Deep Learning-Based System for Early Symptoms Recognition of Grapevine Red Blotch and Leafroll Diseases and Its Implementation on Edge Computing Devices
Abstract
1. Introduction
2. Related Work
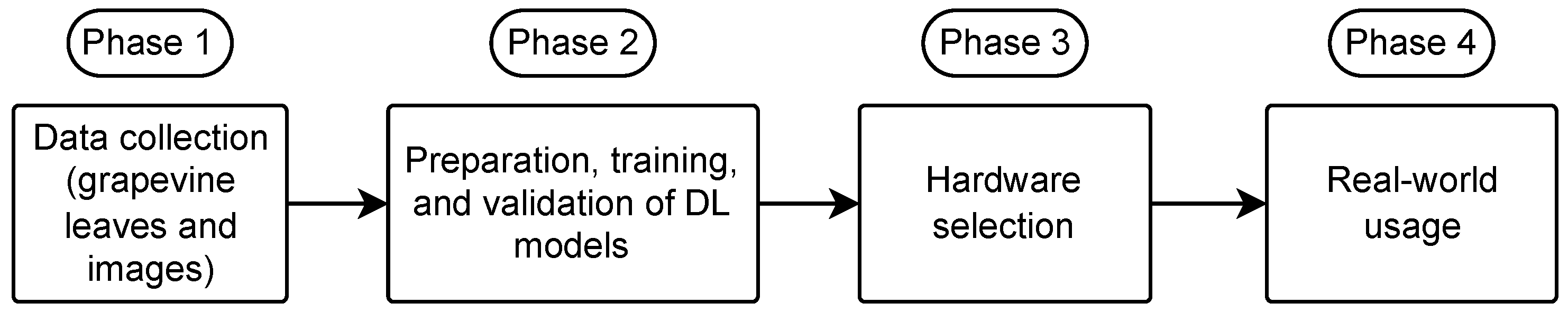
3. Materials and Methods
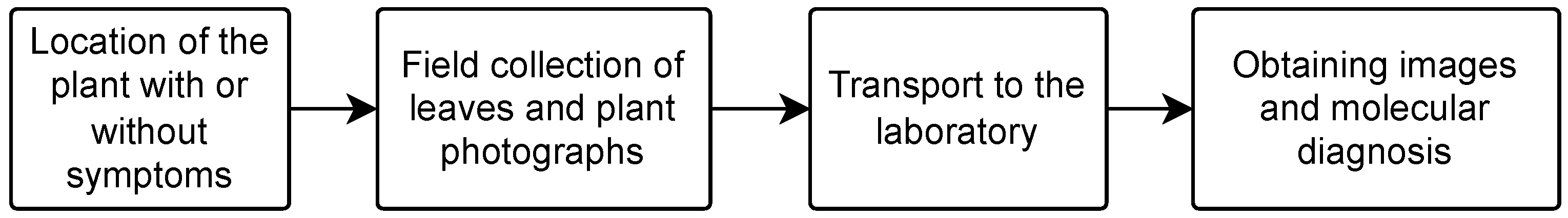
3.1. Data Collection
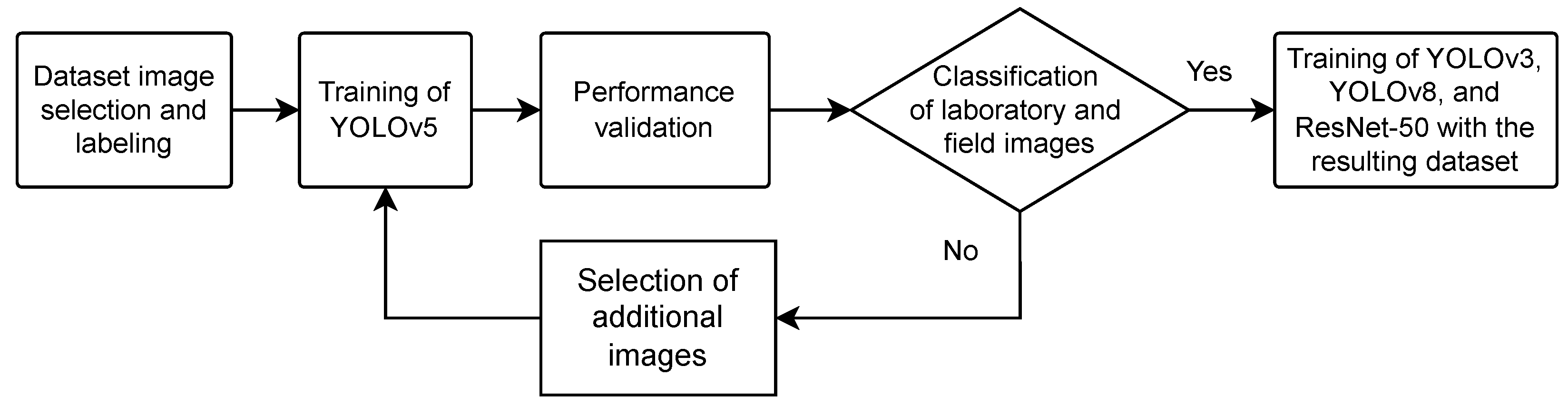
3.2. DL Models Training
3.3. Hardware Selection
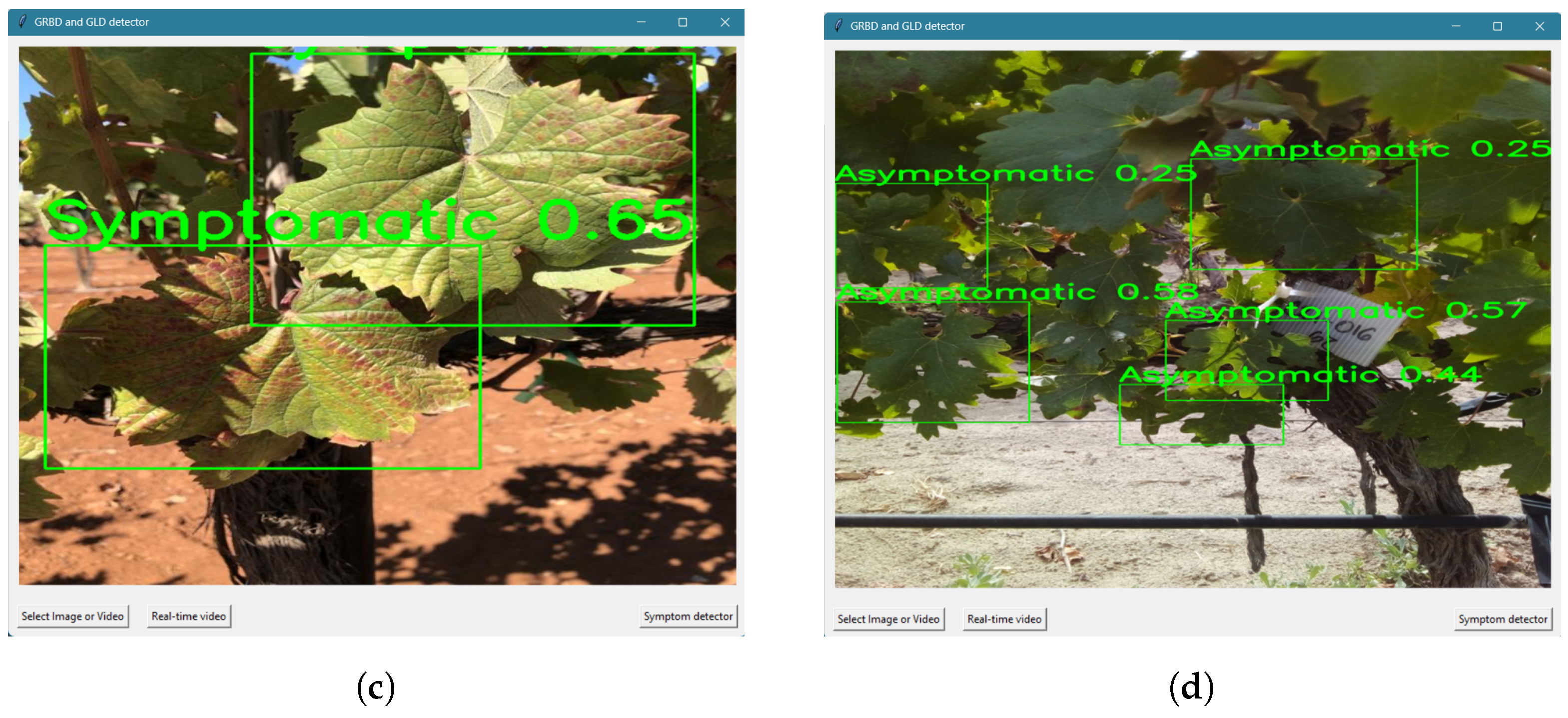
3.4. Real-World Usage
4. Results
4.1. Data Collection and Preparation
4.2. Training and Validation of DL Models
4.3. Hardware Selection
4.4. Real-World Usage Scenario
5. Discussion
6. Conclusions
Future Work
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| AI | Artificial intelligence |
| ANN | Artificial Neural Network |
| CBAM | Convolutional Block Attention Module |
| CNN | Convolutional Neural Network |
| DL | Deep learning |
| GLD | Grapevine leafroll disease |
| GLRaVs | Grapevine leafroll-associated viruses |
| GRBD | Grapevine red blotch disease |
| GRBV | Grapevine red blotch virus |
| LS-SVM | Least squares support vector machine |
| ML | Machine learning |
| RT-PCR | Reverse transcription polymerase chain reaction |
| ResNet | Residual network |
| RF | Random Forest |
| YOLO | You Only Look Once |
References
- Gatou, P.; Tsiara, X.; Spitalas, A.; Sioutas, S.; Vonitsanos, G. Artificial Intelligence Techniques in Grapevine Research: A Comparative Study with an Extensive Review of Datasets, Diseases, and Techniques Evaluation. Sensors 2024, 24, 6211. [Google Scholar] [CrossRef] [PubMed]
- International Organisation of Vine and Wine. 2022. Available online: https://www.oiv.int/sites/default/files/documents/OIV_Actualidad_de_la_coyuntura_del_sector_vitivinicola_mundial_en_2022_0.pdf (accessed on 2 February 2024).
- Instituto Nacional de Estadística y Geografía (INEGI). Economy and Productive Sectors: Agriculture. 2025. Available online: https://www.inegi.org.mx/temas/agricultura/ (accessed on 20 February 2025).
- Grape Production in Mexico 2022. Available online: https://www.gob.mx/siap/documentos/produccion-de-uva-en-mexico-2022 (accessed on 19 December 2023).
- Sembradas 4,365 hectáreas con vid en la Zona Costa de baja California: Agricultura. 2023. Available online: https://www.gob.mx/agricultura/bajacalifornia/articulos/sembradas-4-365-hectareas-con-vid-en-la-zona-costa-de-baja-california-agricultura (accessed on 29 January 2024).
- Yepes, L.M.; Cieniewicz, E.; Krenz, B.; McLane, H.; Thompson, J.R.; Perry, K.L.; Fuchs, M. Causative Role of Grapevine Red Blotch Virus in Red Blotch Disease. Phytopathology® 2018, 108, 902–909. [Google Scholar] [CrossRef] [PubMed]
- Gasperin-Bulbarela, J.; Licea-Navarro, A.F.; Pino-Villar, C.; Hernández-Martínez, R.; Carrillo-Tripp, J. First Report of Grapevine Red Blotch Virus in Mexico. Plant Dis. 2019, 103, 381. [Google Scholar] [CrossRef]
- Krenz, B.; Fuchs, M.; Thompson, J.R. Grapevine red blotch disease: A comprehensive Q&A guide. PLoS Pathog. 2023, 19, e1011671. [Google Scholar] [CrossRef]
- ICTV. Genus: Grablovirus. 2010. Available online: https://ictv.global/report/chapter/geminiviridae/geminiviridae/grablovirus (accessed on 19 December 2023).
- Fiallo-Olivé, E.; Lett, J.M.; Martin, D.P.; Roumagnac, P.; Varsani, A.; Zerbini, F.M.; Navas-Castillo, J. ICTV Virus Taxonomy Profile: Geminiviridae 2021. J. Gen. Virol. 2021, 102, 001696. [Google Scholar] [CrossRef] [PubMed]
- Fuchs, M.; Bar-Joseph, M.; Candresse, T.; Maree, H.J.; Martelli, G.P.; Melzer, M.J.; Menzel, W.; Minafra, A.; Sabanadzovic, S.; ICTV Report Consortium. ICTV Virus Taxonomy Profile: Closteroviridae. J. Gen. Virol. 2020, 101, 364–365. [Google Scholar] [CrossRef]
- Hommay, G.; Beuve, M.; Herrbach, E. Transmission of Grapevine Leafroll-Associated Viruses and Grapevine Virus A by Vineyard-Sampled Soft Scales (Parthenolecanium corni, Hemiptera: Coccidae). Viruses 2022, 14, 2679. [Google Scholar] [CrossRef]
- Cabaleiro, C.; Pesqueira, A.M.; Segura, A. Planococcus ficus and the spread of grapevine leafroll disease in vineyards: A 30-year-long case study in north-West Spain. Eur. J. Plant Pathol. 2022, 163, 733–747. [Google Scholar] [CrossRef]
- Comité Estatal de Sanidad Vegetal de Baja California. Plagas de vid. 2023. Available online: https://www.cesvbc.org/copia-de-manejo-fitosanitario-de-fr (accessed on 29 January 2024).
- Herrbach, E.; Alliaume, A.; Prator, C.A.; Daane, K.M.; Cooper, M.L.; Almeida, R.P.P. Vector Transmission of Grapevine Leafroll-Associated Viruses. In Grapevine Viruses: Molecular Biology, Diagnostics and Management; Springer International Publishing: Cham, Switzerland, 2017; pp. 483–503. [Google Scholar] [CrossRef]
- Atallah, S.S.; Gómez, M.I.; Fuchs, M.F.; Martinson, T.E. Economic Impact of Grapevine Leafroll Disease on Vitis vinifera cv. Cabernet franc in Finger Lakes Vineyards of New York. Am. J. Enol. Vitic. 2012, 63, 73–79. [Google Scholar] [CrossRef]
- Gao, Z.; Khot, L.R.; Naidu, R.A.; Zhang, Q. Early detection of grapevine leafroll disease in a red-berried wine grape cultivar using hyperspectral imaging. Comput. Electron. Agric. 2020, 179, 105807. [Google Scholar] [CrossRef]
- Burger, J.T.; Maree, H.J.; Gouveia, P.; Naidu, R.A. Grapevine leafroll-associated virus3. In Grapevine Viruses: Molecular Biology, Diagnostics and Management; Springer International Publishing: Cham, Switzerland, 2017; pp. 167–195. [Google Scholar] [CrossRef]
- Bahder, B.W.; Zalom, F.G.; Jayanth, M.; Sudarshana, M.R. Phylogeny of Geminivirus Coat Protein Sequences and Digital PCR Aid in Identifying Spissistilus festinus as a Vector of Grapevine red blotch-associated virus. Phytopathology® 2016, 106, 1223–1230. [Google Scholar] [CrossRef] [PubMed]
- Blanco-Ulate, B.; Hopfer, H.; Figueroa-Balderas, R.; Ye, Z.; Rivero, R.M.; Albacete, A.; Pérez-Alfocea, F.; Koyama, R.; Anderson, M.M.; Smith, R.J.; et al. Red blotch disease alters grape berry development and metabolism by interfering with the transcriptional and hormonal regulation of ripening. J. Exp. Bot. 2017, 68, 1225–1238. [Google Scholar] [CrossRef] [PubMed]
- Rumbaugh, A.C.; Sudarshana, M.R.; Oberholster, A. Grapevine Red Blotch Disease Etiology and Its Impact on Grapevine Physiology and Berry and Wine Composition. Horticulturae 2021, 7, 552. [Google Scholar] [CrossRef]
- Ricketts, K.D.; Gómez, M.I.; Fuchs, M.F.; Martinson, T.E.; Smith, R.J.; Cooper, M.L.; Moyer, M.M.; Wise, A. Mitigating the Economic Impact of Grapevine Red Blotch: Optimizing Disease Management Strategies in U.S. Vineyards. Am. J. Enol. Vitic. 2017, 68, 127–135. [Google Scholar] [CrossRef]
- Sudarshana, M.R.; Perry, K.L.; Fuchs, M.F. Grapevine Red Blotch-Associated Virus, an Emerging Threat to the Grapevine Industry. Phytopathology® 2015, 105, 1026–1032. [Google Scholar] [CrossRef] [PubMed]
- Lee, L.; Reynolds, A.; Lan, Y.; Meng, B. Identification of unique electromagnetic signatures from GLRaV-3 infected grapevine leaves in different stages of virus development. Smart Agric. Technol. 2024, 8, 100464. [Google Scholar] [CrossRef]
- Calvi, B.L. Effects of Red-leaf Disease on Cabernet Sauvignon at the Oakville Experimental Vineyard and Mitigation by Harvest Delay and Crop Adjustment. Master’s Thesis, University of California, Davis, CA, USA, 2011. [Google Scholar]
- Krenz, B.; Thompson, J.R.; McLane, H.L.; Fuchs, M.; Perry, K.L. Grapevine red blotch-associated virus Is Widespread in the United States. Phytopathology® 2014, 104, 1232–1240. [Google Scholar] [CrossRef] [PubMed]
- Naidu, R.A.; Maree, H.J.; Burger, J.T. Grapevine Leafroll Disease and Associated Viruses: A Unique Pathosystem. Annu. Rev. Phytopathol. 2015, 53, 613–634. [Google Scholar] [CrossRef] [PubMed]
- Naidu, R.; Rowhani, A.; Fuchs, M.; Golino, D.; Martelli, G.P. Grapevine Leafroll: A Complex Viral Disease Affecting a High-Value Fruit Crop. Plant Dis. 2014, 98, 1172–1180. [Google Scholar] [CrossRef]
- Martelli, G. Directory of virus and virus-like diseases of the grapevine and their agents. J. Plant Pathol. 2014, 96, 1–136. [Google Scholar]
- Sekharamantry, P.K.; Rao, M.S.; Srinivas, Y.; Uriti, A. PSR-LeafNet: A Deep Learning Framework for Identifying Medicinal Plant Leaves Using Support Vector Machines. Big Data Cogn. Comput. 2024, 8, 176. [Google Scholar] [CrossRef]
- Sawyer, E.; Laroche-Pinel, E.; Flasco, M.; Cooper, M.L.; Corrales, B.; Fuchs, M.; Brillante, L. Phenotyping grapevine red blotch virus and grapevine leafroll-associated viruses before and after symptom expression through machine-learning analysis of hyperspectral images. Front. Plant Sci. 2023, 14, 1117869. [Google Scholar] [CrossRef] [PubMed]
- Maeda Gutiérrez, V.; Guerrero Méndez, C.; Olvera Olvera, C.A.; Araiza Esquivel, M.A.; Espinoza García, G.; Bordón López, R. Convolutional neural networks for detection and classification of plant diseases based on digital imagenes. Rev. BiolóGico Agropecu. Tuxpan 2018. [Google Scholar] [CrossRef]
- Kunduracioglu, I.; Pacal, I. Advancements in deep learning for accurate classification of grape leaves and diagnosis of grape diseases. J. Plant Dis. Prot. 2024, 131, 1061–1080. [Google Scholar] [CrossRef]
- Elsherbiny, O.; Elaraby, A.; Alahmadi, M.; Hamdan, M.; Gao, J. Rapid Grapevine Health Diagnosis Based on Digital Imaging and Deep Learning. Plants 2024, 13, 135. [Google Scholar] [CrossRef] [PubMed]
- Pinheiro, I.; Moreira, G.; Queirós da Silva, D.; Magalhães, S.; Valente, A.; Moura Oliveira, P.; Cunha, M.; Santos, F. Deep Learning YOLO-Based Solution for Grape Bunch Detection and Assessment of Biophysical Lesions. Agronomy 2023, 13, 1120. [Google Scholar] [CrossRef]
- Wang, C.; Wang, Y.; Ma, G.; Bian, G.; Ma, C. Identification of Grape Diseases Based on Improved YOLOXS. Appl. Sci. 2023, 13, 5978. [Google Scholar] [CrossRef]
- Schieck, M.; Krajsic, P.; Loos, F.; Hussein, A.; Franczyk, B.; Kozierkiewicz, A.; Pietranik, M. Comparison of deep learning methods for grapevine growth stage recognition. Comput. Electron. Agric. 2023, 211, 107944. [Google Scholar] [CrossRef]
- Foundation Plant Services, UC Davis. FPS Grape Program—Sample Collection. 2025. Available online: http://fps.ucdavis.edu/samplecollection.cfm (accessed on 24 January 2025).
- García, K.; Carrillo Tripp, J. Grapevine Virus and Symptom Database (GVS-DB); Mendeley Data. 2024. Available online: https://data.mendeley.com/datasets/wkbd3wsjpj/1 (accessed on 5 December 2024).
- Gambino, G.; Perrone, I.; Gribaudo, I. A Rapid and effective method for RNA extraction from different tissues of grapevine and other woody plants. Phytochem. Anal. 2008, 19, 520–525. [Google Scholar] [CrossRef]
- Osman, F.; Leutenegger, C.; Golino, D.; Rowhani, A. Real-time RT-PCR (TaqMan) assays for the detection of Grapevine Leafroll associated viruses 1-5 and 9. J. Virol. Methods 2007, 141, 22–29. [Google Scholar] [CrossRef]
- Jocher, G. YOLOv5 by Ultralytics. 2020. [Google Scholar] [CrossRef]
- Liu, H.; Sun, F.; Gu, J.; Deng, L. SF-YOLOv5: A Lightweight Small Object Detection Algorithm Based on Improved Feature Fusion Mode. Sensors 2022, 22, 5817. [Google Scholar] [CrossRef]
- Ultralytics. Data Augmentation—Tools and Libraries. 2024. Available online: https://www.ultralytics.com/glossary/data-augmentation#tools-and-libraries (accessed on 18 January 2024).
- Redmon, J.; Farhadi, A. YOLOv3: An Incremental Improvement. arXiv 2018. [Google Scholar] [CrossRef]
- Reis, D.; Kupec, J.; Hong, J.; Daoudi, A. Real-Time Flying Object Detection with YOLOv8. arXiv 2023. [Google Scholar] [CrossRef]
- Jocher, G.; Chaurasia, A.; Qiu, J. Ultralytics YOLOv8. 2023. [Google Scholar] [CrossRef]
- He, K.; Zhang, X.; Ren, S.; Sun, J. Deep Residual Learning for Image Recognition. arXiv 2015. [Google Scholar] [CrossRef]
- Bendjillali, R.I.; Beladgham, M.; Merit, K.; Taleb-Ahmed, A. Illumination-robust face recognition based on deep convolutional neural networks architectures. Indones. J. Electr. Eng. Comput. Sci. 2020, 18, 1015–1027. [Google Scholar] [CrossRef]
- Dwyer, B.; Nelson, J.; Hansen, T. Roboflow, Version 1.0. 2024. Available online: https://roboflow.com (accessed on 18 January 2024).
- Galarza-Falfan, J.; García-Guerrero, E.E.; Aguirre-Castro, O.A.; López-Bonilla, O.R.; Tamayo-Pérez, U.J.; Cárdenas-Valdez, J.R.; Hernández-Mejía, C.; Borrego-Dominguez, S.; Inzunza-Gonzalez, E. Path Planning for Autonomous Mobile Robot Using Intelligent Algorithms. Technologies 2024, 12, 82. [Google Scholar] [CrossRef]
- Ju, Y.L.; Yue, X.F.; Zhao, X.F.; Zhao, H.; Fang, Y.L. Physiological, micro-morphological and metabolomic analysis of grapevine (Vitis vinifera L.) leaf of plants under water stress. Plant Physiol. Biochem. 2018, 130, 501–510. [Google Scholar] [CrossRef] [PubMed]
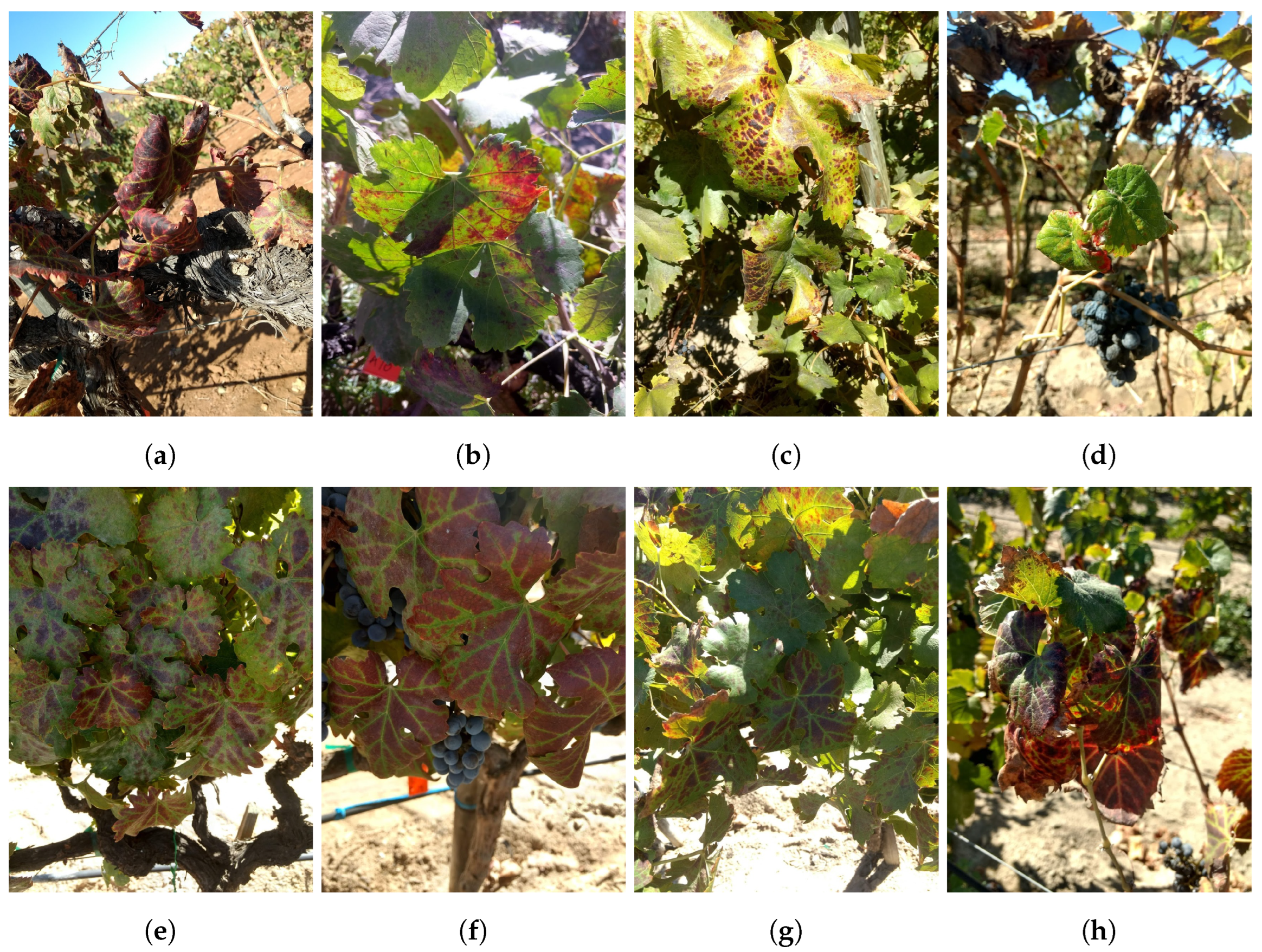
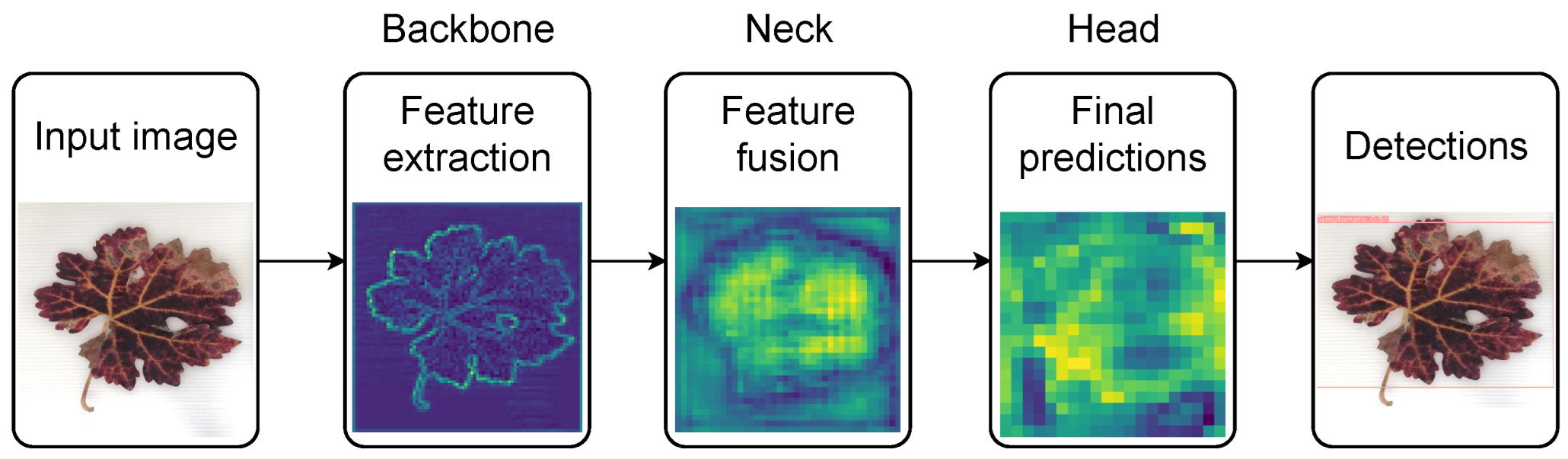




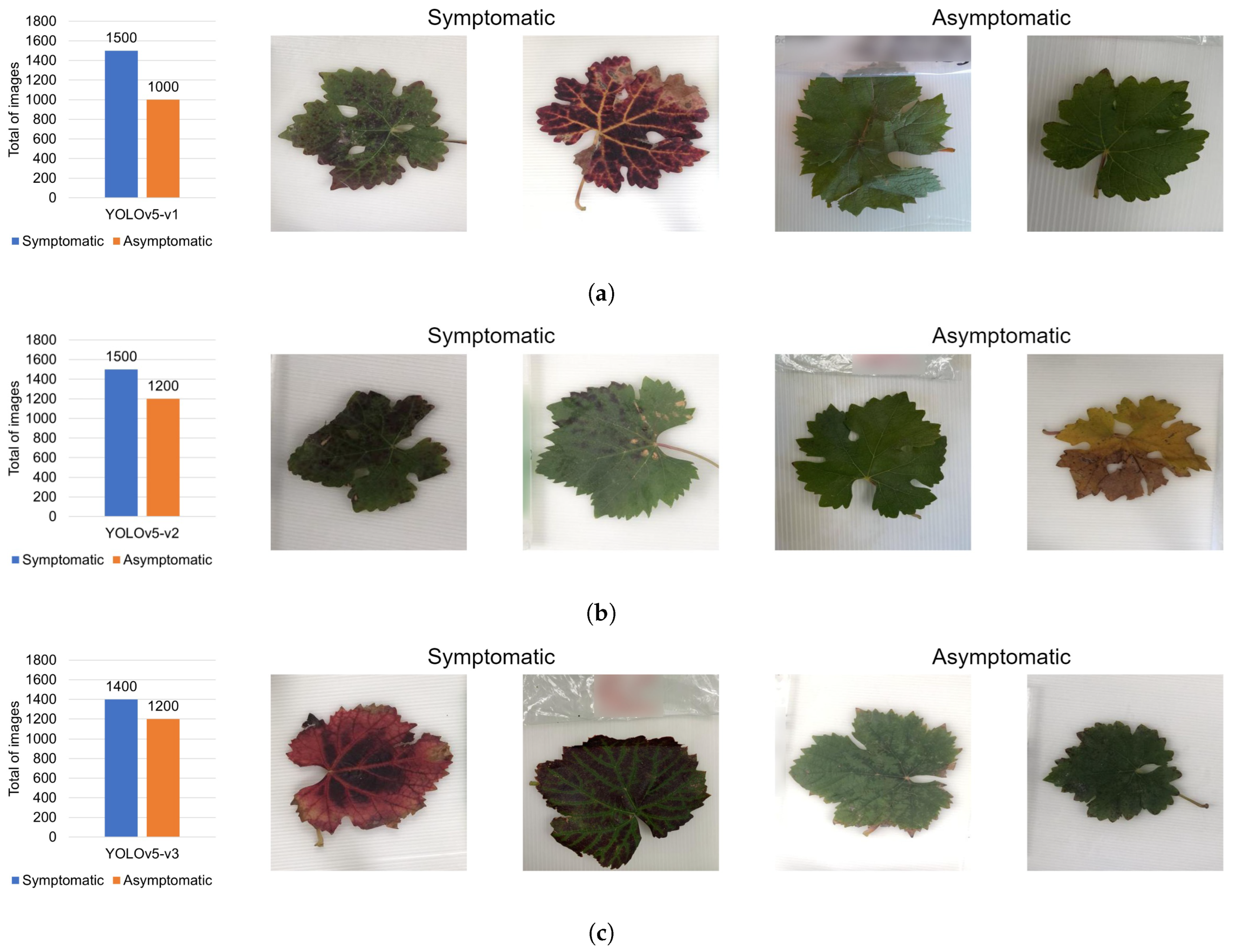





| Reference | Contributions | Algorithm/Model | Dataset | Results | Year |
|---|---|---|---|---|---|
| This work | Identification of symptoms related to GLD and GRBD in grapevines (Vitis vinifera) | DL, CNN and YOLOv5 | 3198 grapevine leaf images | YOLOv5 achieved an Accuracy of 95.36%, Overall Recall 95.77%, and F1-score 95.56% | 2025 |
| Kunduracioglu et al. [33] | Accurate classification of grapevine leaves and diagnosis of grape diseases | Performance comparison of 14 CNN and 17 vision Transformer models | 4062 images from the PlantVillage dataset and 500 images from the Grapevine dataset | 4 models reached an Accuracy of 100% for both datasets | 2024 |
| Elsherbiny et al. [34] | Rapid grapevine diagnosis using DL | CNN, LSTM, DNN, transfer learning with VGG16, VGG19, ResNet50, and ResNet101V2 | 295 images from the PlantVillage dataset | Validation Accuracy, Precision, Recall, and F1-score of 96.6% and an intersection over union of 93.4% | 2024 |
| Sawyer et al. [31] | Detection of GLD and GRBD in grapevine leaves | RF and 3D CNN | 500 hyperspectral images | The CNN model performed better, with an average Precision of 87% against 82.8% from the RF model | 2023 |
| Pinheiro et al. [35] | Grape bunch detection and identification of biophysical lesions | YOLOv5x6, YOLOv7-E6E, and YOLOR-CSP-X | 910 images | YOLOv7 achieved the best results with a Precision of 98%, a Recall of 90%, an F1-score of 94%, and a mAP of 77% | 2023 |
| Wang et al. [36] | Identification of 15 grape diseases | Improved YOLOXS and Convolutional Block Attention Module (CBAM) | China State Key Laboratory of Plant Pest Biology dataset | Average Precision of 99.1% | 2023 |
| Schieck et al. [37] | Grapevine growth stage recognition using DL models | ResNet, DenseNet, and InceptionV3 | Grapevine growth stage dataset (BBCH 71-79) | ResNet achieved the best classification results with an average Accuracy of 88.1% | 2023 |
| Gao et al. [17] | Identification of GLRaV-3 virus during asymptomatic and symptomatic stages of GLD | Least squares support vector machine (LS-SVM) | 500 hyperspectral images | Classifier Precision between 66.67% and 89.93% | 2020 |
| Edge Computing Device | CPU | GPU | RAM | Cost [USD] |
|---|---|---|---|---|
| Personal computer (laptop) | Ryzen 7 5800H | RTX 3050 | 40 GB | 1000.00 |
| Jetson Nano | Quad-core ARM Cortex-A57 | 128-core Maxwell | 2 GB LPDDR4 | 149.00 |
| Raspberry Pi 4 | Quad-core ARM Cortex-A72 | Broadcom VideoCore VI | 4 GB LPDDR4 | 72.80 |
| Year | Number of Images | Grapevine Cultivar | Leaves with Molecular Diagnosis Photographed |
|---|---|---|---|
| 2023 | 989 | Tempranillo, Syrah, Cabernet Sauvignon, Malbec, Nebbiolo, Barbera, Chenin blanc, Thompson, Crimson, Grenache, Red globe, Sauvignon blanc, and Mision | 347 |
| 2022 | 1044 | Tempranillo, Syrah, Cabernet Sauvignon, Chenin blanc, Colombard, Malbec, Nebbiolo, Merlot, Chardonnay, Grenache, Red globe, Carignan, and Petite Syrah | 453 |
| 2021 | 1142 | Cabernet Sauvignon, Nebbiolo italiana, Merlot, and Nebbiolo | 0 |
| 2019 | 13 | Gamay, Nebbiolo, Mounedre, Petit verdot, Merlot, Cabernet Sauvignon, Mision, and Crimson | 0 |
| 2018 | 10 | Nebbiolo, Temporal, Chardonnay, and Tempranillo | 0 |
| Total | 3198 | 23 different cultivars | 800 |
| Model | Resolution | Wide Angle Aperture | Ultra-Wide Angle Aperture | Telephoto Lens | Image Format |
|---|---|---|---|---|---|
| iPhone 8 | 12 MP | ƒ/1.8 | NA | NA | HEIF and JPEG |
| iPhone 10 | 12 MP | ƒ/1.8 | NA | ƒ/2.4 lens aperture | HEIF and JPEG |
| iPhone 13 | 12 MP | ƒ/1.6 | ƒ/2.4 lens aperture, 120° field of view | NA | HEIF and JPEG |
| iPhone 14 | 12 MP | ƒ/1.5 | ƒ/2.4 lens aperture, 120° field of view | NA | HEIF and JPEG |
| Hyperparameter | Value |
| Image size (–img) | 416 |
| Batch size (–batch) | 5 |
| Number of epochs (–epochs) | 30 |
| Data configuration file (–data) | data.yaml |
| Pre-trained weights (–weights) | yolov5s.pt |
| Experiment name (–name) | yolov5s_results_EN |
| Device (–device) | 1 |
| Cache images (–cache) | Enabled |
| Learning rate | 0.01 (default initial value) |
| Optimizer | SGD (Stochastic Gradient Descent) |
| Data Augmentation Hyperparameter | Value |
| HSV Hue | 0.015 |
| HSV Saturation | 0.7 |
| HSV Value | 0.4 |
| Translate | 0.1 |
| Scale | 0.5 |
| Flip left–right | 0.5 |
| Mosaic | 1 |
| Asymptomatic | Symptomatic | Total | |
|---|---|---|---|
| RT-PCR diagnosis | 200 | 600 | 800 |
| Visual symptoms diagnosis | 1335 | 1063 | 2398 |
| Total | 1535 | 1663 | 3198 |
| Metrics | YOLOv5-v1 | YOLOv5-v2 | YOLOv5-v3 | YOLOv5-v4 | YOLOv5-v5 |
|---|---|---|---|---|---|
| Asymptomatic class Precision | 95.52% | 95.92% | 93.93% | 97.76% | 94.85% |
| Asymptomatic class error | 4.48% | 4.08% | 6.07% | 2.24% | 5.15% |
| Symptomatic class Precision | 88.06% | 94.12% | 92.94% | 95.05% | 95.87% |
| Symptomatic class error | 11.94% | 5.88% | 7.06% | 4.95% | 4.13% |
| Accuracy | 91.41% | 95.00% | 93.43% | 96.37% | 95.36% |
| Classification of individual leaves | Yes | Yes | Yes | Yes | Yes |
| Classification of asymptomatic grapevine leaves | No | No | Yes | Yes | Yes |
| Classification of symptomatic grapevine leaves | No | No | No | Yes | Yes |
| Classification of low-resolution images | No | No | No | No | Yes |
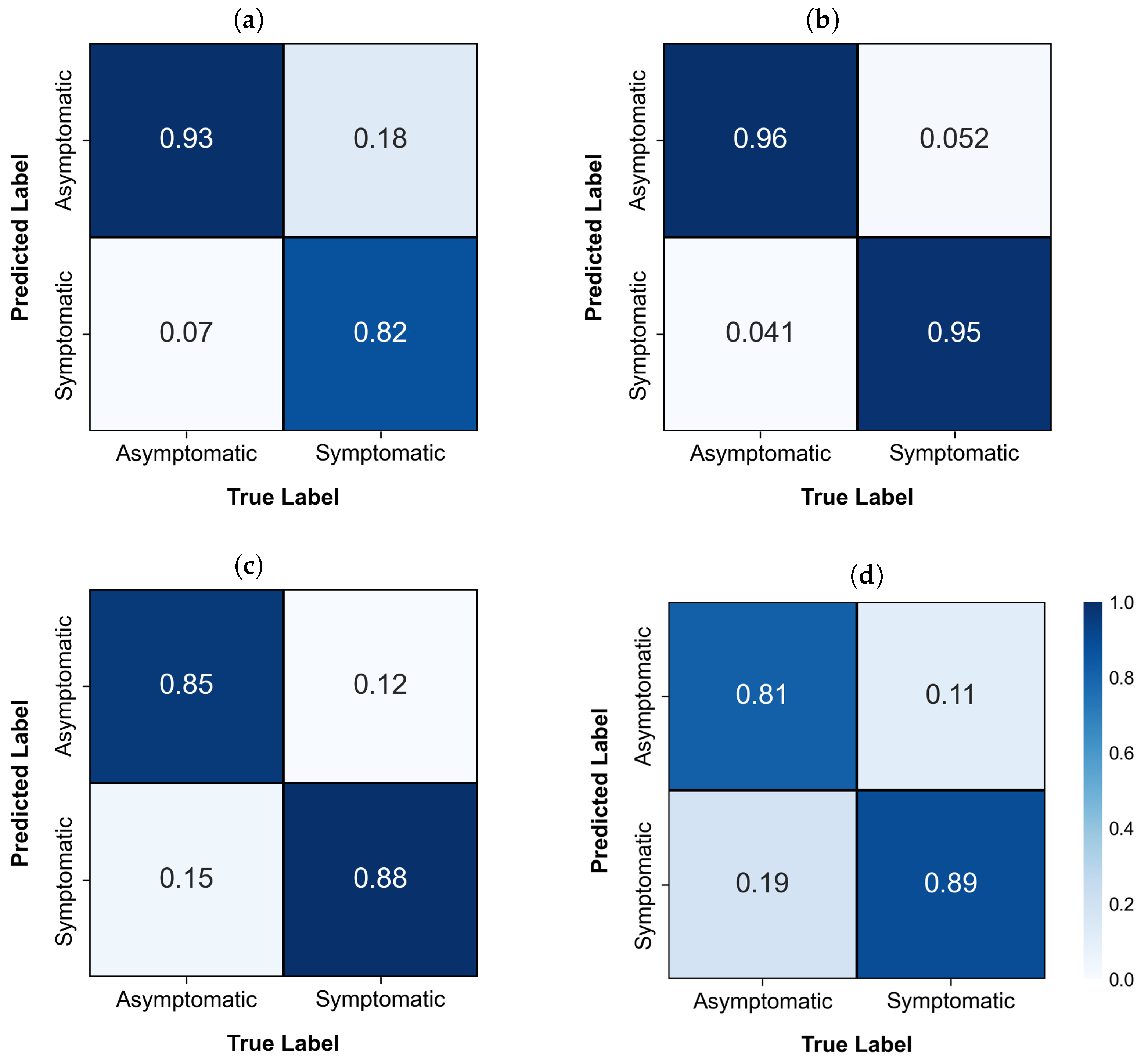
| Model | Classes | Accuracy | Precision | 1-Precision | Recall | 1-Recall | F1-Score |
|---|---|---|---|---|---|---|---|
| YOLOv3 | Asymptomatic | 0.8750 | 0.8378 | 0.1622 | 0.9300 | 0.0700 | 0.8815 |
| Symptomatic | 0.9213 | 0.0787 | 0.8200 | 0.1800 | 0.8677 | ||
| YOLOv5 | Asymptomatic | 0.9536 | 0.9485 | 0.0515 | 0.9592 | 0.0408 | 0.9538 |
| Symptomatic | 0.9587 | 0.0413 | 0.9479 | 0.0521 | 0.9533 | ||
| YOLOv8 | Asymptomatic | 0.8650 | 0.8763 | 0.1237 | 0.8500 | 0.1500 | 0.8629 |
| Symptomatic | 0.8544 | 0.1456 | 0.8800 | 0.1200 | 0.8670 | ||
| ResNet-50 | Asymptomatic | 0.8516 | 0.8799 | 0.1201 | 0.8143 | 0.1857 | 0.8459 |
| Symptomatic | 0.8272 | 0.1728 | 0.8889 | 0.1111 | 0.8569 |
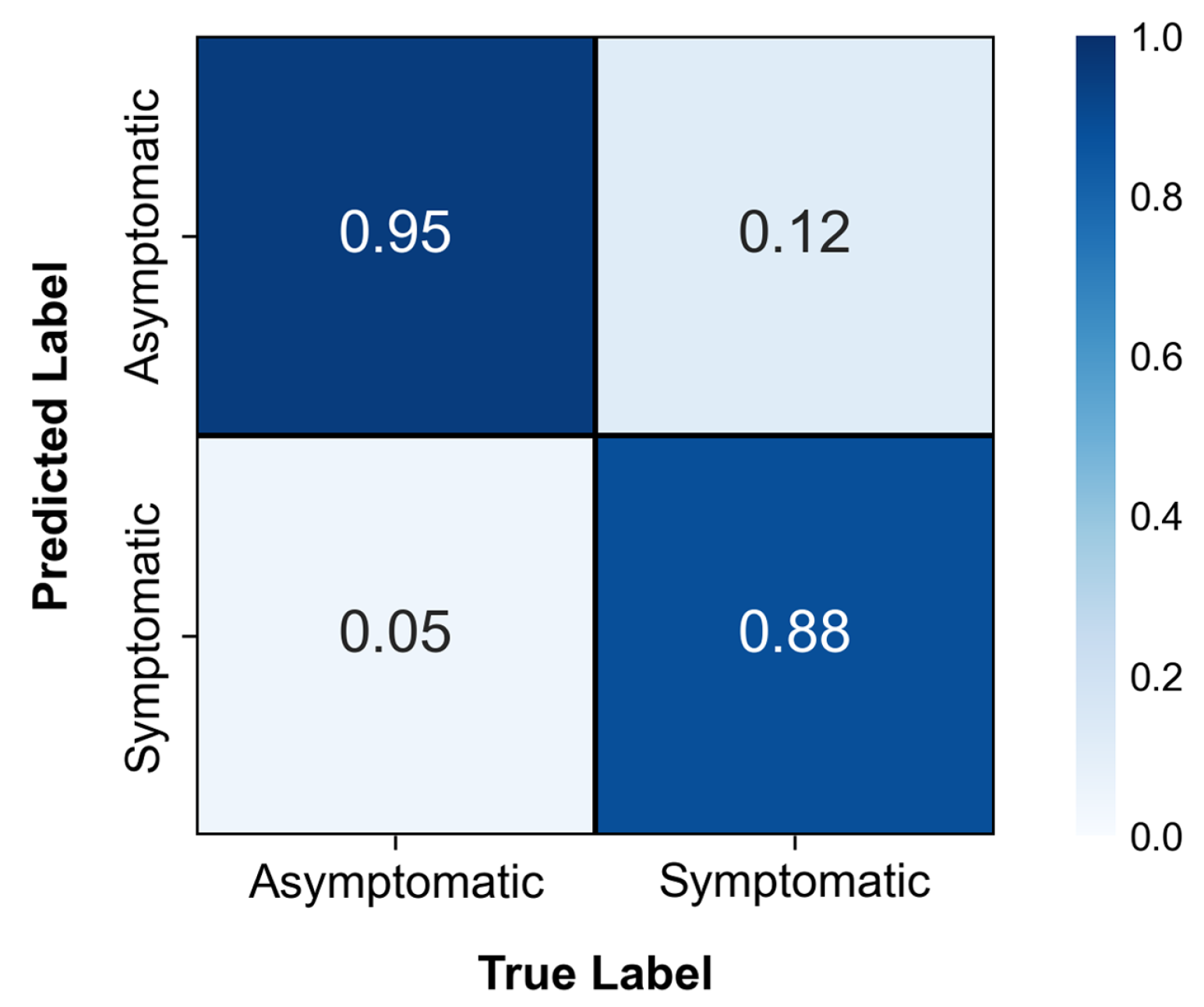
| Image Resolution | Classes | Accuracy | Precision | 1-Precision | Recall | 1-Recall | F1-Score |
|---|---|---|---|---|---|---|---|
| 240 × 240 | Asymptomatic | 0.8763 | 0.8309 | 0.1691 | 0.9450 | 0.0550 | 0.8843 |
| Symptomatic | 0.9362 | 0.0638 | 0.8077 | 0.1923 | 0.8672 | ||
| 480 × 480 | Asymptomatic | 0.9125 | 0.8837 | 0.1163 | 0.9500 | 0.0500 | 0.9157 |
| Symptomatic | 0.9459 | 0.0541 | 0.8750 | 0.1250 | 0.9091 | ||
| 640 × 640 | Asymptomatic | 0.9300 | 0.8909 | 0.1091 | 0.9800 | 0.0200 | 0.9333 |
| Symptomatic | 0.9778 | 0.0222 | 0.8800 | 0.1200 | 0.9263 |
| Edge Computing Device | Inference Time Based on Image Resolution [ms] | FPS | ||||
|---|---|---|---|---|---|---|
| 240 × 240 | 480 × 480 | 640 × 640 | 240 × 240 | 480 × 480 | 640 × 640 | |
| Raspberry Pi | 521.4 | 1309.8 | 2160.7 | 1.8181 | 0.9012 | 0.5554 |
| Jetson NANO | 315.2 | 757.3 | 1277.4 | 3.9682 | 1.8181 | 1.0204 |
| Personal computer (laptop) | 10.4 | 10.4 | 10.5 | 114.9425 | 96.15384 | 78.74015 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lazcano-García, C.; García-Resendiz, K.G.; Carrillo-Tripp, J.; Inzunza-Gonzalez, E.; García-Guerrero, E.E.; Cervantes-Vasquez, D.; Galarza-Falfan, J.; Lopez-Mercado, C.A.; Aguirre-Castro, O.A. Deep Learning-Based System for Early Symptoms Recognition of Grapevine Red Blotch and Leafroll Diseases and Its Implementation on Edge Computing Devices. AgriEngineering 2025, 7, 63. https://doi.org/10.3390/agriengineering7030063
Lazcano-García C, García-Resendiz KG, Carrillo-Tripp J, Inzunza-Gonzalez E, García-Guerrero EE, Cervantes-Vasquez D, Galarza-Falfan J, Lopez-Mercado CA, Aguirre-Castro OA. Deep Learning-Based System for Early Symptoms Recognition of Grapevine Red Blotch and Leafroll Diseases and Its Implementation on Edge Computing Devices. AgriEngineering. 2025; 7(3):63. https://doi.org/10.3390/agriengineering7030063
Chicago/Turabian StyleLazcano-García, Carolina, Karen Guadalupe García-Resendiz, Jimena Carrillo-Tripp, Everardo Inzunza-Gonzalez, Enrique Efrén García-Guerrero, David Cervantes-Vasquez, Jorge Galarza-Falfan, Cesar Alberto Lopez-Mercado, and Oscar Adrian Aguirre-Castro. 2025. "Deep Learning-Based System for Early Symptoms Recognition of Grapevine Red Blotch and Leafroll Diseases and Its Implementation on Edge Computing Devices" AgriEngineering 7, no. 3: 63. https://doi.org/10.3390/agriengineering7030063
APA StyleLazcano-García, C., García-Resendiz, K. G., Carrillo-Tripp, J., Inzunza-Gonzalez, E., García-Guerrero, E. E., Cervantes-Vasquez, D., Galarza-Falfan, J., Lopez-Mercado, C. A., & Aguirre-Castro, O. A. (2025). Deep Learning-Based System for Early Symptoms Recognition of Grapevine Red Blotch and Leafroll Diseases and Its Implementation on Edge Computing Devices. AgriEngineering, 7(3), 63. https://doi.org/10.3390/agriengineering7030063










