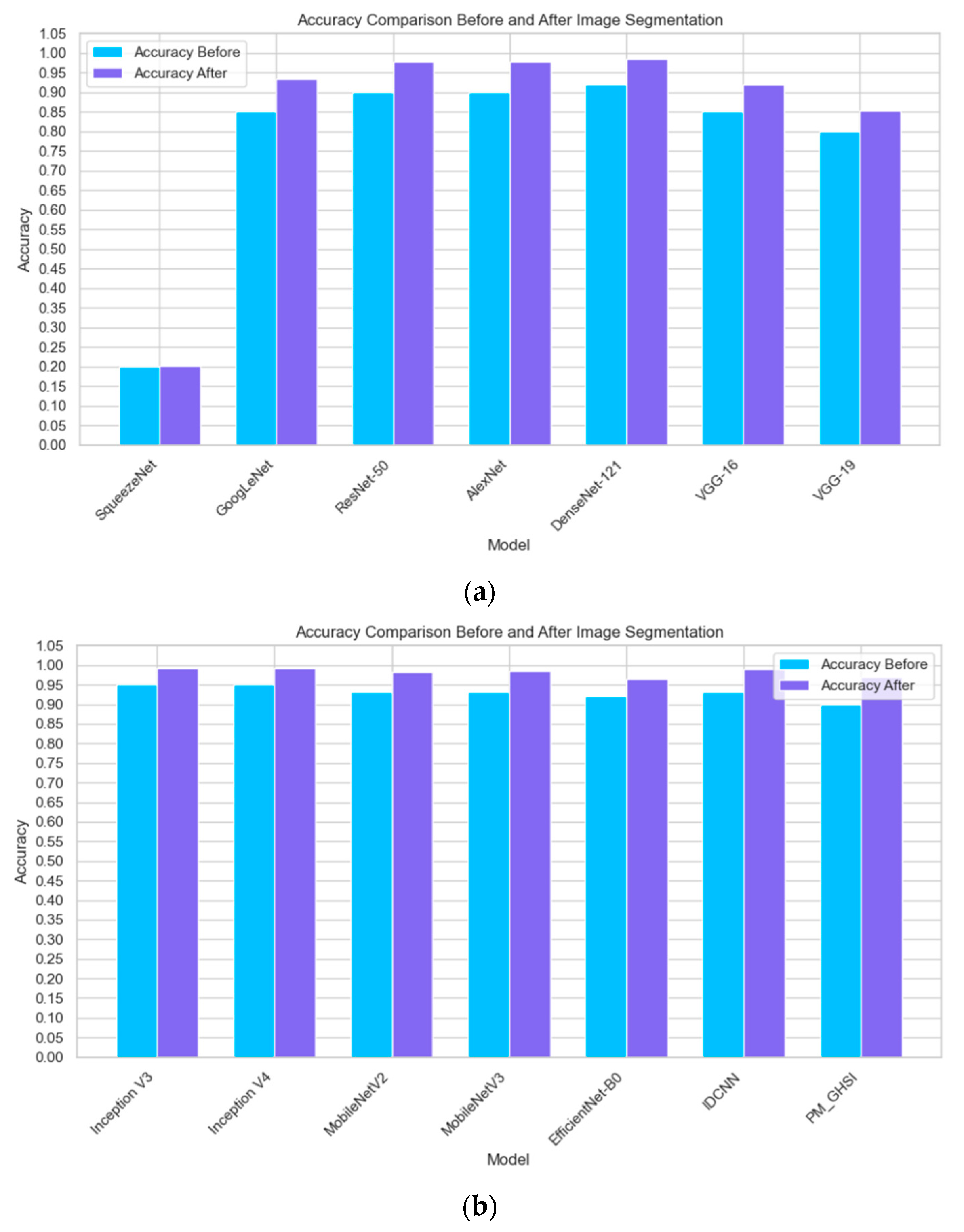
3.2. Effect of Image Segmentation
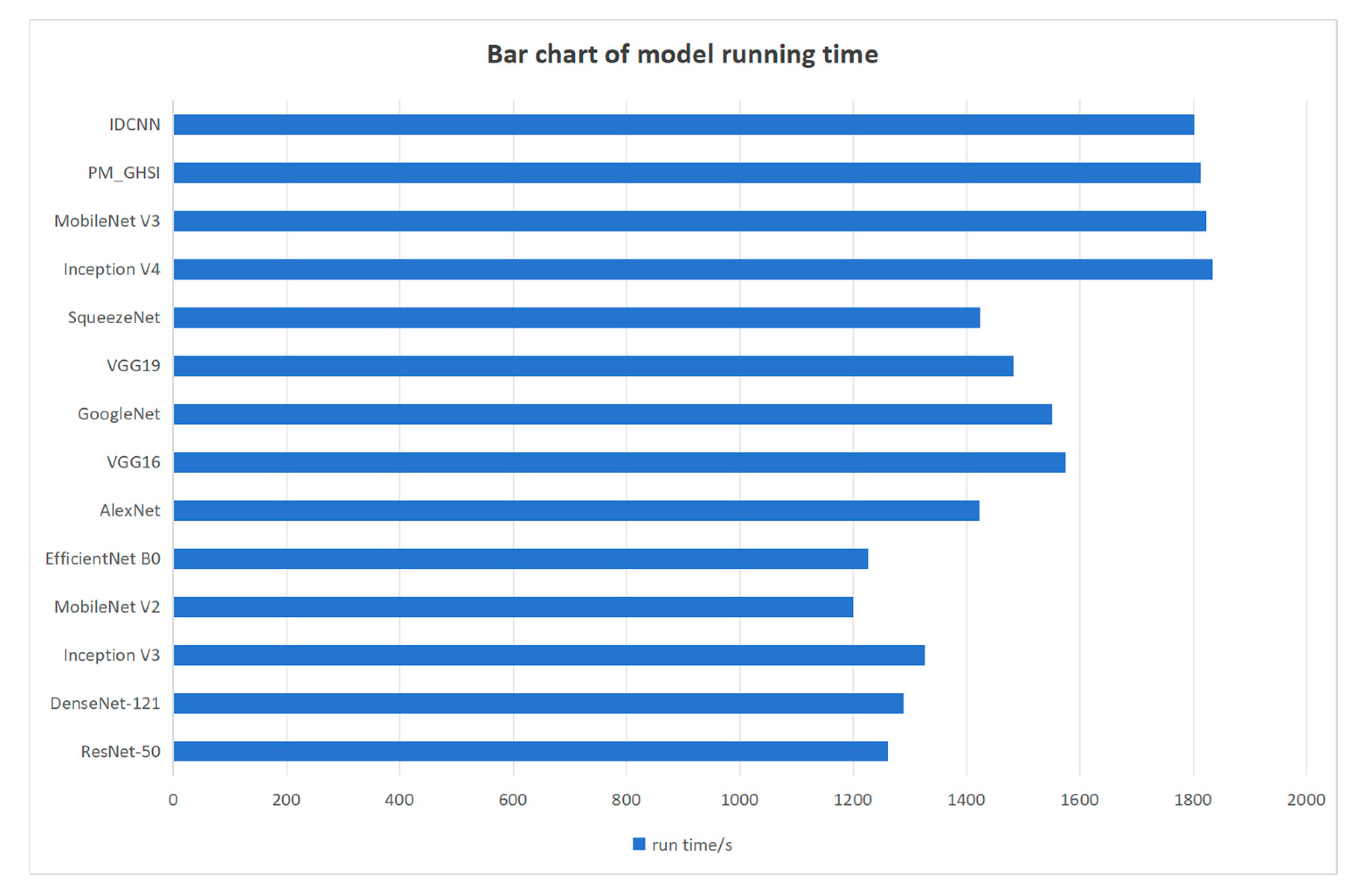
3.3. Inference Time Comparison
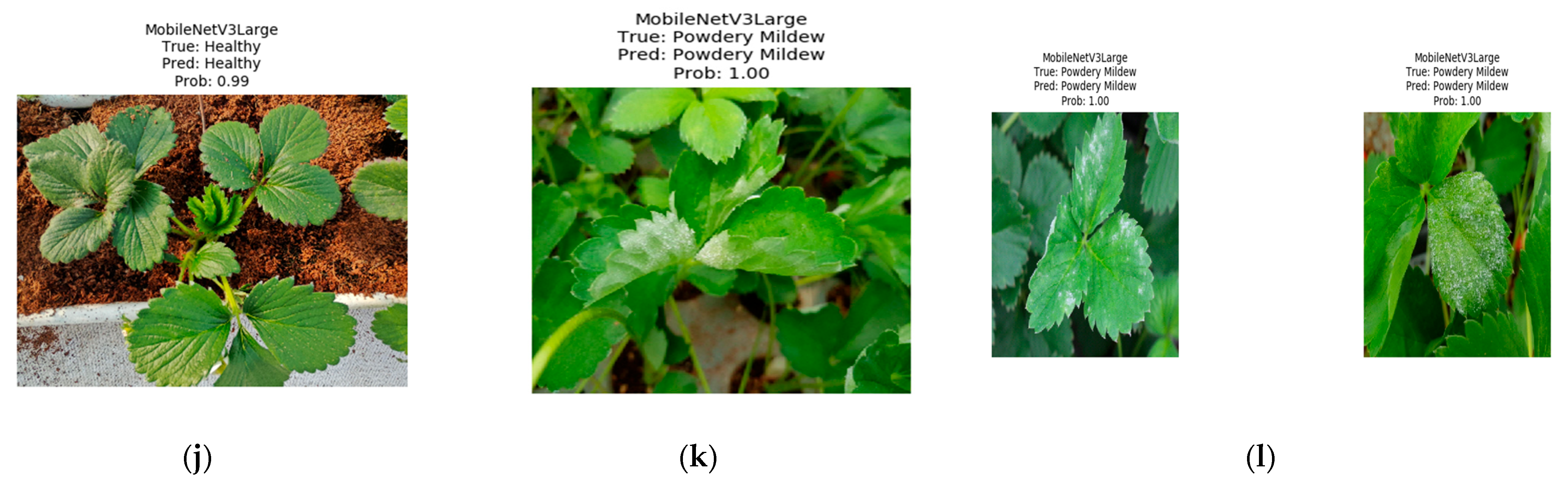
3.4. Visual Comparison of Representative Models on Healthy and Infected Leaves
4. Discussion
4.1. Experimental Results
4.2. Comparison with Traditional Machine Learning Approaches
4.3. Prospects for Agricultural Applications
5. Conclusions
- We constructed and annotated a dedicated image dataset of strawberry powdery mildew captured in real-world, complex agricultural environments. This dataset addresses a lack of publicly available, high-quality data for this specific disease under field conditions and lays a solid foundation for future studies.
- To enhance disease feature extraction from noisy and cluttered backgrounds, we introduced an HSV-based segmentation preprocessing method. This approach improves the visibility of disease regions and contributes to better model performance in complex scenes.
- We systematically evaluated and compared the performance of 14 widely used deep learning models, covering a diverse range of network architectures. The benchmarking results offer valuable guidance for selecting appropriate models based on trade-offs between accuracy and computational complexity (TFLOPs), facilitating informed decisions in real-world deployments.
- Our comparative experiments revealed how different architectures vary in their sensitivity to background interference and disease feature patterns. This insight is essential for understanding model robustness and for transferring these findings to other crop disease scenarios.
- Dataset expansion and fine-grained annotation
- 2.
- Lightweight model optimization
- 3.
- Multimodal data fusion
- 4.
- Field deployment and user interface design
- 5.
- Transferability and generalization
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Skrovankova, S.; Sumczynski, D.; Mlcek, J.; Jurikova, T.; Sochor, J. Bioactive compounds and antioxidant activity of different types of berries. Int. J. Mol. Sci. 2015, 16, 24673–24706. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; Chen, J.; Zhang, D.; Sun, Y.; Nanehkaran, Y.A. Using deep transfer learning for image-based plant disease identification. Comput. Electron. Agric. 2020, 173, 105393. [Google Scholar] [CrossRef]
- Khan, A.I.; Quadri, S.M.K.; Banday, S.; Shah, J.L. Deep diagnosis: A real-time apple leaf disease detection system based on deep learning. Comput. Electron. Agric. 2022, 198, 107093. [Google Scholar] [CrossRef]
- Vielba-Fernández, A.; Polonio, Á.; Ruiz-Jiménez, L.; de Vicente, A.; Pérez-García, A.; Fernández-Ortuño, D. Fungicide Resistance in Powdery Mildew Fungi. Microorganisms 2020, 8, 1431. [Google Scholar] [CrossRef]
- Xu, X.-M.; Robinson, J.D. Effects of Temperature on the Incubation and Latent Periods of Hawthorn Powdery Mildew (Podosphaera clandestina). Plant Pathol. 2000, 49, 791–797. [Google Scholar] [CrossRef]
- Mohanty, S.P.; Hughes, D.P.; Salathé, M. Using deep learning for image-based plant disease detection. Front. Plant Sci. 2016, 7, 1419. [Google Scholar] [CrossRef]
- Li, Y.; Zhang, L.; Wang, Y. Classification of Multi Diseases in Apple Plant Leaves Using HSV Color Space and LBP Texture Features. Available online: https://www.diva-portal.org/smash/record.jsf?pid=diva2:1380974 (accessed on 9 April 2025).
- Hassan, M.M.; Islam, M.T.; Rahman, M.A.; Hasan, M.M. A Comprehensive Survey on Deep Learning Approaches for Plant Disease Detection. Artif. Intell. Rev. 2023, 56, 1–35. [Google Scholar]
- Sai, A. Deep Learning for Plant Disease Detection: A Review. Artif. Intell. Rev. 2021, 54, 1–20. [Google Scholar]
- Yu, R.; Luo, Y.; Zhou, Q.; Zhang, X.; Wu, D.; Ren, L. Early detection of pine wilt disease using deep learning algorithms and UAV-based multispectral imagery. For. Ecol. Manag. 2021, 497, 119493. [Google Scholar] [CrossRef]
- Hughes, D.P.; Salathé, M. An open access repository of images on plant health to enable the development of mobile disease diagnostics. Comput. Electron. Agric. 2015, 145, 311–318. [Google Scholar]
- Lee, S.; Arora, A.S.; Yun, C.M. Detecting strawberry diseases and pest infections in the very early stage with an ensemble deep-learning model. Front. Plant Sci. 2022, 13, 991134. [Google Scholar] [CrossRef]
- Atila, M.; Yildirim, M.; Yildirim, Y.; Yildirim, A.; Yildirim, M.; Yildirim, S. Plant leaf disease classification using EfficientNet deep learning model. Comput. Electron. Agric. 2024, 186, 106145. [Google Scholar] [CrossRef]
- Argüeso, D.; Picon, A.; Irusta, U.; Medela, A.; San-Emeterio, M.G.; Bereciartua, A.; Alvarez-Gila, A. Few-Shot Learning Approach for Plant Disease Classification Using Images Taken in the Field. Comput. Electron. Agric. 2020, 175, 105542. [Google Scholar] [CrossRef]
- He, K.; Zhang, X.; Ren, S.; Sun, J. Deep residual learning for image recognition. Proc. IEEE Conf. Comput. Vis. Pattern Recognit. 2016, 2016, 770–778. [Google Scholar] [CrossRef]
- Howard, A.G.; Zhu, M.; Chen, B.; Kalenichenko, D.; Wang, W.; Weyand, T.; Andreetto, M.; Adam, H. MobileNets: Efficient convolutional neural networks for mobile vision applications. arXiv 2017, arXiv:1704.04861. Available online: https://arxiv.org/abs/1704.04861 (accessed on 5 April 2025).
- Krizhevsky, A.; Sutskever, I.; Hinton, G.E. Imagenet classification with deep convolutional neural networks. Commun. ACM 2017, 60, 84–90. [Google Scholar] [CrossRef]
- Liu, Y.; Zhang, C.; Wang, Z. Image segmentation evaluation: A survey of performance measures. Pattern Recognit. 2021, 116, 107963. [Google Scholar]
- Long, J.; Shelhamer, E.; Darrell, T. Fully convolutional networks for semantic segmentation. Proc. IEEE Conf. Comput. Vis. Pattern Recognit. 2015, 2015, 3431–3440. [Google Scholar] [CrossRef]
- Picon, A.; Alvarez-Gila, A.; Seitz, M.; Ortiz-Barredo, A.; Echazarra, J.; Johannes, A. Deep convolutional neural networks for mobile capture device-based crop disease classification in the wild. Comput. Electron. Agric. 2019, 161, 280–290. [Google Scholar] [CrossRef]
- Ronneberger, O.; Fischer, P.; Brox, T. U-Net: Convolutional networks for biomedical image segmentation. Lect. Notes Comput. Sci. 2015, 9351, 234–241. [Google Scholar]
- Sandler, M.; Howard, A.; Zhu, M.; Zhmoginov, A.; Chen, L.C. MobileNetV2: Inverted residuals and linear bottlenecks. Proc. IEEE Conf. Comput. Vis. Pattern Recognit. 2018, 2018, 4510–4520. [Google Scholar] [CrossRef]
- Simonyan, K.; Zisserman, A. Very deep convolutional networks for large-scale image recognition. arXiv 2014, arXiv:1409.1556. Available online: https://arxiv.org/abs/1409.1556 (accessed on 8 April 2025).
- Singh, A.; Ganapathysubramanian, B.; Singh, A.K.; Sarkar, S. Machine learning for high-throughput stress phenotyping in plants. Trends Plant Sci. 2016, 21, 110–124. [Google Scholar] [CrossRef] [PubMed]
- Szegedy, C.; Liu, W.; Jia, Y.; Sermanet, P.; Reed, S.; Anguelov, D.; Erhan, D.; Vanhoucke, V.; Rabinovich, A. Going deeper with convolutions. Proc. IEEE Conf. Comput. Vis. Pattern Recognit. 2015, 2015, 1–9. [Google Scholar] [CrossRef]
- Tan, M.; Le, Q. Efficientnet: Rethinking model scaling for convolutional neural networks. Proc. 36th Int. Conf. Mach. Learn. (ICML) 2019, 97, 6105–6114. [Google Scholar]
- Too, E.C.; Yujian, L.; Njuki, S.; Yingchun, L. A comparative study of fine-tuning deep learning models for plant disease identification. Comput. Electron. Agric. 2019, 161, 272–279. [Google Scholar] [CrossRef]
- Wang, G.; Sun, Y.; Wang, J. Automatic image-based plant disease severity estimation using deep learning. Comput. Intell. Neurosci. 2017, 2017, 2917536. [Google Scholar] [CrossRef]
- Wang, X.; Zhou, Q.; Ji, L.; Zhai, Y. Research on corn disease identification method based on deep convolutional neural network. Comput. Electron. Agric. 2020, 179, 105824. [Google Scholar] [CrossRef]
- Woo, S.; Park, J.; Lee, J.Y.; Kweon, I.S. CBAM: Convolutional block attention module. Proc. Eur. Conf. Comput. Vis. (ECCV) 2018, 2018, 3–19. [Google Scholar] [CrossRef]
- Zhang, Q.; Yang, L.T.; Chen, Z.; Li, P. A survey on deep learning for big data. Inf. Fusion 2018, 42, 146–157. [Google Scholar] [CrossRef]
- Liu, W.; Anguelov, D.; Erhan, D.; Szegedy, C.; Reed, S.; Fu, C.Y.; Berg, A.C. SSD: Single shot multibox detector. Proc. Eur. Conf. Comput. Vis. 2016, 9905, 21–37. [Google Scholar]
- Redmon, J.; Divvala, S.; Girshick, R.; Farhadi, A. You only look once: Unified, real-time object detection. Proc. IEEE Conf. Comput. Vis. Pattern Recognit. 2016, 2016, 779–788. [Google Scholar] [CrossRef]
- Ren, S.; He, K.; Girshick, R.; Sun, J. Faster R-CNN: Towards real-time object detection with region proposal networks. Adv. Neural Inf. Process. Syst. 2015, 28. Available online: https://proceedings.neurips.cc/paper_files/paper/2015/file/14bfa6bb14875e45bba028a21ed38046-Paper.pdf (accessed on 10 April 2025). [CrossRef] [PubMed]
- Lin, T.Y.; Goyal, P.; Girshick, R.; He, K.; Dollar, P. Focal loss for dense object detection. Proc. IEEE Int. Conf. Comput. Vis. 2017, 2017, 2980–2988. [Google Scholar] [CrossRef]
- Kingma, D.P.; Ba, J. Adam: A method for stochastic optimization. arXiv 2014, arXiv:1412.6980. Available online: https://arxiv.org/abs/1412.6980 (accessed on 6 April 2025).
- Deng, J.; Dong, W.; Socher, R.; Li, L.J.; Li, K.; Fei-Fei, L. Imagenet: A large-scale hierarchical image database. Proc. IEEE Conf. Comput. Vis. Pattern Recognit. 2009, 2009, 248–255. [Google Scholar] [CrossRef]
- Iandola, F.N.; Han, S.; Moskewicz, M.W.; Ashraf, K.; Dally, W.J.; Keutzer, K. SqueezeNet: AlexNet-level accuracy with 50x fewer parameters and <0.5 MB model size. arXiv 2016, arXiv:1602.07360. Available online: https://arxiv.org/abs/1602.07360 (accessed on 13 April 2025).
- Hatuwal, B.K.; Shakya, A.; Joshi, B. Plant Leaf Disease Recognition Using Random Forest, KNN, SVM and CNN. Polibits 2020, 62, 13–19. [Google Scholar]
- Rajpal, K. Machine Learning for Leaf Disease Classification: Data, Techniques and Applications. Artif. Intell. Rev. 2023, 56, 3571–3616. [Google Scholar]
- Dang-Ngoc, T.T.; Nguyen, T.T.; Nguyen, T.T.; Le, T.T.; Nguyen, T.T. An Optimal Hybrid Multiclass SVM for Plant Leaf Disease Detection Using Spatial Fuzzy C-Means Model. Expert Syst. Appl. 2023, 214, 118989. [Google Scholar]
- Rumy, S.H.; Sultana, S.; Rahman, M.M.; Hossain, M.S. A Comparative Analysis of Efficacy of Machine Learning Techniques for Disease Detection in Some Economically Important Crops. Crop Prot. 2025, 190, 107093. [Google Scholar]
- Alhwaiti, Y.; Ishaq, M.; Siddiqi, M.H.; Waqas, M.; Alruwaili, M.; Alanazi, S.; Khan, A.; Khan, F. Early Detection of Late Blight Tomato Disease Using Histogram Oriented Gradient Based Support Vector Machine. arXiv 2023, arXiv:2306.08326. [Google Scholar]




| Model | Convolutional Layers | Image Size | Model Features |
|---|---|---|---|
| SqueezeNet | 23 | 224 × 224 | Through extrusion and excitation mechanisms, the accuracy is maintained, and the weight of the model is realized. |
| GoogLeNet | 22 | 224 × 224 | The Inception module is introduced, which uses convolutional kernels of different scales for parallel computing. |
| ResNet-50 | 50 | 224 × 224 | The residual connection is proposed to solve the problem of gradient disappearance and degradation with the increase of neural network layers. |
| AlexNet | 5 | 227 × 227 | The first deep convolutional neural network successfully applied to large-scale image classification, using the ReLU activation function and the Dropout technique. |
| DenseNet-121 | 121 | 224 × 224 | Dense connections are used to connect the feature maps of all the previous layers to the later layers. |
| VGG-16/19 | 16/19 | 224 × 224 | Stacking multiple small 3 × 3 convolutional kernels instead of large convolutional kernels increases the nonlinearity of the network. |
| Inception V3/V4 | 42/57 | 229 × 229 | Continuously optimize the Inception module by introducing more convolutional layers and optimizing structures. |
| MobileNetV2/V3 | 19/54 | 224 × 224 | The use of deep separable convolution greatly reduces the parameters and computational cost of the model. |
| EfficientNet-B0 | 53 | 224 × 224 | Through joint optimization of the width, depth, and resolution of the network. |
| IDCNN | 4 | 224 × 224 | The ReLU activation function introduces nonlinear, pooled layer dimensionality reduction. |
| PM_GHSI | 4 | 224 × 224 | The ReLU activation function introduces nonlinear, pooled layer dimensionality reduction. |
| Model | Accuracy | F1-Score | Recall | TFLOPs |
|---|---|---|---|---|
| SqueezeNet | 20.17% | 0.12 | 0.14 | 0.0002 |
| GoogLeNet | 93.35% | 0.96 | 0.95 | 0.0016 |
| ResNet-50 | 97.87% | 0.98 | 0.97 | 0.0041 |
| AlexNet | 92.73% | 0.97 | 0.97 | 0.0007 |
| DenseNet-121 | 98.54% | 0.98 | 0.99 | 0.0002 |
| VGG-16 | 91.89% | 0.96 | 0.95 | 0.0155 |
| VGG-19 | 85.38% | 0.91 | 0.89 | 0.0196 |
| Inception V3 | 99.14% | 0.98 | 0.98 | 0.0006 |
| Inception V4 | 99.23% | 0.99 | 0.99 | 0.0002 |
| MobileNetV2 | 98.12% | 0.92 | 0.92 | 0.0013 |
| MobileNetV3 | 98.43% | 0.98 | 0.98 | 0.0007 |
| EfficientNet-B0 | 96.57% | 0.96 | 0.96 | 0.0121 |
| IDCNN | 98.88% | 0.99 | 0.98 | 0.0022 |
| PM_GHSI | 97.05% | 0.97 | 0.96 | 0.0003 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, J.; Li, J.; Meng, F. Recognition of Strawberry Powdery Mildew in Complex Backgrounds: A Comparative Study of Deep Learning Models. AgriEngineering 2025, 7, 182. https://doi.org/10.3390/agriengineering7060182
Wang J, Li J, Meng F. Recognition of Strawberry Powdery Mildew in Complex Backgrounds: A Comparative Study of Deep Learning Models. AgriEngineering. 2025; 7(6):182. https://doi.org/10.3390/agriengineering7060182
Chicago/Turabian StyleWang, Jingzhi, Jiayuan Li, and Fanjia Meng. 2025. "Recognition of Strawberry Powdery Mildew in Complex Backgrounds: A Comparative Study of Deep Learning Models" AgriEngineering 7, no. 6: 182. https://doi.org/10.3390/agriengineering7060182
APA StyleWang, J., Li, J., & Meng, F. (2025). Recognition of Strawberry Powdery Mildew in Complex Backgrounds: A Comparative Study of Deep Learning Models. AgriEngineering, 7(6), 182. https://doi.org/10.3390/agriengineering7060182





