Artificial Neural Network and Mathematical Modeling to Estimate Losses in the Concentration of Bioactive Compounds in Different Tomato Varieties During Cooking
Abstract
:1. Introduction
2. Materials and Methods
2.1. Fresh Tomatoes
2.2. Processing
2.3. Quality Determination of Fresh Tomatoes and Processed Products
2.4. Experimental Design
2.5. Application of Artificial Neural Networks
2.6. Regression Analysis
3. Results and Discussion
4. Conclusions
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
Nomenclature
| MLP | Multilayer perceptron network |
| ANN | Artificial neural network |
| MSE | Mean square error |
| Yob | Obtained outputs |
| Ydes | Desired outputs |
References
- Capanoglu, E.; Beekwilder, J.; Boyacioglu, D.H.R.; De Vos, R. Changes in antioxidant and metabolite profiles during production of tomato paste. J. Agric. Food Chem. 2008, 56, 964–973. [Google Scholar] [CrossRef] [PubMed]
- Rodriguez-Amaya, D.B. Latin American food sources of carotenoids. Arch. Latinoam. Nutr. 1999, 49, 74S–84S. [Google Scholar] [PubMed]
- Azeez, L.; Segun, A.A.; Oyedeji, A.O.; Adetoro, R.O.; Tijani, K.O. Bioactive compounds’ contents, drying kinetics and mathematical modelling of tomato slices influenced by drying temperatures and time. J. Saudi Soc. Agric. Sci. 2019, 18, 120–126. [Google Scholar] [CrossRef]
- Bramley, P.M. Is lycopene beneficial to human health? Phytochemistry 2000, 54, 233–236. [Google Scholar] [CrossRef] [PubMed]
- Wu, X.; Yu, L.; Pehrsson, P.R. Are processed tomato products as nutritious as fresh tomatoes? Scoping review on the effects of industrial processing on nutrients and bioactive compounds in tomatoes. Adv. Nutr. 2022, 13, 138–151. [Google Scholar] [CrossRef]
- Chang, C.H.; Lin, H.Y.; Chang, C.Y.; Liu, Y.C. Comparações sobre as propriedades antioxidantes de tomates frescos, liofilizados e secos ao ar quente. J. Eng. Aliment. 2006, 77, 478–485. [Google Scholar] [CrossRef]
- Perez-Conesa, D.; Garcia-Alonso, J.; Garcia-Valverde, V.; Iniesta, M.D.; Jacob, K.; Sanchez-Siles, L.M. Changes in bioactive compounds and antioxidant activity during homogenization and thermal processing of tomato puree. Innov. Food Sci. Emerg. Technol. 2009, 10, 179–188. [Google Scholar] [CrossRef]
- Braga, A.P.; Ludermir, T.B.; Carvalho, A.C.P.L.F. Redes Neurais Artificiais Teoria e Aplicações; LTC—Livros Técnicos e Científicos Editora S.A.: Rio de Janeiro, Brazil, 2000. [Google Scholar]
- Soares, F.C.; Robaina, A.D.; Peiter, M.X.; Russi, J.L. Predição da produtividade da cultura do milho utilizando rede neural artificial. Ciênc. Rural 2015, 45, 1987–1993. [Google Scholar] [CrossRef]
- Kakani, V.; Nguyen, V.H.; Kumar, B.P.; Kim, H.; Pasupuleti, V.R. A critical review on computer vision and artificial intelligence in food industry. J. Agric. Food Res. 2020, 2, 100033. [Google Scholar] [CrossRef]
- Velázquez Martí, B.; Bonini Neto, A.; Nuñez Retana, D.; Carrillo Parra, A.; Guerrero-Luzuriaga, S. Determination of biomass drying speed using neural networks. Biomass Bioenergy 2024, 186, 107260. [Google Scholar] [CrossRef]
- Castillo-Girones, S.; Munera, S.; Martínez-Sober, M.; Blasco, R.; Cubero, S.; Gómez-Sanchis, J. Artificial Neural Networks in Agriculture, the core of artificial intelligence: What, When, and Why. Comput. Electron. Agric. 2025, 230, 109938. [Google Scholar] [CrossRef]
- Bonini Neto, A.; Souza, A.V.; Bonini, C.S.B.; Mello, J.M.; Moreira, A. Classification of Banana Ripening Stages by Artificial Neural Networks as a Function of Plant Physical, Physicochemical and Biochemical Parameters. Eng. Agric. 2022, 42, e20210197. [Google Scholar] [CrossRef]
- Praharsha, C.H.; Poulose, A.; Badgujar, C. Comprehensive Investigation of Machine Learning and Deep Learning Networks for Identifying Multispecies Tomato Insect Images. Sensors 2024, 24, 7858. [Google Scholar] [CrossRef]
- López-Correa, J.M.; Moreno, H.; Ribeiro, A.; Andújar, D. Intelligent Weed Management Based on Object Detection Neural Networks in Tomato Crops. Agronomy 2022, 12, 2953. [Google Scholar] [CrossRef]
- Tran, T.-T.; Choi, J.-W.; Le, T.-T.H.; Kim, J.-W. A Comparative Study of Deep CNN in Forecasting and Classifying the Macronutrient Deficiencies on Development of Tomato Plant. Appl. Sci. 2019, 9, 1601. [Google Scholar] [CrossRef]
- Kabaş, A.; Ercan, U.; Kabas, O.; Moiceanu, G. Prediction of Total Soluble Solids Content Using Tomato Characteristics: Comparison Artificial Neural Network vs. Multiple Linear Regression. Appl. Sci. 2024, 14, 7741. [Google Scholar] [CrossRef]
- Kabas, O.; Kayakus, M.; Ünal, İ.; Moiceanu, G. Deformation Energy Estimation of Cherry Tomato Based on Some Engineering Parameters Using Machine-Learning Algorithms. Appl. Sci. 2023, 13, 8906. [Google Scholar] [CrossRef]
- Park, H.; Kim, Y.J.; Shin, Y. Estimation of daily intake of lycopene, antioxidant contents and activities from tomatoes, watermelons, and their processed products in Korea. Appl. Biol. Chem. 2020, 63, 50. [Google Scholar] [CrossRef]
- Monteiro, C.S. Desenvolvimento de Molho de Tomate Lycopersicon esculentum Mill: Formulado com Cogumelo Agarius brasiliensis. Ph.D. Thesis, Doutorado em Tecnologia de Alimentos—Universidade Federal do Paraná, Curitiba, Brazil, 2008; 176p. Available online: https://acervodigital.ufpr.br/handle/1884/15780 (accessed on 10 January 2025).
- Dubois, M.; Gilles, K.A.; Hamilton, J.K.; Rebers, P.A.; Smith, F. Colorimetric method for determination of sugars and related substances. Anal. Chem. 1956, 28, 350–356. [Google Scholar] [CrossRef]
- Instituto Adolfo Lutz. Normas Analíticas do Instituto Adolfo Lutz. In v. 1: Métodos Químicos e Físicos para Análise de Alimentos, 4th ed.; 1st digital edition; IMESP: São Paulo, Brazil, 2008. Available online: https://www.ial.sp.gov.br/ial/publicacoes/livros/metodos-fisico-quimicos-para-analise-de-alimentos (accessed on 11 January 2024).
- Singleton, V.L.; Rossi, J.A. Colorimetry of total phenolics with phosphomolybidic-phosphotungstic acid reagents. Am. J. Enol. Vitic. 1965, 16, 144–158. [Google Scholar] [CrossRef]
- Nagata, M.; Yamashita, I. Simple method for simultaneous determination of chlorophyll and carotenoids in tomato fruit. Nippon Shokuhin Kogyo Gakkaish 1992, 39, 925–928. [Google Scholar] [CrossRef]
- Brand-Williams, W.; Cuvelier, M.E.; Berset, C. Use of free radical method to evaluate antioxidant activity. Lebensm.-Wiss. Technol. 1995, 28, 25–30. [Google Scholar] [CrossRef]
- Benzie, I.F.F.; Strain, J.J. The ferric reducing ability of plasma (FRAP) as a measure of antioxidant power: The FRAP assay. Anal. Biochem. 1996, 239, 70–76. [Google Scholar] [CrossRef]
- Mathworks. Available online: http://www.mathworks.com (accessed on 20 January 2024).
- Bonini Neto, A.; de Queiroz, A.; da Silva, G.G.; Gifalli, A.; de Souza, A.N.; Garbelini, E. Predictive Modeling of Total Real and Reactive Power Losses in Contingency Systems Using Function-Fitting Neural Networks with Graphical User Interface. Technologies 2025, 13, 15. [Google Scholar] [CrossRef]
- Gifalli, A.; Bonini Neto, A.; de Souza, A.N.; de Mello, R.P.; Ikeshoji, M.A.; Garbelini, E.; Neto, F.T. Fault Detection and Normal Operating Condition in Power Transformers via Pattern Recognition Artificial Neural Network. Appl. Syst. Innov. 2024, 7, 41. [Google Scholar] [CrossRef]
- da Silva, G.G.; de Queiroz, A.; Garbelini, E.; dos Santos, W.P.L.; Minussi, C.R.; Bonini Neto, A. Estimation of Total Real and Reactive Power Losses in Electrical Power Systems via Artificial Neural Network. Appl. Syst. Innov. 2024, 7, 46. [Google Scholar] [CrossRef]
- Varga, I.; Radočaj, D.; Jurišić, M.; Kulundžić, A.M.; Antunović, M. Prediction of sugar beet yield and quality parameters with varying nitrogen fertilization using ensemble decision trees and artificial neural networks. Comput. Electron. Agric. 2023, 212, 108076. [Google Scholar] [CrossRef]
- Engel, Y.; Mannor, S.; Meir, R. The kernel recursive least-squares algorithm. IEEE Trans. Signal Process. 2004, 40752, 2275–2285. [Google Scholar] [CrossRef]
- Díaz-Longueira, A.; Rubiños, M.; Arcano-Bea, P.; Calvo-Rolle, J.L.; Quintián, H.; Zayas-Gato, F. An Intelligent Regression-Based Approach for Predicting a Geothermal Heat Exchanger’s Behavior in a Bioclimatic House Context. Energies 2024, 17, 2706. [Google Scholar] [CrossRef]
- Poojari, S.; Acharya, S.; Varun Kumar, S.G.; Serrao, V. Modified least squares ratio estimator for autocorrelated data: Estimation and prediction. J. Comput. Math. Data Sci. 2025, 14, 100109. [Google Scholar] [CrossRef]



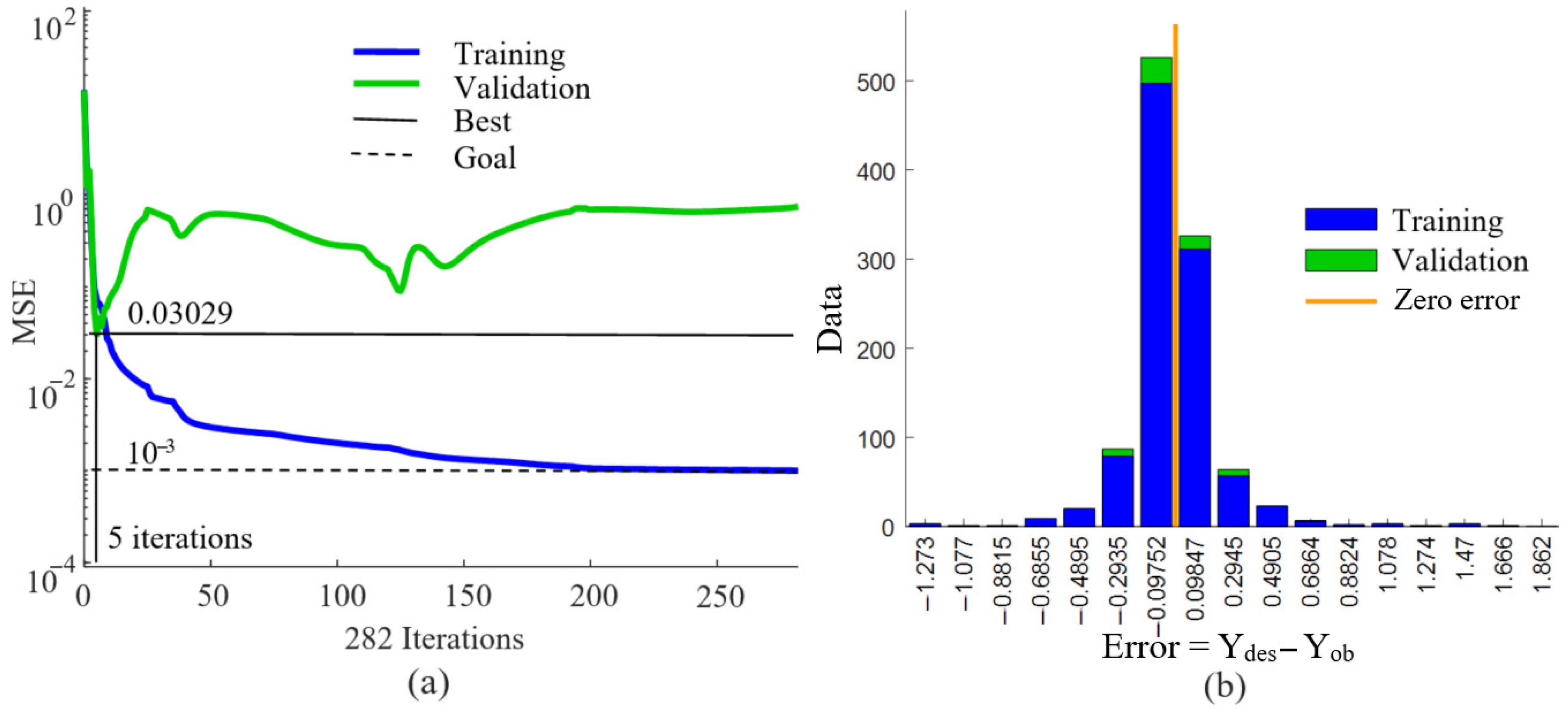




| ANN | Specified Value | Achieved Value |
|---|---|---|
| Iterations | 1000 | 282 |
| Time (s) | 60 | 8 |
| Training performance (MSE) | 0.001 | 0.000999 * |
| Training correlation (R2) | 1.0 | 0.9148 |
| Validation performance (MSE) | 0.001 | 0.03029 |
| Validation correlation (R2) | 1.0 | 0.8609 |
| Best validation performance (iteration) | 100 | 5 |
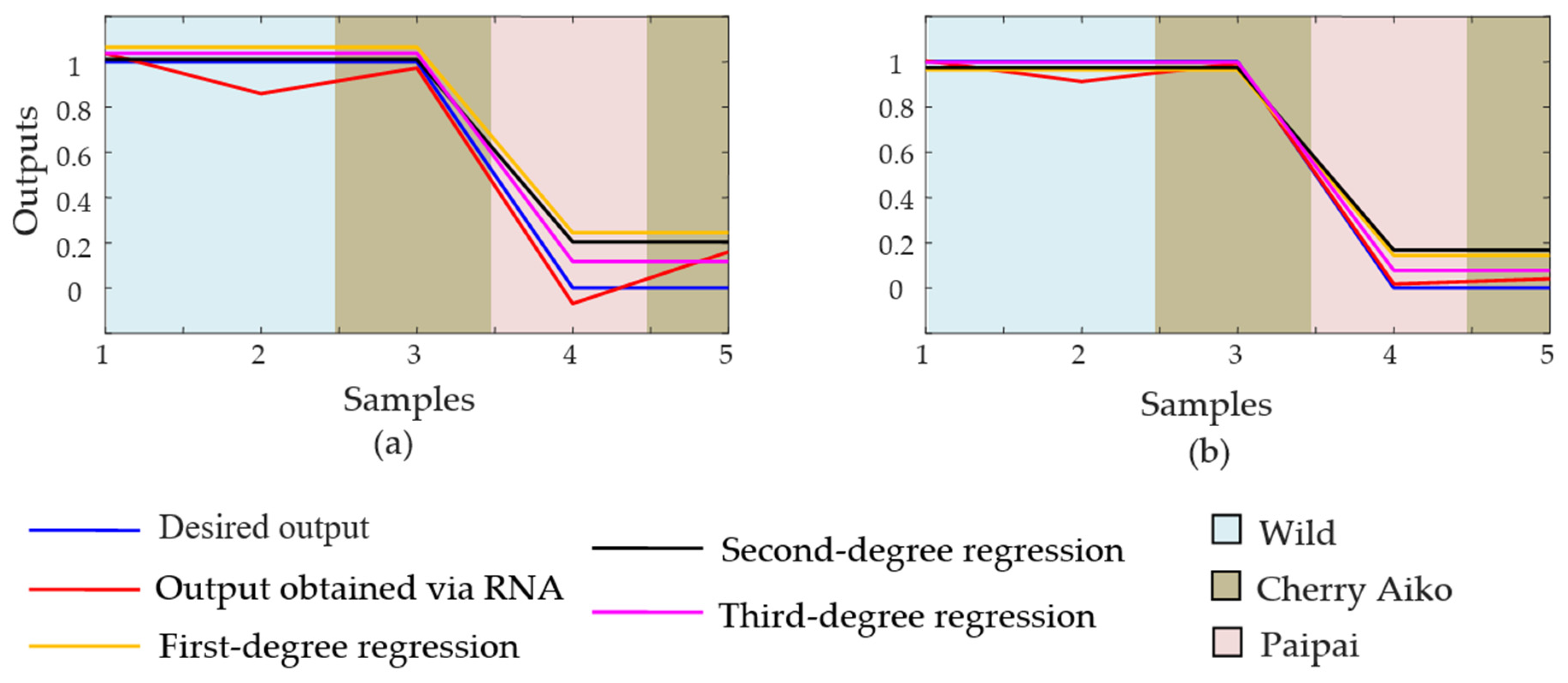
| Correlation (R2) of 100% of samples via ANN | 1.0 | 0.9025 |
| First-degree regression (R2) for 100% of samples | 1.0 | 0.8817 |
| Second-degree regression (R2) for 100% of samples | 1.0 | 0.8819 |
| Third-degree regression (R2) for 100% of samples | 1.0 | 0.8941 |
| Neurons in the Hidden Layer | ||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Neurons in the input layer | m1 | m2 | m3 | m4 | m5 | m6 | m7 | m8 | m9 | m10 | m11 | m12 | m13 | m14 | m15 | |
| R1 | −8.5274 | −6.1899 | 1.7758 | −4.0797 | −6.8135 | 1.9245 | 6.0938 | −0.555 | 4.0853 | −0.5534 | −3.2263 | 1.5964 | 3.1964 | 1.4625 | −3.7006 | |
| R2 | 2.4851 | −1.2733 | 0.9146 | −4.5785 | −5.5716 | 6.5616 | −0.2183 | 0.5956 | −5.3887 | 0.072 | −3.4158 | 0.273 | 1.5518 | 3.0546 | 0.0446 | |
| R3 | 0.5805 | 1.9429 | 3.1154 | 2.4426 | −5.1405 | 3.2454 | 0.5807 | −0.2981 | −1.5411 | 9.2252 | 16.6548 | −4.4227 | −0.840 | −1.6666 | −2.5458 | |
| R4 | −0.2361 | −2.227 | 1.1846 | −3.7793 | 1.1519 | −2.5137 | −0.6701 | 0.1609 | 1.121 | −0.956 | −2.2151 | −1.1961 | 1.0664 | −5.3676 | 1.7935 | |
| R5 | −2.6401 | −2.5067 | −6.7222 | −7.8229 | 9.4098 | −1.6673 | 1.4625 | 0.1137 | 0.4789 | −3.7218 | −6.1581 | −5.7297 | −1.308 | 0.8562 | 1.0316 | |
| R6 | 0.7689 | 0.3875 | 2.2094 | −0.3519 | 1.2378 | −0.0994 | −0.2327 | −0.4243 | −0.0967 | −2.4434 | −0.5536 | 2.0608 | 2.0921 | 4.6 | 0.066 | |
| R7 | 0.1631 | −0.3966 | 2.6101 | 8.5767 | 2.2655 | −1.3073 | −0.0683 | −0.5233 | 0.707 | 2.4967 | 1.9221 | −0.2551 | −0.687 | 6.193 | 1.1104 | |
| R8 | 1.0261 | 0.183 | 2.3229 | 21.0954 | 6.2141 | −1.5754 | −1.6078 | 0.7713 | 0.7821 | 8.7331 | 13.2627 | −0.2488 | 1.4084 | 1.4612 | 1.1786 | |
| R9 | 0.3946 | −2.5208 | 7.8396 | −7.4163 | −3.627 | −7.6013 | −0.919 | −0.1414 | 2.0722 | −1.2292 | −2.3102 | −5.4033 | 7.0866 | −0.3067 | 5.1824 | |
| R10 | 0.1364 | 1.0808 | −4.5368 | −1.9847 | 3.734 | 2.9101 | 1.1764 | 0.1058 | −0.2126 | 1.7894 | 4.9000 | 2.8762 | −3.509 | 1.3923 | −2.8117 | |
| R11 | −1.3407 | 2.1537 | −3.3648 | −2.1224 | 3.9595 | 3.7399 | 0.2315 | 0.2308 | −1.7972 | 2.2608 | 4.3569 | 2.8795 | −4.880 | 1.2431 | −1.9319 | |
| R12 | 1.7784 | 1.8852 | 0.3609 | −11.273 | 2.5423 | −2.2087 | −2.6742 | −0.7855 | 0.2872 | −0.8562 | −1.0191 | 2.4097 | 0.2308 | −1.8845 | 0.1018 | |
| Neurons in the Output Layer | |||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Neurons in the hidden layer | i1 | i2 | i3 | i4 | i5 | i6 | i7 | i8 | i9 | i10 | i11 | i12 | |
| m1 | −1.1279 | −1.1288 | −2.7619 | −2.1548 | 0.584 | −3.0818 | −1.1356 | −1.4516 | 0.0126 | 0.0126 | −1.1279 | −1.1288 | |
| m2 | 2.0195 | 2.7705 | 0.3674 | 4.5635 | 0.3571 | 7.2269 | −8.5052 | −7.6177 | 1.076 | 1.076 | 2.0195 | 2.7705 | |
| m3 | 0.4288 | 0.4193 | 0.0412 | −0.2552 | −0.1448 | 0.0082 | 0.8013 | 0.9234 | −0.0274 | −0.0274 | 0.4288 | 0.4193 | |
| m4 | 0.0283 | 0.051 | 0.1283 | 0.0632 | 0.1161 | −0.0169 | −0.0091 | 0.0071 | 0.0039 | 0.0039 | 0.0283 | 0.051 | |
| m5 | −0.6479 | −0.6358 | −0.0011 | 0.151 | 0.505 | 0.0604 | 0.5412 | 0.4067 | −0.0079 | −0.0079 | −0.6479 | −0.6358 | |
| m6 | 2.5219 | 2.2002 | 2.2449 | 1.1621 | −0.4918 | 3.1965 | 6.0982 | 5.8724 | −0.499 | −0.499 | 2.5219 | 2.2002 | |
| m7 | −0.818 | −0.6751 | −3.1964 | −2.4455 | 0.605 | −3.6152 | −2.5168 | −2.8124 | 0.0875 | 0.0875 | −0.818 | −0.6751 | |
| m8 | −2.4192 | −2.096 | −6.5673 | −7.3913 | 1.4047 | −6.2779 | −2.9087 | −3.4491 | 0.6021 | 0.6021 | −2.4192 | −2.096 | |
| m9 | −2.7672 | −1.8849 | −7.0939 | −4.0533 | 1.6531 | −2.4244 | −11.1474 | −11.1002 | 0.507 | 0.507 | −2.7672 | −1.8849 | |
| m10 | −0.1757 | −0.2555 | −1.3204 | −0.5041 | 0.2582 | −0.1121 | 0.0839 | −0.0589 | 0.0226 | 0.0226 | −0.1757 | −0.2555 | |
| m11 | 0.1534 | 0.2112 | 1.1934 | 0.4441 | −0.3547 | 0.1233 | −0.075 | 0.0521 | −0.0285 | −0.0285 | 0.1534 | 0.2112 | |
| m12 | 6.1687 | 6.1369 | 1.2407 | 4.4115 | −2.1753 | 0.7779 | 0.3561 | 0.741 | −0.4564 | −0.4564 | 6.1687 | 6.1369 | |
| m13 | 1.0069 | 1.6743 | −0.291 | 3.1263 | 0.1878 | 3.6519 | −8.8392 | −8.1134 | 0.9586 | 0.9586 | 1.0069 | 1.6743 | |
| m14 | 0.944 | 0.9111 | −0.0213 | −1.3963 | 2.2297 | −0.1665 | −0.4242 | −0.4378 | −0.0927 | −0.0927 | 0.944 | 0.9111 | |
| m15 | 4.5652 | 3.5979 | 5.9615 | 3.0844 | −1.327 | 3.0518 | 13.5429 | 13.0881 | −0.9047 | −0.9047 | 4.5652 | 3.5979 | |
| Neurons in the Hidden Layer (15 × 1) | Neurons in the Output Layer (12 × 1) | ||
|---|---|---|---|
| 1 | 1.9415 | 1 | −0.3373 |
| 2 | 5.6420 | ||
| 3 | −8.9695 | 2 | 0.1225 |
| 4 | −16.5084 | 3 | −0.3217 |
| 5 | 6.6865 | 4 | −0.7471 |
| 6 | −5.3725 | 5 | −0.3581 |
| 7 | −2.2595 | 6 | 0.0826 |
| 8 | 4.6028 | ||
| 9 | −6.1423 | 7 | 0.9655 |
| 10 | −0.7085 | 8 | 1.1458 |
| 11 | −2.6891 | 9 | −0.3927 |
| 12 | −11.2968 | 10 | 0.2316 |
| 13 | 7.0877 | 11 | 0.1225 |
| 14 | −9.1538 | 12 | −0.3217 |
| 15 | 4.3167 | ||
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Canato, V.; Bonini Neto, A.; Montagnani, J.C.R.; de Mello, J.M.; Fávaro, V.F.d.S.; Souza, A.V.d. Artificial Neural Network and Mathematical Modeling to Estimate Losses in the Concentration of Bioactive Compounds in Different Tomato Varieties During Cooking. AgriEngineering 2025, 7, 130. https://doi.org/10.3390/agriengineering7050130
Canato V, Bonini Neto A, Montagnani JCR, de Mello JM, Fávaro VFdS, Souza AVd. Artificial Neural Network and Mathematical Modeling to Estimate Losses in the Concentration of Bioactive Compounds in Different Tomato Varieties During Cooking. AgriEngineering. 2025; 7(5):130. https://doi.org/10.3390/agriengineering7050130
Chicago/Turabian StyleCanato, Vinícius, Alfredo Bonini Neto, Julio Cesar Rocha Montagnani, Jéssica Marques de Mello, Vitória Ferreira da Silva Fávaro, and Angela Vacaro de Souza. 2025. "Artificial Neural Network and Mathematical Modeling to Estimate Losses in the Concentration of Bioactive Compounds in Different Tomato Varieties During Cooking" AgriEngineering 7, no. 5: 130. https://doi.org/10.3390/agriengineering7050130
APA StyleCanato, V., Bonini Neto, A., Montagnani, J. C. R., de Mello, J. M., Fávaro, V. F. d. S., & Souza, A. V. d. (2025). Artificial Neural Network and Mathematical Modeling to Estimate Losses in the Concentration of Bioactive Compounds in Different Tomato Varieties During Cooking. AgriEngineering, 7(5), 130. https://doi.org/10.3390/agriengineering7050130











