Abstract
Agricultural production is a critical sector that directly impacts the economy and social life of any society. The identification of plant disease in a real-time environment is a significant challenge for agriculture production. For accurate plant disease detection, precise detection of plant leaves is a meaningful and challenging task for developing smart agricultural systems. Most researchers train and test models on synthetic images. So, when using that model in a real-time scenario, it does not give a satisfactory result because when a model trained on images of leaves is fed with the image of the plant, then its accuracy is affected. In this research work, we have integrated two models, the Segment Anything Model (SAM) with YOLOv8, to detect the tomato leaf of a tomato plant, mask the leaf, and extract the leaf in a real-time environment. To improve the performance of leaf disease detection in plant leaves in a real-time environment, we need to detect leaves accurately. We developed a system that will detect the leaf, mask the leaf, extract the leaf, and then detect the disease in that specific leaf. For leaf detection, the modified YOLOv8 is used, and for masking and extraction of the leaf images from the tomato plant, the Segment Anything Model (SAM) is used. Then, for that specific leaf, an image is provided to the deep neural network to detect the disease.
1. Introduction
In recent years, the integration of computer vision and machine learning techniques in agriculture has revolutionized crop management and disease detection [1]. Amongst the various applications, particularly in tomato plants, the exact segmentation and analysis of plant leaves have gained significant attention because of their importance in early disease detection, nutrient deficiency identification, and overall plant health monitoring [2]. Plant leaf segmentation is an important aspect of computer vision and agricultural technology. It supports various applications, including disease monitoring, growth monitoring, and yield prediction. However, there has been a great improvement in the deep learning domain, due to which the methods and results of leaf segmentation processes have become more efficient and effective [3].
Tomato (Solanum lycopersicum) happens to be one of the most widely cultivated as well as economically important vegetable crops across the globe. However, tomato plants are affected by different diseases and environmental conditions that lower the yield and quality of the crop. Therefore, timely notice and correct assessment of plant health conditions are essential to enable quick response actions for minimization of the loss of the crops [4]. Most traditional methods of plant monitoring are based on physical observations or inspections, which are time-consuming, labor-intensive, and are also error-prone because of human factors. New computer vision approaches can help solve some of these problems because they provide a way to evaluate the state of crops quickly, correctly, and at large scales [5].
The authors in [6] stated that constructing an object detection problem in terms of a regression task was possible because of the introduction of the YOLO architecture. YOLOv8, which was issued by Ultralytics in 2023, is a very advanced real-time object detection and image segmentation model. YOLO is a very speedy and accurate framework, but version 8 offers some improvements in architecture and the training processes of the model, which makes it very effective for speed-dependent tasks. In other works, refs. [7,8,9] the authors have conducted several studies that showed the high efficacy of YOLOv8 in the image segmentation tasks and further object detection tasks.
Key improvements of YOLOv8 include:
- Improved backbone and neck architecture for better feature extraction.
- Improved loss functions and data augmentation methods.
- Native support, for instance, segmentation tasks.
- Improved performance on small object detection.
Likewise, the authors in [10] have also proposed the Segment Anything Model (SAM), which has allowed for significant advancement in image segmentation as it has the ability to segment any object in an image based on various types of prompts, including points, boxes, and even text. SAM is envisioned as a promptable segmentation system that can create a mask for every target that can be found on an image, all based on the type of text prompt it receives. The main features of SAM include:
- SAM can segment objects accurately without prior training.
- SAM has input prompts flexibility such as accepting points, masks, text, or boxes as prompts.
- SAM is trained on 11 million images and 1.1 billion masks.
- For interactive applications, SAM can accurately generate masks in real time.
It is worth noting that SAM was not specifically developed for agricultural uses; however, its potential functionality can be proved useful in other tasks, such as leaf segmentation. The authors in [11] showed how SAM can be utilized in crop monitoring by examining the crop features in detail and making attempts to obtain such features for the leaves of the plants. Advanced models for tomato leaf segmentation have immense research potential due to reconciling the intricacies of agriculture image analysis with the advancements in computer vision technologies. Leaf segmentation is very important for the later steps of the analysis, such as in the case of disease detection, growth monitoring, and yield prediction [12]. Individual leaf extraction from complex backgrounds which include other parts of the plant, soil, and agricultural equipment, is considered a complex task. These models have the potential to provide a robust foundation for more specialized agricultural AI applications.
The identification of plant disease in a real-time environment is a major challenge for agriculture production [13]. For the accurate detection of plant disease, accurate detection of plant leaves is very important for the development of smart agriculture systems. A majority of the researchers train and test the models on synthetic images. Therefore, when using that model in a real-time scenario, it does not give a satisfactory result because the model trained with leaf images does not perform well with plant images, and its precision suffers.
For this purpose, we have used YOLOv8 and SAM, two recent deep-learning models. In this work, we have focused on tomato leaf detection, segmentation, and extraction. The objectives of this study are to understand the capabilities of both YOLOv8 and SAM regarding tomato leaf detection and segmentation. We measure their performance around accuracy, efficiency, and how well they cope with changes in their surroundings. Apart from this, we have investigated the potential of these models in this study to handle the inherent challenges in agricultural imaging, such as varying lighting conditions, natural variability in leaf shapes and sizes, and occlusions. The results of this work are very useful for creating automated tomato plant monitoring systems in the agricultural sector. We aim to expand the boundaries of precision agriculture by utilizing the best features of YOLOv8 and SAM, which will allow farmers and researchers to make data-based crop management and disease control decisions.
The proposed work comprises six sections. We outline the structure of this paper as follows: In Section 2, the literature review has been discussed in detail; Section 3 presents the materials and methods; in this section, the methodology of the proposed work has been discussed in detail; Section 4 consists of the results which demonstrate the simulation results using YOLOv8 and SAM; Section 5 consists of a discussion on the strengths and limitations of the proposed model along with the comparison of our work with the previous studies; Section 6 presents the conclusion.
2. Related Works
Tomato leaf detection and segmentation are very important for accurate tomato leaf disease detection. The integration of cutting-edge technologies such as YOLOv8 and SAM has played an important role in the improvement of these tasks. Current research suggest some effective methods for plant leaf segmentation. This study begins with a review of studies on the usage of YOLOv8 and SAM for different purposes to better comprehend the existing literature on their identification. This section focuses on the contribution of this study using other studies in this field.
In this study, the authors proposed a modified YOLOv8s-Seg network for the segmentation of tomato fruits and leaves in real-time, tackling problems emanating from weather conditions and surface characteristics. The proposed model has a mean average precision (mAP) of 92.2%, which outperforms previous models and offers technical support for tomato health monitoring and intelligent harvesting [14].
The authors integrated a new channel attention model into the advanced YOLOv8 model that makes it possible to add the Efficient Channel Attention mechanisms to improve the feature extraction of the model designed for tomato leaf disease detection. The experiments showed that the modified model outperformed the baseline by approximately 1.8% increase in mAP50, while the parameter size was reduced by 74%, which means a more accurate and less computational cost detection system [15].
This work aims to analyze and evaluate different segmentation models in detecting single and multiple diseases in tomato leaves. The accuracy of model B, which is Hybrid-DSCNN, a model combining U-Net and SegNet, was 98.24%, and it proved to be the fastest model, completing 1004 images in 30 nanoseconds [16].
The primary objective of this work is to come up with a deep neural network to detect tomato leaf diseases based on the YOLOv8 architecture. It uses multi-head self-attention to improve feature learning, which in turn improves detection accuracy and makes the model more robust to environmental changes [17].
The authors in this work used YOLOv8 and RoboFlow to detect tomato leaf disease with nine classes and achieved an average precision of 98.9% mAP. The results provided by the authors in this research have the potential for detecting and classifying tomato leaf disease at early stages [18].
The authors in this work stated the deep learning models for classifying and segmenting tomato leaf diseases, focusing on the importance of accurate segmentation in disease detection. The authors in this work highlight the efficiency of models such as U-Net in segmenting diseased areas, contributing to precision agriculture practices [19].
By incorporating SAM along with post-processed steps of segmentation, the author can achieve a zero-shot segmentation of potato leaves without using any supplementary training data. The approach is not exclusive to tomatoes but provides some insight into the segmentation tasks specific to tomato leaves [20].
In this research, the authors have used different CNN architectures such as VGG, ResNet, and DenseNet for tomato leaf disease detection with 10 classes, including healthy ones. The authors have merged the two datasets. After checking the accuracy of these models, the authors developed their own custom CNN model with 10 layers and compared the accuracy of their model with the other models. Their custom-deployed model has an accuracy of over 99% on both datasets [21].
The authors have conducted research on analyzing tomato leaf for disease detection. The authors have used a hybrid system in which they integrated the machine learning algorithm, Exponential Discriminant Analysis (EDA), with transfer learning techniques such as ResNet50, DenseNet201, EfficientNetB0, and DarkNet53. The authors have used the Taiwan and Plant–Village tomato leaf datasets. The results in this work demonstrate that the authors have achieved the mean accuracies of 98.09% and 98.29% on these datasets, respectively [22].
This research meaningfully improves YOLOv8-Seg models towards real-time instance segmentation of plant diseases using the Tomato Leaf Disease dataset as a test case. The modified models are very effective in the segmentation of the diseased regions, and sequentially, disease control can be carried out promptly [23].
The DS-DETR model presented in [9] integrates improvements to the detection transformer (DETR) to enhance its efficiency in the segmentation of tomato leaves infected with diseases. It also promotes the efficient management of diseases by quickly generating accurate disease damage assessments since it achieves an AP mask of 0.6823, which is above state-of-the-art models [24].
In this work, a YOLOV8s-based model for detecting diseases in wilted tomato leaves is presented, yielding an mAP of 92.5%. The model is expected to perform better than YOLOV5 and Faster R-CNN, showing a capability of real-time disease detection [25].
In response to the limitation of deploying detection algorithms based on deep learning on embedded devices, this paper presents an optimization of the YOLOv8n model. The algorithm has been able to perform reliable and efficient real-time detection of tomato leaf diseases and has a low model size, which can be utilized in embedded systems [26].
This study attempts to detect the tomato leaf miner pest using the YOLOv8-Seg models. The research clearly emphasizes the ability of the model to detect even those tomato leaves of low density for effective pest management measures. Towards this end, the authors go ahead to modify the YOLOv8-seg model to include the Ghost and BiFPN modules which improves the segmentation of leaves. Five test datasets were used to test the model for leaf segmentation with a score of 86.4%, which is high compared with other methods reported in the literature [27].
An approach was taken in this study where a framework based on Mask R-CNN is used for the detection and segmentation of plant leaf disease. The model had a mAP of 76.94%, which indicates its efficiency in segmenting the diseased area of different kinds of plants, including tomatoes [28].
3. Materials and Methods
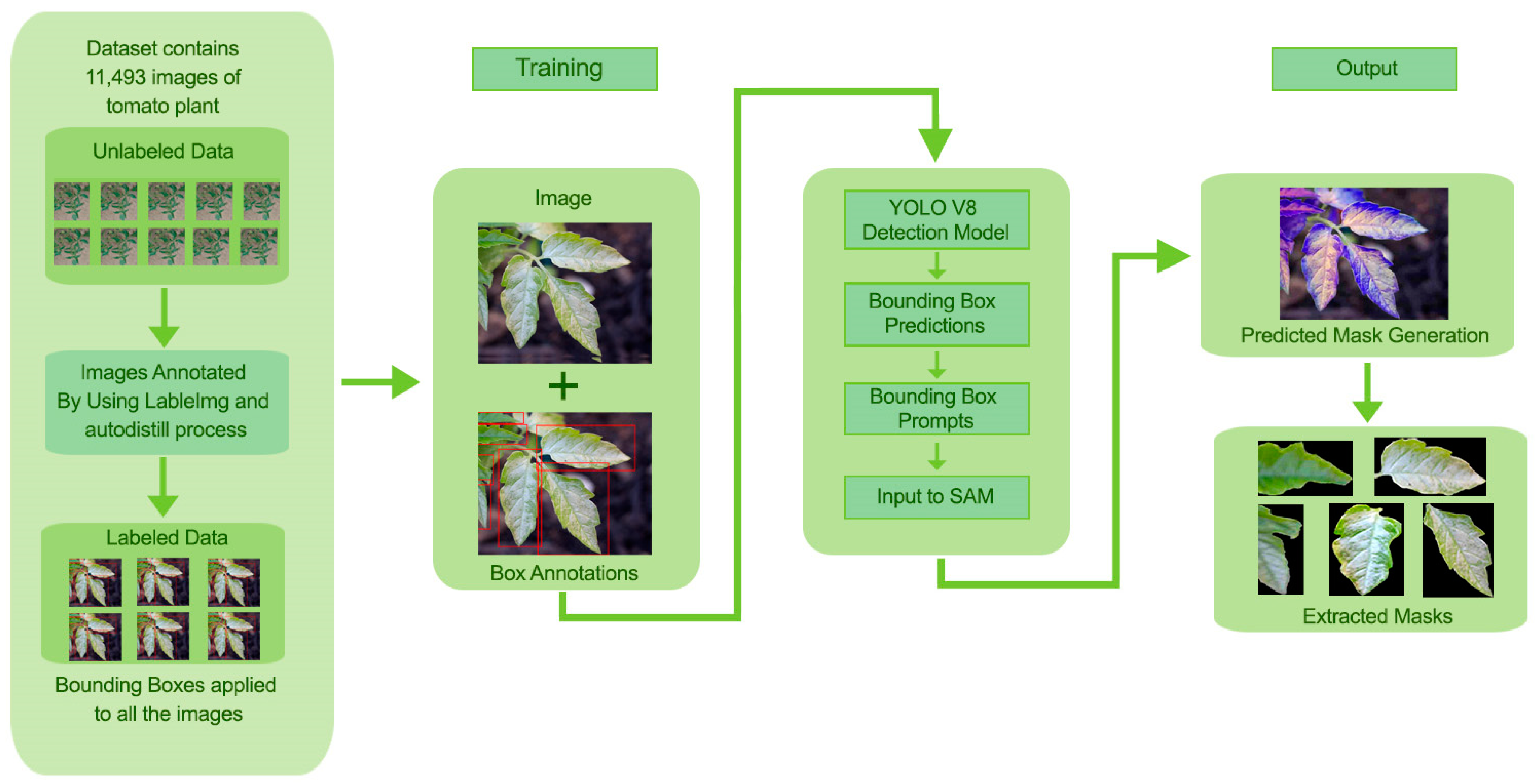
In this section, a detailed methodology for leaf detection, segmentation, and extraction is discussed by integrating YOLOv8 for object detection and SAM for leaf segmentation. The proposed approach influences the strengths of YOLOv8 for efficient tomato leaf detection and SAM for high-quality segmentation within detected bounding boxes. The detailed methodology of the proposed approach is shown in Figure 1.
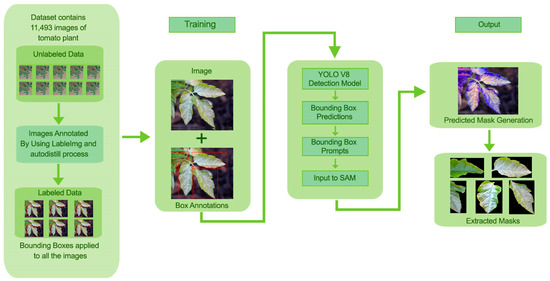
Figure 1.
Methodology for Tomato Leaf Detection, Segmentation, and Masks Extraction.
3.1. Dataset Preparation
The dataset consists of high-resolution images of tomato plants from the tomato village [29]. The images were obtained using contributions from a wide range of publicly available sources to ensure variability in light conditions, the angle of leaves, and the stage of development of the plants. The images were larger, originally comprising 640 × 640 pixels.
To achieve image consistency for model training and testing, all images were remapped to a single resolution of 640 × 640 pixels. The model used was also augmented by performing simple augmentations such as random rotation, horizontal flip, and changing brightness. After collecting the dataset was annotated using the autodistill process. The dataset comprises 11,493 images. These annotated data are divided into training (70%), validation (20%) and testing (10%) sets. This dataset is further provided to YOLOv8.
3.2. YOLOv8 for Leaf Detection
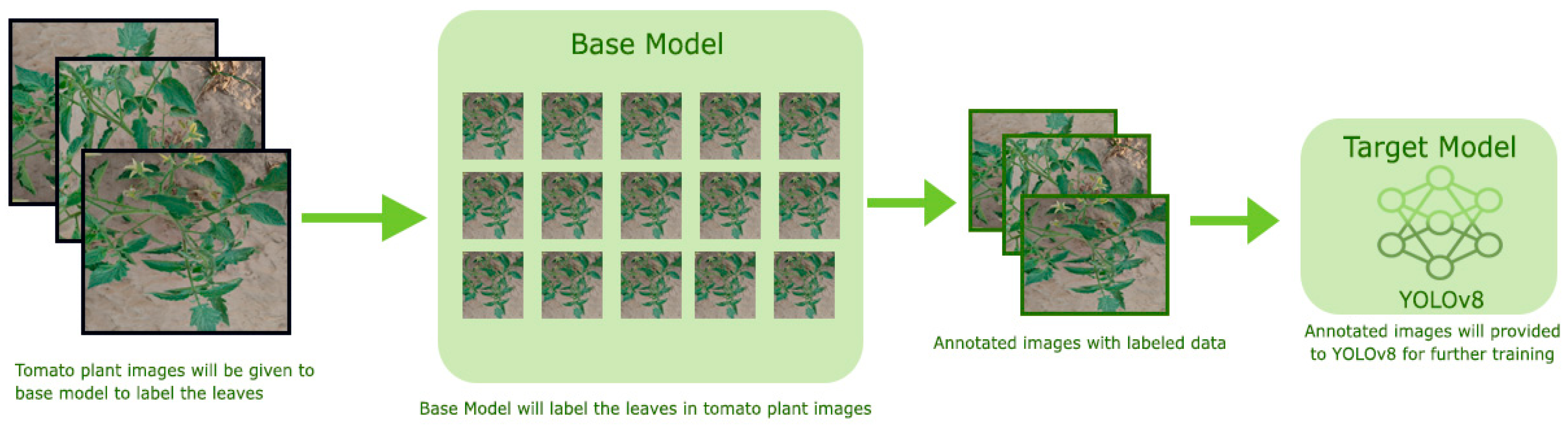
For detecting tomato leaves in tomato plant images, we employed the YOLOv8 model. YOLOv8 was preferred because of its ability to detect objects in real time with high accuracy and low computational cost. The boxes were automatically encased around the tomato leaves themselves by using YOLOv8, which was later refined through SAM. YOLOv8 was employed for detection tasks. The network was initialized using the pre-trained weights from the COCO dataset and then adjusted to the tomato leaf dataset. Unlabeled data for YOLOv8 were annotated using the autodistill. With the help of autodistill, we can train small, faster-supervised models with big slower foundation models. Using autodistill we can create labeled data to inference on a custom model running on the edge. For using the autodistill, the unlabeled data is given to the base model, which uses ontology to mark the dataset labeled that is used to train the targeted model, which outputs a distilled model fine-tuned to perform a specific task. The autodistill process is shown in Figure 2.
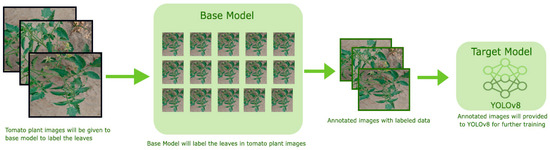
Figure 2.
Autodistill Process for auto-labeling unlabeled data.
Autodistill comprises the following steps:
Task: The Task explains what will be predicted by the target model. For each component, such as the base, ontology, and target model of the autodistill pipeline it must match and be compatible with each other.
Base Model: A base model is a large foundation and a multimodal, such as GoundedSAM, that can perform many tasks. We create the dataset from the unlabeled data along with the ontology by using the base model.
Ontology: An ontology explains the prompted labels, what your dataset will describe, and what your target model will predict. A simple Ontology is CaptionOntology, which prompts a Base Model with text captions and maps them to class names. Other Ontologies may, for instance, use a CLIP vector or example images instead of a text caption. This research work is focused on detecting the tomato leaf, so CaptionOntology was like “leaf” = “tomato leaf”.
Dataset: The dataset here includes labeled data, which is the output generated by the base model and that can be used to train the target model, which in our case is the YOLOv8 model.
Target Model: The target model is the supervised model that will consume the labeled dataset generated by the base model, and that is ready for implementation. For performing the specific task, the target model is fined-tuned, and that is usually small and fast. In our case, the target model is YOLOv8.
Distilled Model: A distilled model is the final output of the autodistill process; it is a set of weights fine-tuned for your task that can be deployed to obtain predictions. It is the final outcome of the autodistill. This is a set of fine-tuned weights for the specific task and can be implemented to obtain predictions.
After obtaining the labeled data, it is divided into training (70%), validation (20%), and testing (10%) sets. These sets provided YOLOv8 for training, which is the target model. The resolution of the images was reduced to 640 × 640 pixels for this purpose before feeding to YOLOv8.
The training parameters are as follows:
- -
- Batch Size: 16
- -
- Epochs: 100
- -
- Learning Rate: 0.001
- -
- Optimizer: The Adam optimizer is used to minimize the loss function.
Output: YOLOv8 has given the output of bounding box coordinates representing each leaf identified together with their respective confidence scores. YOLOv8 has trained well enough to recognize distinctive leaves in the tomato plant images, more so in instances where leaves may be overlapping or occluded. SAM received bounding boxes obtained from YOLOv8 as preliminary data for the generation of final segmentation masks.
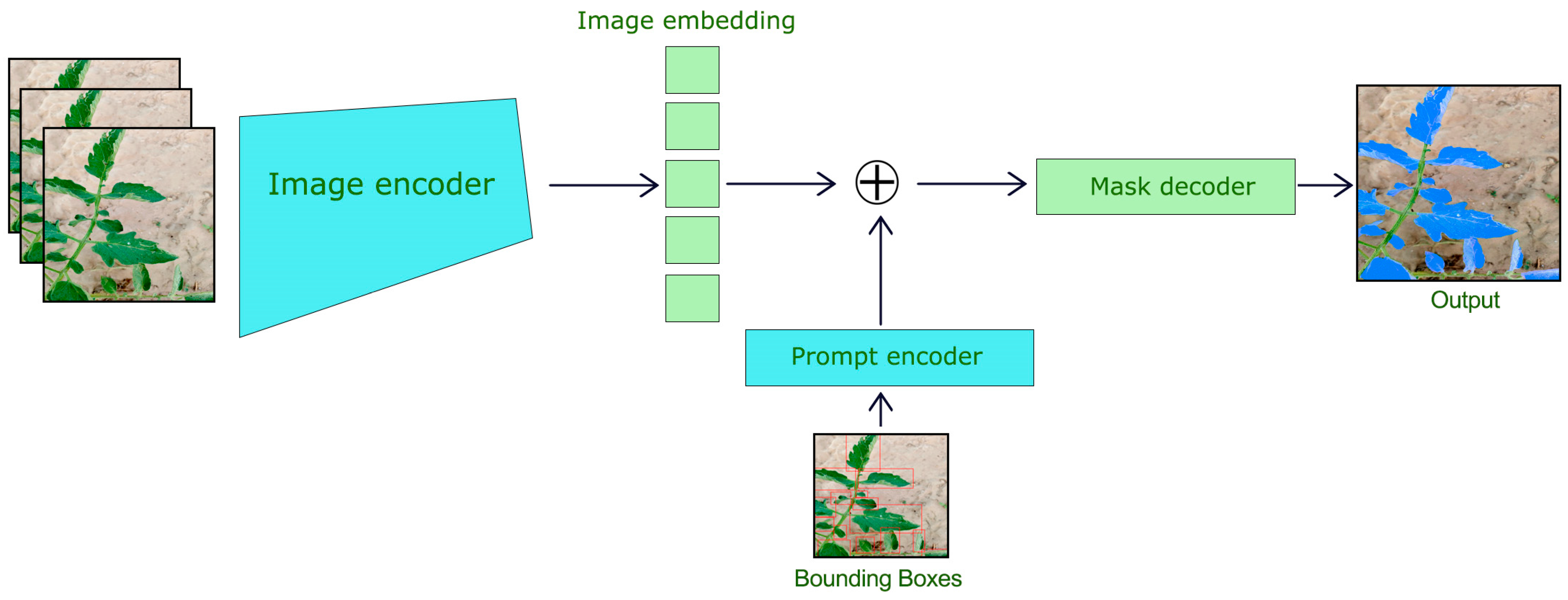
3.3. Segment Anything Model (SAM) for Leaf Segmentation
Segment Anything Model (SAM) is used to segment the tomato leaves within YOLOv8 bounding boxes. The SAM has also been designed as a prompt-based segmentation model, whereby once a bounding box or points are provided as prompts to it, fine segmentation occurs. Our focus was targeted segmentation, so we implemented SAM ViT-H (Vision Transformer with a hierarchical structure) and trained on large-scale datasets amenable to segmentation tasks. We picked up a pre-trained checkpoint (“sam_vit_h_4b8939.pth”), which was tailored to our task. For speed of inference, the model was executed on 2 GPU 12GB NVIDIA GeForce GTX 1080 Ti. For generating high-quality segmentation masks, SAM is used for each detected bounding box from YOLOv8, as shown in Figure 3.
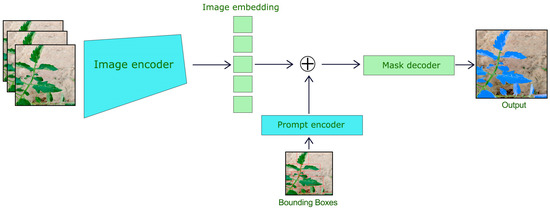
Figure 3.
Segment Anything Model for Tomato Leaf Segmentation.
The segmentation process consists of the following steps as shown:
- Input Image and Bounding Box Setup: The SAM was fed with the original RGB image along with the bounding box from YOLOv8.
- Image Preprocessing: The image was normalized and resized per the input specification of SAM.
- Segmentation Output: SAM provided binary masks portraying the segmented leaf areas inside the defined bounding boxes. SAM’s features along with YOLOv8, have the ability to work around the complex boundaries and the occlusions, each mask was an accurate representation of the leaf’s shape.
The segmented leaves, after the segmentation process, were then extracted from the image for further analysis or to be used in subsequent applications such as disease detection. For every segmented leaf, the following post-processing techniques were implemented:
- Mask Refinement: The binary masks were refined by erosion and dilation such as morphological techniques to smoothen the object boundaries.
- Leaf Extraction: The mask was used to cut out each leaf from the image, while the coordinates of the bounding box were used to define the area of the leaf.
- Resizing: The size of the cutout leaf areas was resized to 640 × 640 pixels using the bilinear interpolation method so that the samples remained consistent.
The extracted leaves were saved and transferred to a designated folder for further analysis. The extracted leaf masks were exported in a PNG format whereas a corresponding mask was recorded for use in the performance evaluation.
The performance of YOLOv8 is assessed through the computation of precision, recall, and F1 confidence score. The performance of SAM is assessed with standard segmentation metrics, which include the Intersection over Union (IoU), Dice coefficient, and qualitative visual assessments. We also report the computational efficiency of how well YOLOv8 and SAM perform together to segment the tomato leaves. For the tomato leaf detection, the performance evaluation metrics are listed below (1)–(4).
where:
- FP: False Positives (incorrectly predicted positive cases)
- TP: True Positives (correctly predicted positive cases)
- FN: False Negatives (incorrectly predicted negative cases)
4. Results
This section presents the results of the experiments conducted for leaf detection using YOLOv8 and the SAM for segmentation in the images of tomato plants, focusing on the segmentation of the tomato leaves.
The tomato leaves were detected in the tomato plant images using the YOLOv8 model. The bounding boxes that Yolov8 had created were provided as input to the SAM for precise segmentation. The performance of YOLOv8 + SAM for detection and segmentation of the tomato leaves was assessed based on the following parameters:
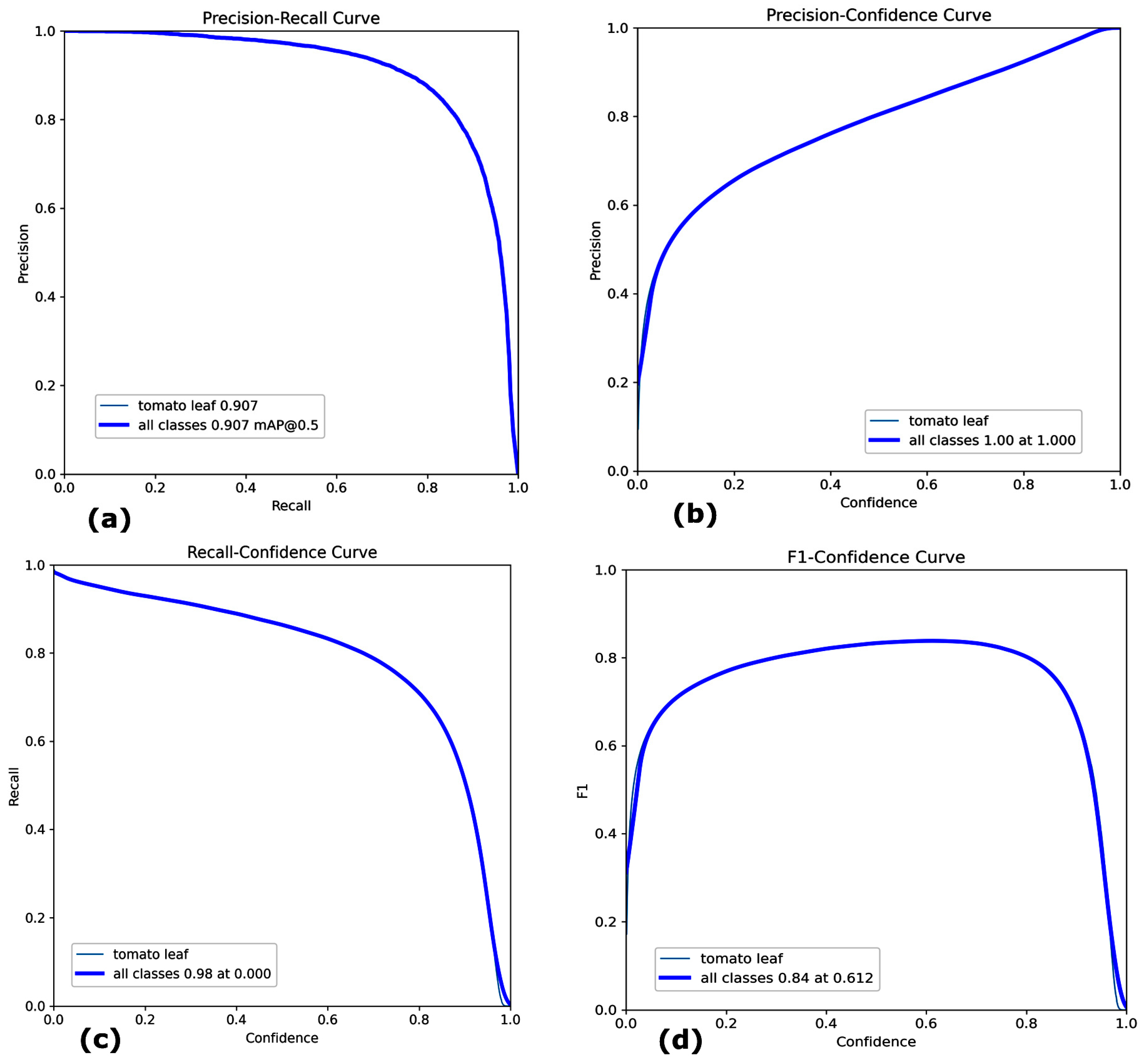
- Mean Average Precision (mAP): The model achieved a mAP of 90.7% with a confidence threshold of 0.5. This is shown in Figure 4a, which indicates the high accuracy of detection of tomato leaves on different tomato plant images.
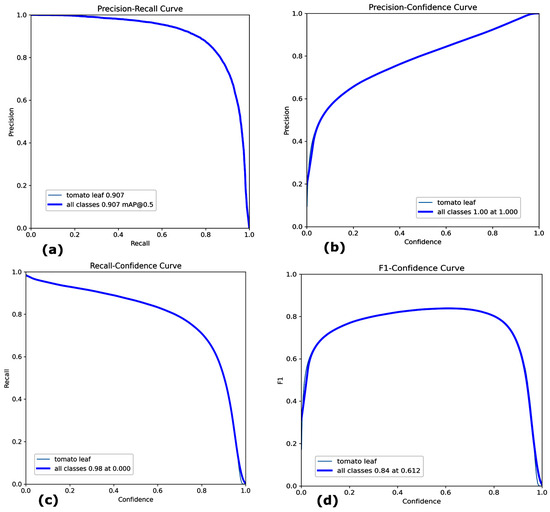 Figure 4. Performance Metrics of YOLOv8 (a) Precision–Recall Curve (b) Precision–Confidence Curve (c) Recall–Confidence Curve (d) F1–Confidence Curve.
Figure 4. Performance Metrics of YOLOv8 (a) Precision–Recall Curve (b) Precision–Confidence Curve (c) Recall–Confidence Curve (d) F1–Confidence Curve. - Precision, Recall, and F1 Score: To assess the quality of the segmentation masks, precision, recall, and the F1 score were determined for every image at the pixel level. The Precision curve measures the proportion of correctly predicted positive pixels. The Recall curve measures the proportion of actual positive pixels that were correctly predicted. The F1 Score is the harmonic mean of precision and recall. Precision was 100% shown in Figure 4b, while recall was 98%, as shown in Figure 4c, reflecting the model’s capability to detect the tomato leaves and minimize false positives correctly.
The Dice Coefficient (or F1 score for segmentation) was used to evaluate the similarity between the predicted masks and ground truth. Figure 4d shows the F1 confidence curve of 84% at 0.612.
To evaluate the performance of the YOLOv8 + SAM framework, we divided the dataset into a training set (70%), validation set (20%), and test set (10%), as shown in Table 1.

Table 1.
Training, validation, and testing results of our model for tomato leaf class on the tomato village dataset.
All segmentation results were compared against the ground truth masks using the evaluation metrics mentioned above.
After detecting the tomato leaves using YOLOv8, the SAM model was used to segment the leaves based on the bounding boxes. SAM’s ability to provide fine-grained segmentation of leaf boundaries was critical in improving the precision of the leaf regions, particularly for complex shapes and overlapping leaves. IoU measures the overlap between the predicted segmentation mask and the ground truth mask. We reported IoU scores for individual leaves and calculated the average IoU over the entire test set.
The average Dice coefficient was 88%, which is a strong indicator of the overlap between the predicted and ground truth masks. This suggests that SAM accurately segmented the leaf regions, particularly around the edges, where precision is essential for identifying healthy and diseased leaves. The overall pixel accuracy of the segmentation process was 80.78%, showing that the model correctly identified the majority of leaf pixels while minimizing false positives for the background or non-leaf regions.
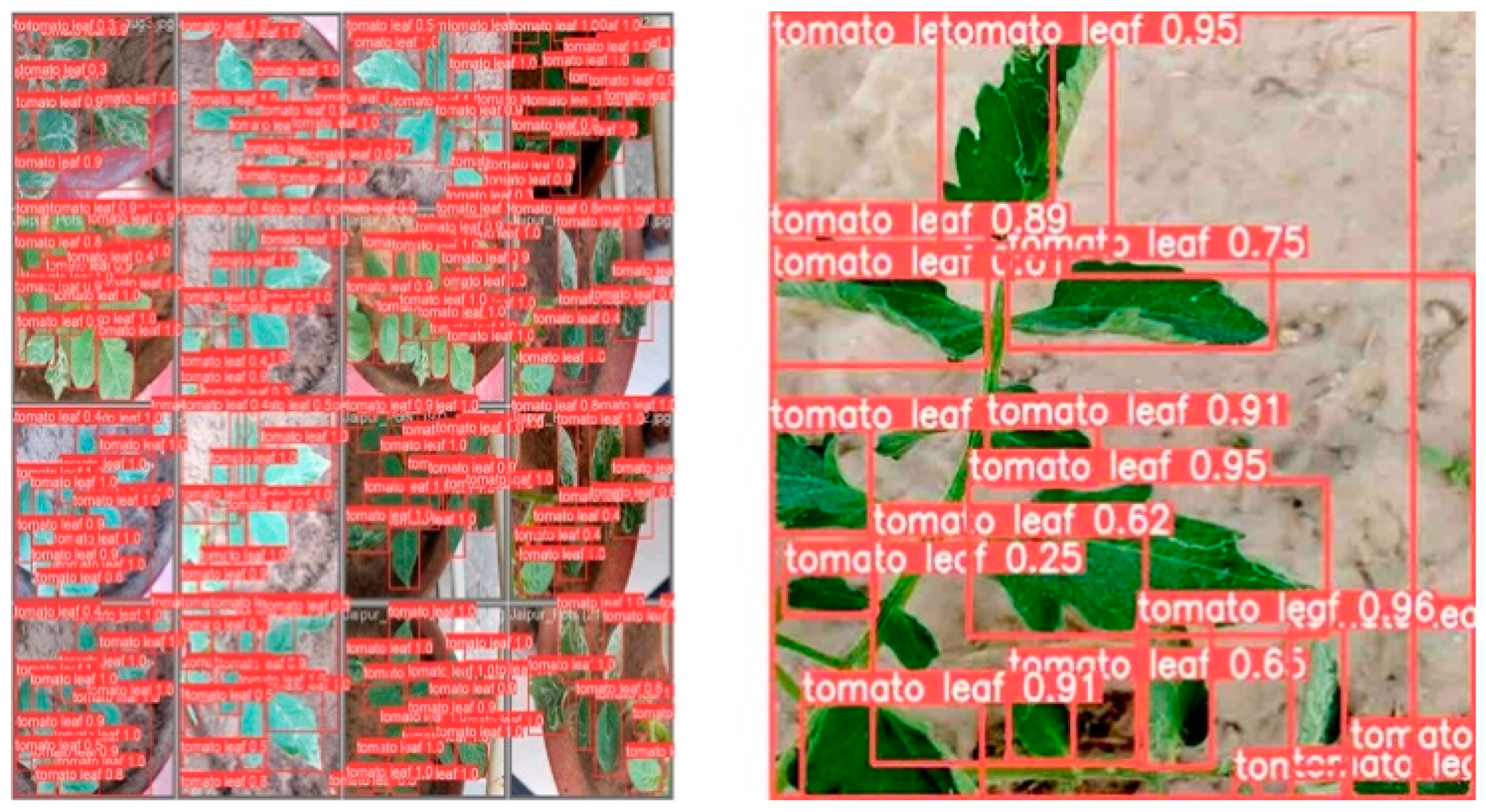
Alongside the quantitative metrics, the segmentation quality was assessed through visual analysis. Figure 5 and Figure 6 shows examples of the original image, YOLOv8-detected bounding boxes, and the final segmented masks from SAM. In these images, the bounding boxes produced by YOLOv8 successfully identified the main leaf structures.

Figure 5.
Leaf Detection within Bounding Boxes.

Figure 6.
Leaf Masking Using SAM.
SAM further refined the segmentation, providing precise boundaries, even in the presence of overlapping leaves. The segmented masks clearly delineated the tomato leaves from the background, with SAM accurately capturing the leaf edges. The visual inspection confirms that SAM performs well on complex images, handling variations in lighting, occlusions, and leaf orientation.
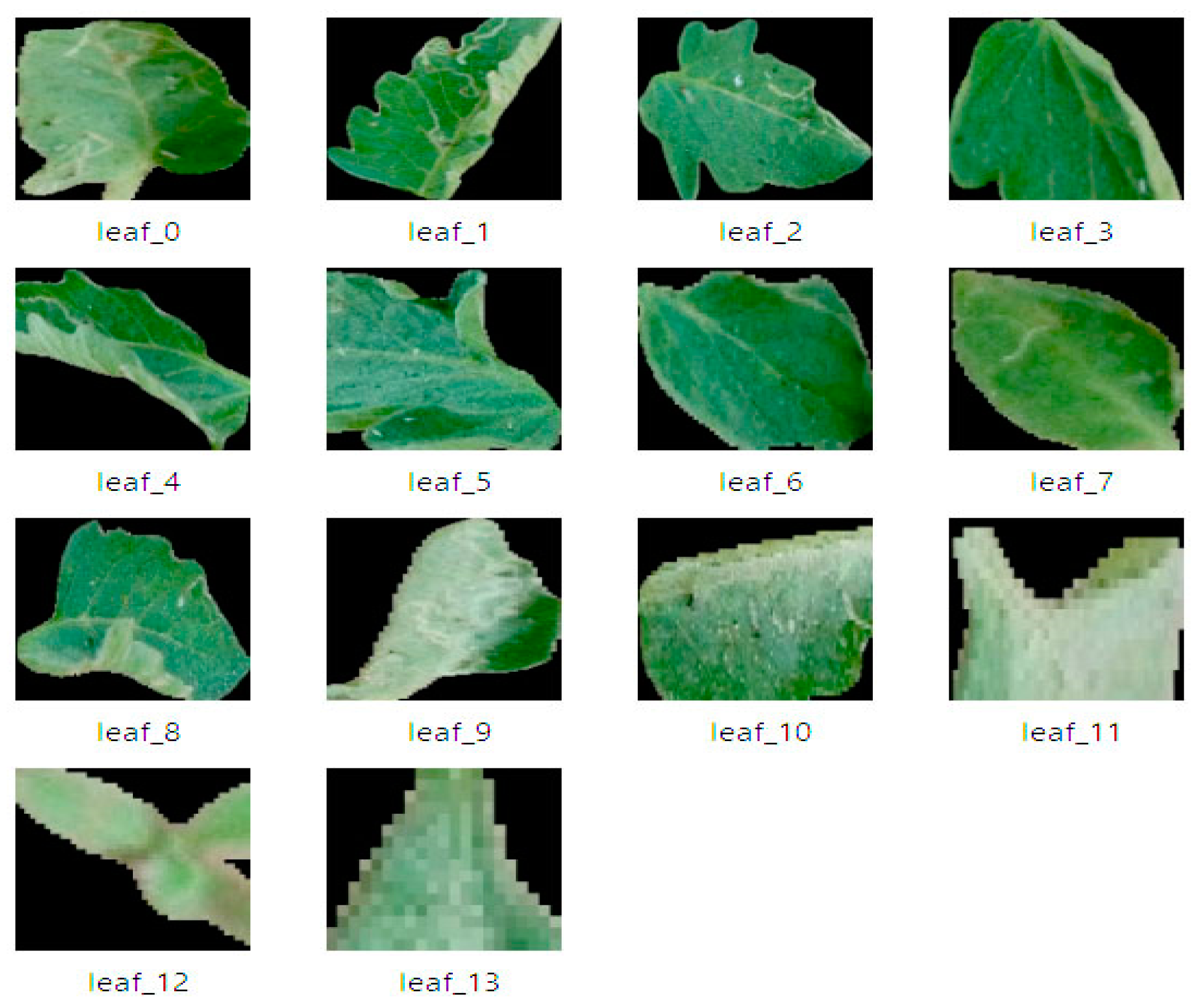
The masked leaves are then extracted and stored and a separate folder for further analysis, such as disease detection, as shown in Figure 7. By integrating SAM with YOLOv8, one of the key improvements reflected was the enhancement of leaf edges. YOLOv8′s bounding boxes and masks provided a rough approximation of the leaf area, but SAM was able to capture the finer details of the leaf shapes, including serrated edges and irregularities.

Figure 7.
Leaf Masks Extraction to a separate folder.
For plant disease detection tasks this is very crucial to precisely segment and identify the small lesions or abnormalities on the leaves. Instead of using YOLOv8 and SAM models alone, the integration of these two exhibited significant enhancements in the tomato leaf segmentation. A notable increase can be seen in the IoU and Dice coefficient by using SAM to refine the YOLOv8-detected regions. Table 1 shows the training, validation, and testing performance of our integrated model in terms of precision, recall score, and mAP@50. During validation, our model has a precision score of 99.7%, a recall score of 99.5%, and an mAP@50 of 89.4%. In comparison, the inference time during validation is 144 milliseconds. The testing accuracy of our model shows a precision score of 100% and a recall score of 98%, whereas the inference time during testing was 142 milliseconds. Further analysis confirmed the effectiveness of SAM in handling complex leaf structures and occlusions.
This work shows the effectiveness of YOLOv8 and SAM for accurate tomato leaf detection and extraction. We achieved a precision of 100%, a recall of 98%, and an F1 score of 84%. These findings have several practical implications for agriculture. First, our system can enable the accurate identification and extraction of individual tomato leaves. This system can be integrated with a tomato disease detection model for further disease analysis. This can lead to minimizing crop losses by early disease detection in tomato plants. Second, our findings can contribute to precision agriculture practices by enabling targeted interventions based on individual tomato leaf characteristics. Third, our system can reduce farmer costs by automating leaf counting and measurement. Moreover, it has the potential for real-time applications; it can be integrated into a drone-based monitoring system. We believe that our work can contribute to the development of more efficient and sustainable agricultural practices.
5. Discussion
In the given research, the tomato leaves were segmented using the images of tomato plants with the help of the YOLOv8 and the Segment Anything Model (SAM) model. The research aimed to evaluate the accuracy of the segmentation of tomato leaves by both models as it is important for many agricultural techniques which include plant monitoring, disease detection, and farming automation. This discussion also examines both models, their integration as well as the strengths and limitations of models to further precision agriculture objectives.
During experiments, YOLOv8 demonstrated a very good ability to detect tomato leaves even in cases such as many disorganized overlapping leaves where multiple leaves were dispersed and or alongside each other. The bounding boxes generated by YOLOv8 provided an adequate preliminary stage for various segmentation tasks as they included all the individual leaves. Additionally, the high precision and recall of the model, measured by its mAP capacity of the model to perform numerous overlapping tasks, enabled the model to detect objects within complicated environments accurately. However, there are still some performance limitations of YOLOv8 in terms of pixel-level segmentation accuracy which is designed only for target bounding box detection. In more complex cases, such as heavily shadowed leaves or leaves that were in the background, the bounding box was typically considered too broad. The limitations of YOLOv8 necessitated the use of a more refined segmentation approach, such as SAM, for achieving finer pixel-level accuracy. The regions identified by YOLOv8 were provided as input to SAM for producing detailed segmentation masks for each detected leaf. This method allowed for a more precise delineation of leaf boundaries, especially in cases where YOLOv8′s bounding boxes were too coarse. SAM was particularly effective in handling the complex shapes and contours of tomato leaves, which is most important for accurate phenotypic analysis in precision agriculture.
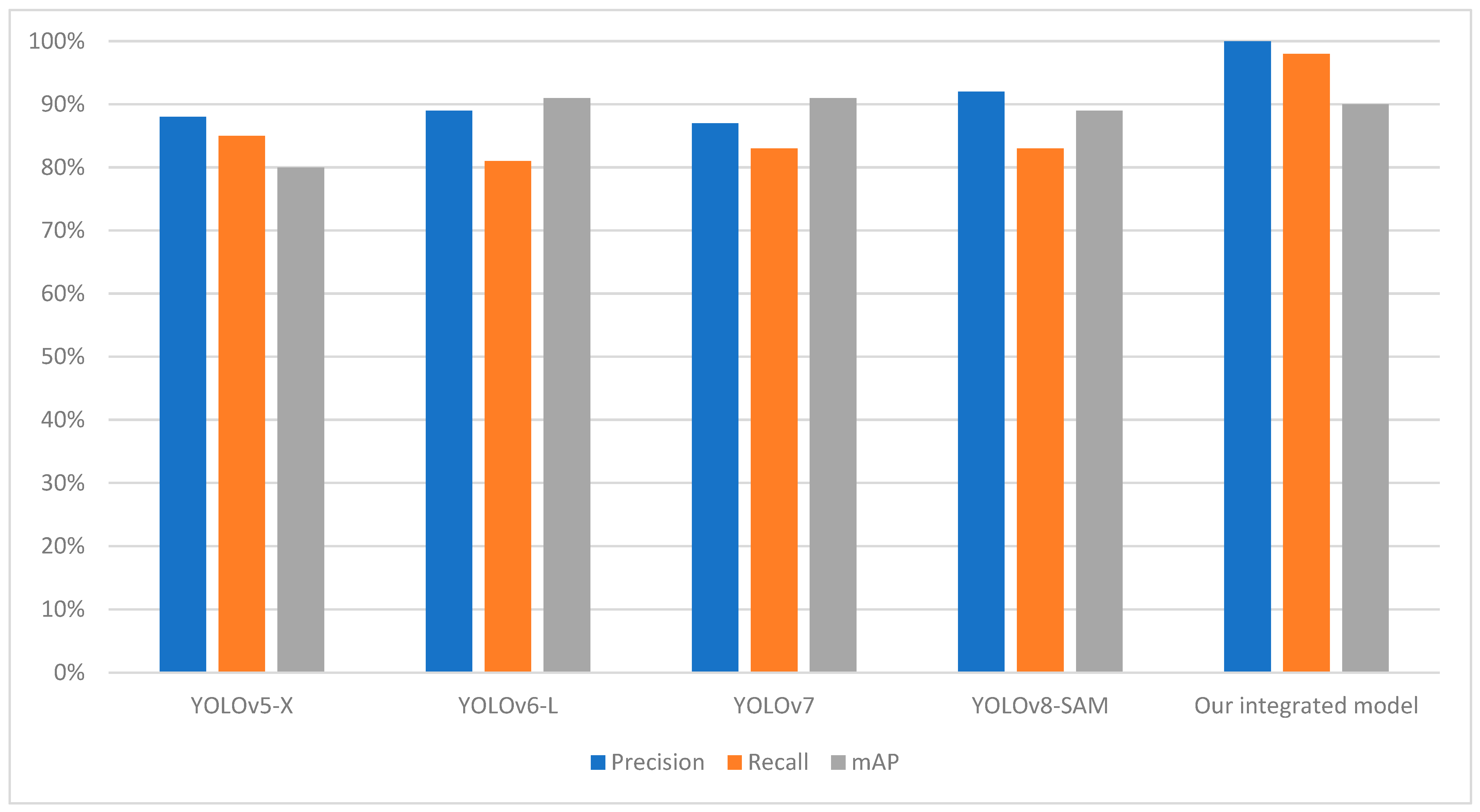
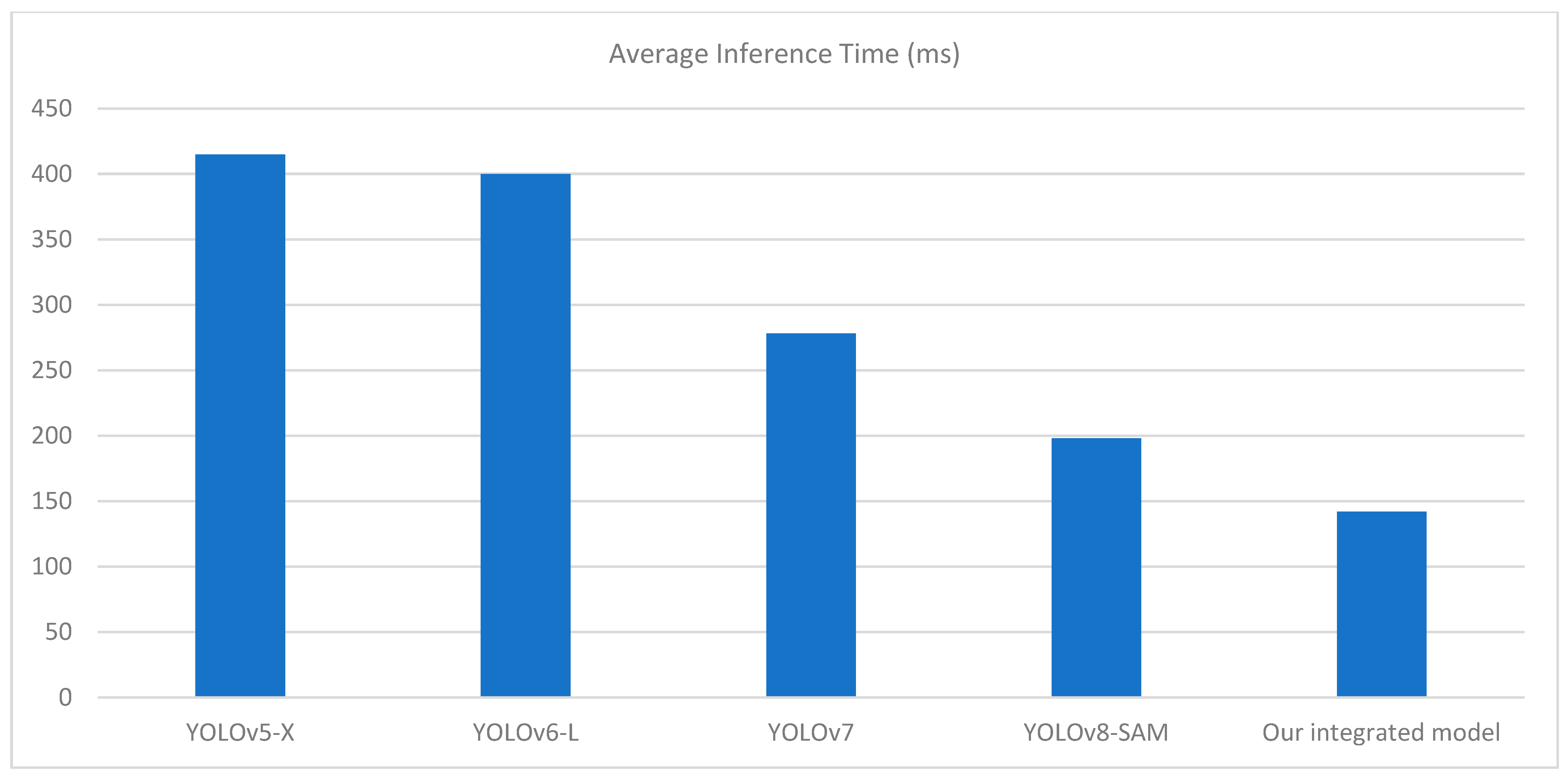
Table 2 shows that our integrated model outperforms other segmentation models in terms of mAP, Precision, and Recall score. The most significant advancement is observed in the mAP, where our model achieves 0.90%, outperforming YOLOv8-SAM, the previous best performer. The precision score of our integrated model is estimated at 100%, meaning that when it identifies a tomato leaf, the chances of actually detecting it are exceptionally high. The average inference time of our integrated model over 100 runs is quite better than previous models which is 142 milliseconds with a GPU usage of 71.19%. This helps to mitigate false positives, which, in most cases, highly depend on the precision of the model’s detection, especially in real-time applications. A recall rate of 98% reveals that the model has a high probability of detecting most of the tomato leaves in the provided image.

Table 2.
Comparison of our integrated model with different detection algorithm models.
This demonstrates the performance of the model under different conditions, such as the presence of small leaves and light conditions, which is a common feature in real-world problems. Our proposed approach, integrating YOLOv8 and SAM, has shown convincing performance in terms of detecting, masking, and extracting tomato leaves. It is important to note that other models, such as YOLOv5-X [30], YOLOv6-L [31], YOLOv7 [32], and YOLOv8-SAM [33], has its merits, but the proposed combination of YOLOv8 and SAM has shown a comparable level of effectiveness, as indicated in Table 2. The visualization of comparatives analysis with the previous model with respect to performance metrics and average inference time is shown in Figure 8 and Figure 9.
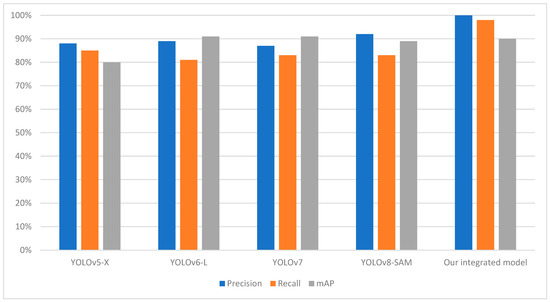
Figure 8.
Comparison chart with previous models based on precision, recall, and accuracy.
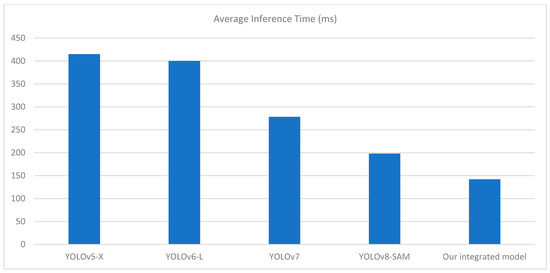
Figure 9.
Comparison chart with previous models based on average inference time (ms).
Regardless of the overall success of the integrated YOLOv8 and SAM approach, we faced several limitations during this work. A major problem was the computational cost of performing both models in sequence. One can appreciate YOLOv8 for its lower threshold time, but the segmentation of SAM is more taxing being its use in high-resolution images or large batches of data. There is a need to find a good balance between segmentation precision, accuracy, and efficiency in real-time systems, especially in field-based robotic systems or hand-held agricultural devices.
Another constraint noted was in instances where tiny or semi-occluded leaves were not detected by YOLOv8. Since SAM depended on input provided by the bounding boxes of the objects detected by YOLOv8, thus in instances when YOLOv8 missed some targets, leaves that were supposed to be segmented were not masked by the SAM. This emphasizes the need to work on enhancing object detection in special cases where the plant is in a complex habitat.
Additionally, there were also times when it was hard for SAM to tell apart leaves that were on top of each other or were in close proximity. In this type of situation, segmentation masks tended together the adjacent leaves making false segmentation possible.
From this research work, several possibilities for future research emerge. Future improvements could involve the use of multimodal inputs or additional preprocessing steps to enhance the separation of individual leaves. Apart from this an integration of hyperspectral and multispectral imaging can improve the accuracy of both detection and segmentation of leaves, especially in cases where visual cues alone are inadequate to distinguish between plant parts. Apart from this, for real-time applications, a more efficient pipeline approach can be a significant direction. The use of edge devices or the quantization of these models can significantly reduce the computational cost associated with both these models (YOLOv8 + SAM) in sequence.
Overall, this system has the potential for real-time applications; it can be integrated into a drone-based monitoring system. We believe that this work can contribute to the development of more efficient and sustainable precision agricultural practices.
6. Conclusions
In this research work YOLOv8 and SAM have been analyzed for the segmenting of tomato leaves in tomato plant images. The main aim of this study is to evaluate the efficiency and performance of YOLOv8 and SAM in detecting and segmenting tomato leaves, which is the most important step in detecting plant diseases in precision agriculture. The integration of using YOLOv8 for object detection and SAM for fine-grained segmentation proved notable strengths and some areas where improvements can be considered. The integration of YOLOv8 and SAM poses a highly effective approach for tomato leaf segmentation, taking advantage of YOLOv8′s rapid object detection and SAM’s high-precision segmentation competencies. Despite challenges, specifically regarding computational cost, efficiency, and small object detection, the results of this research work present a strong foundation for future research in agricultural imaging and automated plant monitoring systems. The potential applications of this work in disease detection, phenotyping, and precision farming highlight its relevance to the future of agricultural technology.
Author Contributions
S.U.I. conceived the presented idea. S.U.I. designed and developed the model and the computational framework and analyzed data. G.F. encouraged and supervised the findings of this work. G.F. and V.P. review this work. All authors have read and agreed to the published version of the manuscript.
Funding
This research received no external funding.
Data Availability Statement
The dataset used in this research work is publicly available and has been obtained from the following link: https://github.com/mamta-joshi-gehlot/Tomato-Village, accessed on 31 March 2025.
Conflicts of Interest
The authors declare that they have no conflicts of interest to report regarding the present study.
References
- Kalinaki, K.; Shafik, W.; Gutu, T.J.; Malik, O.A. Computer vision and machine learning for smart farming and agriculture practices. In Artificial Intelligence Tools and Technologies for Smart Farming and Agriculture Practices; IGI Global: Hershey, PA, USA, 2023; Volume 10, pp. 79–100. [Google Scholar]
- Wang, Y.; Liu, Q.; Yang, J.; Ren, G.; Wang, W.; Zhang, W.; Li, F. A Method for Tomato Plant Stem and Leaf Segmentation and Phenotypic Extraction Based on Skeleton Extraction and Supervoxel Clustering. Agronomy 2024, 14, 198. [Google Scholar] [CrossRef]
- Yang, T.; Zhou, S.; Xu, A.; Ye, J.; Yin, J. An Approach for Plant Leaf Image Segmentation Based on YOLOV8 and the Improved DEEPLABV3+. Plants 2023, 12, 3438. [Google Scholar] [CrossRef] [PubMed]
- Alzahrani, M.S.; Alsaade, F.W. Transform and Deep Learning Algorithms for the Early Detection and Recognition of Tomato Leaf Disease. Agronomy 2023, 13, 1184. [Google Scholar] [CrossRef]
- Kouadio, L.; El Jarroudi, M.; Belabess, Z.; Laasli, S.-E.; Roni, M.Z.K.; Amine, I.D.I.; Mokhtari, N.; Mokrini, F.; Junk, J.; Lahlali, R. A Review on UAV-Based Applications for Plant Disease Detection and Monitoring. Remote Sens. 2023, 15, 4273. [Google Scholar] [CrossRef]
- Han, X.; Chang, J.; Wang, K.J. You only look once: Unified, real-time object detection. Procedia Comput. Sci. 2021, 1, 61–72. [Google Scholar] [CrossRef]
- Vilar, A.M.; García, L.; Garcia, A.J.; Asorey, C.R.; Garcia, H.J. Enhancing Precision Agriculture Pest Control: A generalized Deep Learning Approach with YOLOv8-based Insect Detection. IEEE Access 2024, 12, 84420. [Google Scholar] [CrossRef]
- Singh, P.; Krishnamurthi, R. IoT-based real-time object detection system for crop protection and agriculture field security. J. Real-Time Image Process. 2024, 21, 106. [Google Scholar] [CrossRef]
- Pativada, P.K. Real-Time Detection and Classification of Plant Seeds Using YOLOv8 Object Detection Model. Ph.D. Thesis, Kansas State University, Manhattan, KS, USA, 2024. [Google Scholar]
- Kirillov, A.; Mintun, E.; Ravi, N.; Mao, H.; Rolland, C.; Gustafson, L.; Xiao, T.; Whitehead, S.; Berg, A.C.; Lo, W.Y.; et al. Segment anything. In Proceedings of the IEEE/CVF International Conference on Computer Vision, Paris, France, 4–6 October 2023; pp. 4015–4026. [Google Scholar]
- Song, B.; Yang, H.; Wu, Y.; Zhang, P.; Wang, B.; Han, G. A multispectral remote sensing crop segmentation method based on Segment Anything Model using Multi-stage Adaptation Fine-tuning. IEEE Trans. Geosci. Remote Sens. 2024, 62, 4408818. [Google Scholar] [CrossRef]
- Itakura, K.; Hosoi, F. Automatic Leaf Segmentation for Estimating Leaf Area and Leaf Inclination Angle in 3D Plant Images. Sensors 2018, 18, 3576. [Google Scholar] [CrossRef]
- Buja, I.; Sabella, E.; Monteduro, A.G.; Chiriacò, M.S.; De Bellis, L.; Luvisi, A.; Maruccio, G. Advances in Plant Disease Detection and Monitoring: From Traditional Assays to In-Field Diagnostics. Sensors 2021, 21, 2129. [Google Scholar] [CrossRef]
- Yue, X.; Qi, K.; Na, X.; Zhang, Y.; Liu, Y.; Liu, C. Improved YOLOv8-Seg Network for Instance Segmentation of Healthy and Diseased Tomato Plants in the Growth Stage. Agriculture 2023, 13, 1643. [Google Scholar] [CrossRef]
- Zhang, R.; Ge, Y.; Li, Q.; Meng, L. An Improved YOLOv8 Tomato Leaf Disease Detector Based on the Efficient-Net backbone. In Proceedings of the 6th International Symposium on Advanced Technologies and Applications in the Internet of Things, Shiga, Japan, 19–22 August 2024; p. 3748. [Google Scholar]
- Kaur, P.; Harnal, S.; Gautam, V.; Singh, M.P.; Singh, S.P. Performance analysis of segmentation models to detect leaf diseases in tomato plant. Multimed. Tools Appl. 2024, 83, 16019–16043. [Google Scholar] [CrossRef]
- Liu, X.; Lei, H.; Zhou, Y.; Feng, J.M.; Niu, G.; Zhou, Y. Tomato leaf disease detection based on improved YOLOv8. In Proceedings of the IEEE 6th International Conference on Internet of Things Automation and Artificial Intelligence (IoTAAI), Guangzhou, China, 26–28 July 2024; pp. 145–150. [Google Scholar]
- Brucal, S.G.; Jesus, L.C.; Peruda, S.R.; Samaniego, L.A.; Yong, E.D. Development of tomato leaf disease detection using YoloV8 model via RoboFlow 2.0. In Proceedings of the IEEE 12th Global Conference on Consumer Electronics (GCCE), Nara, Japan, 10–13 October 2023; pp. 692–694. [Google Scholar]
- Shoaib, M.; Hussain, T.; Shah, B.; Ullah, I.; Shah, S.M.; Ali, F.; Park, S.H. Deep learning-based segmentation and classification of leaf images for detection of tomato plant disease. Front. Plant Sci. 2022, 13, 1031748. [Google Scholar] [CrossRef]
- Williams, D.; Macfarlane, F.; Britten, A. Leaf only SAM: A segment anything pipeline for zero-shot automated leaf segmentation. Smart Agric. Technol. 2024, 8, 100515. [Google Scholar] [CrossRef]
- Islam, S.U.; Zaib, S.; Ferraioli, G.; Pascazio, V.; Schirinzi, G.; Husnain, G. Enhanced Deep Learning Architecture for Rapid and Accurate Tomato Plant Disease Diagnosis. AgriEngineering 2024, 6, 375–395. [Google Scholar] [CrossRef]
- Chouchane, A.; Ouamanea, A.; Himeur, Y.; Amira, A. Deep learning-based leaf image analysis for tomato plant disease detection and classification. In Proceedings of the IEEE International Conference on Image Processing (ICIP), Abu Dhabi, United Arab Emirates, 27–30 October 2024; pp. 2923–2929. [Google Scholar]
- Wu, J.; Wen, C.; Chen, H.; Ma, Z.; Zhang, T.; Su, H.; Yang, C. DS-DETR: A Model for Tomato Leaf Disease Segmentation and Damage Evaluation. Agronomy 2022, 12, 2023. [Google Scholar] [CrossRef]
- Abdullah, A.; Amran, G.A.; Tahmid, S.M.A.; Alabrah, A.; AL-Bakhrani, A.A.; Ali, A. A Deep-Learning-Based Model for the Detection of Diseased Tomato Leaves. Agronomy 2024, 14, 1593. [Google Scholar] [CrossRef]
- Liu, W.; Bai, C.; Tang, W.; Xia, Y.; Kang, J. A Lightweight Real-Time Recognition Algorithm for Tomato Leaf Disease Based on Improved YOLOv8. Agronomy 2024, 14, 2069. [Google Scholar] [CrossRef]
- Uygun, T.; Ozguven, M.M. Determination of tomato leafminer: Tuta absoluta (Meyrick) (Lepidoptera: Gelechiidae) damage on tomato using deep learning instance segmentation method. Eur. Food Res. Technol. 2024, 250, 1837–1852. [Google Scholar] [CrossRef]
- Wang, P.; Deng, H.; Guo, J.; Ji, S.; Meng, D.; Bao, J.; Zuo, P. Leaf Segmentation Using Modified YOLOv8-Seg Models. Life 2024, 14, 780. [Google Scholar] [CrossRef]
- Bondre, S.; Patil, D. Crop disease identification segmentation algorithm based on Mask-RCNN. Agron. J. 2024, 116, 1088–1098. [Google Scholar] [CrossRef]
- Gehlot, M.; Saxena, R.K.; Gandhi, G.C. “Tomato-Village”: A dataset for end-to-end tomato disease detection in a real-world environment. Multimed. Syst. 2023, 29, 3305–3328. [Google Scholar] [CrossRef]
- Peserico, G.; Morato, A. Performance Evaluation of YOLOv5 and YOLOv8 Object Detection Algorithms on Resource-Constrained Embedded Hardware Platforms for Real-Time Applications. In Proceedings of the IEEE 29th International Conference on Emerging Technologies and Factory Automation (ETFA), Padova, Italy, 10–13 September 2024; pp. 1–7. [Google Scholar]
- Li, C.; Li, L.; Jiang, H.; Weng, K.; Geng, Y.; Li, L.; Ke, Z.; Li, Q.; Cheng, M.; Nie, W.; et al. YOLOv6: A single-stage object detection framework for industrial applications. Comput. Vis. Pattern Recognit. 2022, 7, 2209. [Google Scholar]
- Wang, C.Y.; Bochkovskiy, A.; Liao, H.Y. YOLOv7: Trainable bag-of-freebies sets new state-of-the-art for real-time object detectors. In Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition, Vancouver, BC, Canada, 17–24 June 2023; pp. 7464–7475. [Google Scholar]
- Khatua, A.; Bhattacharya, A.; Goswami, A.K.; Aithal, B.H. Developing approaches in building classification and extraction with synergy of YOLOV8 and SAM models. Spat. Inf. Res. 2024, 32, 511–530. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).