Normalized Difference Vegetation Index Prediction for Blueberry Plant Health from RGB Images: A Clustering and Deep Learning Approach
Abstract
1. Introduction
| MS Sensor | Company | Spectral Bands | Price (EUR) | Reference |
|---|---|---|---|---|
| Parrot Sequoia+ | SenseFly, Parrot Group, Paris, France | Green, Red, Red Edge, NIR, and RGB | ~4000.00 | [26] |
| Sentera 6x Thermal | Sentera Sensors & Drones, St. Paul, MN, USA | Green, Red, Red Edge, NIR, Thermal, and RGB | ~20,000.00 | [27] |
| Altum-PT | AgEagle Aerial System Inc., Wichita, KS, USA | Blue, Green, Red, Red Edge NIR, and Panchromatic | ~18,000.00 | [28] |
| RedEdge-P | AgEagle Aerial System Inc., Wichita, KS, USA | Blue, Green, Red, Red Edge NIR, and Panchromatic | ~10,000.00 | [29] |
| MAIA S2 | SAL Engineering, Russi, Italy | Violet, Blue, Green, Red, RedEdge-1, 2, and NIR-1, 2, 3 | ~18,000.00 | [30] |
| Toucan | SILIOS Technologies, Peynier, France | 10 Narrow Bands | ~15,000.00 | [31] |
| A7Rxx quad | Agrowing Ltd., Rishon LeZion, Israel | 10 Narrow Bands and Wide RGB | ~15,000.00 | [32] |
2. Materials and Methods
2.1. Data Acquisition and Platform
2.2. Clustering for Bush Identification
2.3. NDVI Computation
2.4. Deep Learning Model Development
2.4.1. Model Architecture
2.4.2. Model Training and Evaluation
3. Results and Discussion
3.1. Plant Bush Cluster Identification
3.2. Model Performance Evaluation
4. Conclusions
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Foley, J.A.; Ramankutty, N.; Brauman, K.A.; Cassidy, E.S.; Gerber, J.S.; Johnston, M.; Mueller, N.D.; O’Connell, C.; Ray, D.K.; West, P.C.; et al. Solutions for a Cultivated Planet. Nature 2011, 478, 337–342. [Google Scholar] [CrossRef] [PubMed]
- Blesh, J.; Hoey, L.; Jones, A.D.; Friedmann, H.; Perfecto, I. Development Pathways toward “Zero Hunger”. World Dev. 2019, 118, 1–14. [Google Scholar] [CrossRef]
- Zhang, N.; Wang, M.; Wang, N. Precision Agriculture a Worldwide Overview. Comput. Electron. Agric. 2002, 36, 113–132. [Google Scholar] [CrossRef]
- Gebbers, R.; Adamchuk, V.I. Precision Agriculture and Food Security. Science 2010, 327, 828–831. [Google Scholar] [CrossRef] [PubMed]
- Talebpour, B.; Türker, U.; Yegül, U. The Role of Precision Agriculture in the Promotion of Food Security. Int. J. Agric. Food Res. 2015, 4, 1–23. [Google Scholar] [CrossRef]
- Kriegler, F.J.; Malila, W.A.; Nalepka, R.F.; Richardson, W. Preprocessing Transformations and Their Effects on Multispectral Recognition. In Proceedings of the Sixth International Symposium on Remote Sensing of Environment, Ann Arbor, MI, USA, 13–16 October 1969; Volume II, p. 97. [Google Scholar]
- Rouse, J.W.; Haas, R.H.; Schell, J.A.; Deering, D.W. Monitoring Vegetation Systems in the Great Plains with ERTS. NASA Spec. Publ. 1974, 351, 309. [Google Scholar]
- Veloso, A.; Mermoz, S.; Bouvet, A.; Le Toan, T.; Planells, M.; Dejoux, J.-F.; Ceschia, E. Understanding the Temporal Behavior of Crops Using Sentinel-1 and Sentinel-2-like Data for Agricultural Applications. Remote Sens. Env. 2017, 199, 415–426. [Google Scholar] [CrossRef]
- Tucker, C.J. Red and Photographic Infrared Linear Combinations for Monitoring Vegetation. Remote Sens. Env. 1979, 8, 127–150. [Google Scholar] [CrossRef]
- King, M.D.; Platnick, S.; Menzel, W.P.; Ackerman, S.A.; Hubanks, P.A. Spatial and Temporal Distribution of Clouds Observed by MODIS Onboard the Terra and Aqua Satellites. IEEE Trans. Geosci. Remote Sens. 2013, 51, 3826–3852. [Google Scholar] [CrossRef]
- Pelta, R.; Beeri, O.; Tarshish, R.; Shilo, T. Sentinel-1 to NDVI for Agricultural Fields Using Hyperlocal Dynamic Machine Learning Approach. Remote Sens. 2022, 14, 2600. [Google Scholar] [CrossRef]
- Berni, J.; Zarco-Tejada, P.J.; Suarez, L.; Fereres, E. Thermal and Narrowband Multispectral Remote Sensing for Vegetation Monitoring from an Unmanned Aerial Vehicle. IEEE Trans. Geosci. Remote Sens. 2009, 47, 722–738. [Google Scholar] [CrossRef]
- Anderson, K.; Gaston, K.J. Lightweight Unmanned Aerial Vehicles Will Revolutionize Spatial Ecology. Front. Ecol. Env. 2013, 11, 138–146. [Google Scholar] [CrossRef]
- Tsouros, D.C.; Bibi, S.; Sarigiannidis, P.G. A Review on UAV-Based Applications for Precision Agriculture. Information 2019, 10, 349. [Google Scholar] [CrossRef]
- Radoglou-Grammatikis, P.; Sarigiannidis, P.; Lagkas, T.; Moscholios, I. A Compilation of UAV Applications for Precision Agriculture. Comput. Netw. 2020, 172, 107148. [Google Scholar] [CrossRef]
- Pedersen, S.M.; Lind, K.M. Precision Agriculture—From Mapping to Site-Specific Application. In Precision Agriculture: Technology and Economic Perspectives; Marcus, P.S., Lind, K.M., Eds.; Springer International Publishing: Cham, Switzerland, 2017; pp. 1–20. [Google Scholar] [CrossRef]
- Kendall, H.; Clark, B.; Li, W.; Jin, S.; Jones, G.D.; Chen, J.; Taylor, J.; Li, Z.; Frewer, L.J. Precision Agriculture Technology Adoption: A Qualitative Study of Small-Scale Commercial “Family Farms” Located in the North China Plain. Precis. Agric. 2022, 23, 319–351. [Google Scholar] [CrossRef]
- Bai, A.; Kovách, I.; Czibere, I.; Megyesi, B.; Balogh, P. Examining the Adoption of Drones and Categorisation of Precision Elements among Hungarian Precision Farmers Using a Trans-Theoretical Model. Drones 2022, 6, 200. [Google Scholar] [CrossRef]
- Radočaj, D.; Šiljeg, A.; Marinović, R.; Jurišić, M. State of Major Vegetation Indices in Precision Agriculture Studies Indexed in Web of Science: A Review. Agriculture 2023, 13, 707. [Google Scholar] [CrossRef]
- Pallottino, F.; Antonucci, F.; Costa, C.; Bisaglia, C.; Figorilli, S.; Menesatti, P. Optoelectronic Proximal Sensing Vehicle-Mounted Technologies in Precision Agriculture: A Review. Comput. Electron. Agric. 2019, 162, 859–873. [Google Scholar] [CrossRef]
- Lowder, S.K.; Sánchez, M.V.; Bertini, R. Which Farms Feed the World and Has Farmland Become More Concentrated? World Dev. 2021, 142, 105455. [Google Scholar] [CrossRef]
- International Food Policy Research Institute. Global Food Policy Report; International Food Policy Research Institute: Washington, DC, USA, 2018; Volume 10, p. 9780896292970. [Google Scholar]
- Ricciardi, V.; Ramankutty, N.; Mehrabi, Z.; Jarvis, L.; Chookolingo, B. How Much of the World’s Food Do Smallholders Produce? Glob. Food Sec. 2018, 17, 64–72. [Google Scholar] [CrossRef]
- Guiomar, N.; Godinho, S.; Pinto-Correia, T.; Almeida, M.; Bartolini, F.; Bezák, P.; Biró, M.; Bjørkhaug, H.; Bojnec, Š.; Brunori, G.; et al. Typology and Distribution of Small Farms in Europe: Towards a Better Picture. Land. Use Policy 2018, 75, 784–798. [Google Scholar] [CrossRef]
- Tisenkopfs, T.; Adamsone-Fiskovica, A.; Kilis, E.; Šūmane, S.; Grivins, M.; Pinto-Correia, T.; Bjørkhaug, H. Territorial Fitting of Small Farms in Europe. Glob. Food Sec. 2020, 26, 100425. [Google Scholar] [CrossRef]
- Parrot. Parrot Sequoia+. Available online: https://www.parrot.com/en/shop/accessories-spare-parts/other-drones/sequoia (accessed on 15 October 2024).
- Drones, S.S. Sentera 6x Thermal. Available online: https://senterasensors.com/hardware/sensors/6x/ (accessed on 15 October 2024).
- AgEagle Aerial Systems Inc. Altum-PT. Available online: https://ageagle.com/drone-sensors/altum-pt-camera/ (accessed on 15 October 2024).
- AgEagle Aerial Systems Inc. RedEdge-P. Available online: https://ageagle.com/drone-sensors/rededge-p-high-res-multispectral-camera/ (accessed on 15 October 2024).
- SAL Engineering. MAIA S2. Available online: https://www.spectralcam.com/maia-tech-2/ (accessed on 15 October 2024).
- SILIOS Technologies. Toucan. Available online: https://www.silios.com/toucan-camera (accessed on 15 October 2024).
- Agrowing Ltd. A7Rxx Quad. Available online: https://agrowing.com/products/alpha-7rxxx-quad/ (accessed on 15 October 2024).
- Mizik, T. How Can Precision Farming Work on a Small Scale? A Systematic Literature Review. Precis. Agric. 2023, 24, 384–406. [Google Scholar] [CrossRef]
- Stamford, J.D.; Vialet-Chabrand, S.; Cameron, I.; Lawson, T. Development of an Accurate Low Cost NDVI Imaging System for Assessing Plant Health. Plant Methods 2023, 19, 9. [Google Scholar] [CrossRef]
- Cucho-Padin, G.; Loayza, H.; Palacios, S.; Balcazar, M.; Carbajal, M.; Quiroz, R. Development of Low-Cost Remote Sensing Tools and Methods for Supporting Smallholder Agriculture. Appl. Geomat. 2020, 12, 247–263. [Google Scholar] [CrossRef]
- Hobbs, S.; Lambert, A.; Ryan, M.J.; Paull, D.J. Preparing for Space: Increasing Technical Readiness of Low-Cost High-Performance Remote Sensing Using High-Altitude Ballooning. Adv. Space Res. 2023, 71, 1034–1044. [Google Scholar] [CrossRef]
- Holman, F.H.; Riche, A.B.; Castle, M.; Wooster, M.J.; Hawkesford, M.J. Radiometric Calibration of ‘Commercial off the Shelf’ Cameras for UAV-Based High-Resolution Temporal Crop Phenotyping of Reflectance and NDVI. Remote Sens. 2019, 11, 1657. [Google Scholar] [CrossRef]
- Barrows, C.; Bulanon, D. Development of a Low-Cost Multispectral Camera for Aerial Crop Monitoring. J. Unmanned Veh. Syst. 2017, 5, 192–200. [Google Scholar] [CrossRef]
- Costa, L.; Nunes, L.; Ampatzidis, Y. A New Visible Band Index (VNDVI) for Estimating NDVI Values on RGB Images Utilizing Genetic Algorithms. Comput. Electron. Agric. 2020, 172, 105334. [Google Scholar] [CrossRef]
- Davidson, C.; Jaganathan, V.; Sivakumar, A.N.; Czarnecki, J.M.P.; Chowdhary, G. NDVI/NDRE Prediction from Standard RGB Aerial Imagery Using Deep Learning. Comput. Electron. Agric. 2022, 203, 107396. [Google Scholar] [CrossRef]
- Moscovini, L.; Ortenzi, L.; Pallottino, F.; Figorilli, S.; Violino, S.; Pane, C.; Capparella, V.; Vasta, S.; Costa, C. An Open-Source Machine-Learning Application for Predicting Pixel-to-Pixel NDVI Regression from RGB Calibrated Images. Comput. Electron. Agric. 2024, 216, 108536. [Google Scholar] [CrossRef]
- Farooque, A.A.; Afzaal, H.; Benlamri, R.; Al-Naemi, S.; MacDonald, E.; Abbas, F.; MacLeod, K.; Ali, H. Red-Green-Blue to Normalized Difference Vegetation Index Translation: A Robust and Inexpensive Approach for Vegetation Monitoring Using Machine Vision and Generative Adversarial Networks. Precis. Agric. 2023, 24, 1097–1115. [Google Scholar] [CrossRef]
- Jeong, U.; Jang, T.; Kim, D.; Cheong, E.J. Classification and Identification of Pinecone Mulching in Blueberry Cultivation Based on Crop Leaf Characteristics and Hyperspectral Data. Agronomy 2024, 14, 785. [Google Scholar] [CrossRef]
- Anku, K.E.; Percival, D.C.; Lada, R.; Heung, B.; Vankoughnett, M. Remote Estimation of Leaf Nitrogen Content, Leaf Area, and Berry Yield in Wild Blueberries. Front. Remote Sens. 2024, 5, 1414540. [Google Scholar] [CrossRef]
- Zaman, A.; Mahmoud, N.; Virro, I.; Liivapuu, O.; Lillerand, T.; Roy, K.; Olt, J. Learning with Small Data: A Novel Framework for Blueberry Root Collar Detection. In Proceedings of the 35th DAAAM International Symposium; Katalinic, B., Ed.; DAAAM International: Vienna, Austria, 24–25 October 2024. [Google Scholar]
- Mahmoud, N.T.A.; Virro, I.; Zaman, A.G.M.; Lillerand, T.; Chan, W.T.; Liivapuu, O.; Roy, K.; Olt, J. Robust Object Detection Under Smooth Perturbations in Precision Agriculture. AgriEngineering 2024, 6, 4570–4584. [Google Scholar] [CrossRef]
- Lillerand, T.; Virro, I.; Maksarov, V.V.; Olt, J. Granulometric Parameters of Solid Blueberry Fertilizers and Their Suitability for Precision Fertilization. Agronomy 2021, 11, 1576. [Google Scholar] [CrossRef]
- Virro, I.; Arak, M.; Maksarov, V.; Olt, J. Precision Fertilisation Technologies for Berry Plantation. Agronomy Research 2020, 18, 2797–2810. [Google Scholar] [CrossRef]
- Davies, D.L.; Bouldin, D.W. A Cluster Separation Measure. IEEE Trans. Pattern Anal. Mach. Intell. 1979, PAMI-1, 224–227. [Google Scholar] [CrossRef]
- Calinski, T.; Harabasz, J. A Dendrite Method for Cluster Analysis. Commun. Stat. Theory Methods 1974, 3, 1–27. [Google Scholar] [CrossRef]





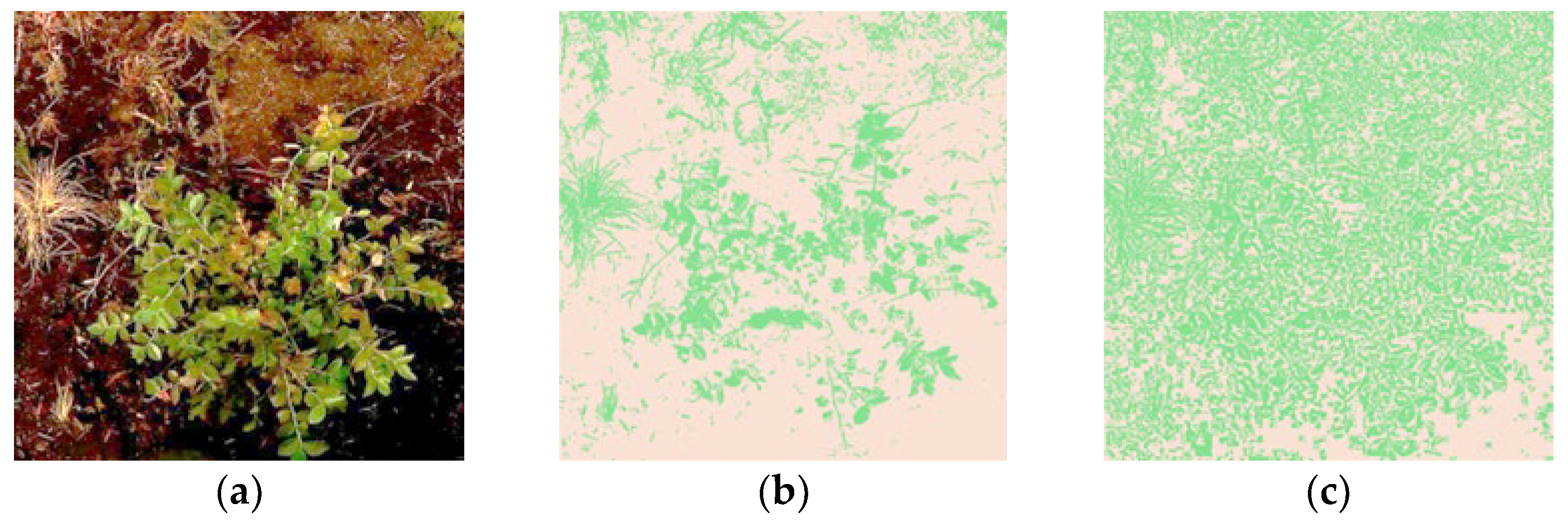






| Clustering Before Adaptive Hue-Based Filtering in HSL | Clustering After Adaptive Hue-Based Filtering in HSL | Improvement After Adaptive Hue-Based Filtering in HSL | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| K-Means | GMM | K-Means | GMM | K-Means | GMM | ||||||||
| Metric | DBI | CHI 1 | DBI | CHI 1 | DBI | CHI 1 | DBI | CHI 1 | DBI (%) | CHI (%) | DBI (%) | CHI (%) | |
| Image | |||||||||||||
| Figure 4a | 0.7959 | 301 K | 1.5704 | 65 K | 0.4040 | 749 K | 0.3658 | 556 K | 49.24 | 149 | 76.71 | 754 | |
| Figure 4b | 0.6872 | 359 K | 1.2164 | 88 K | 0.4352 | 728 K | 0.6226 | 304 K | 36.67 | 103 | 48.81 | 244 | |
| Figure 4c | 0.6437 | 401 K | 1.0613 | 89 K | 0.3535 | 1151 K | 0.6277 | 402 K | 45.08 | 187 | 40.86 | 350 | |
| Average metric value: | 0.3976 | 876 K | 0.5387 | 421 K | |||||||||
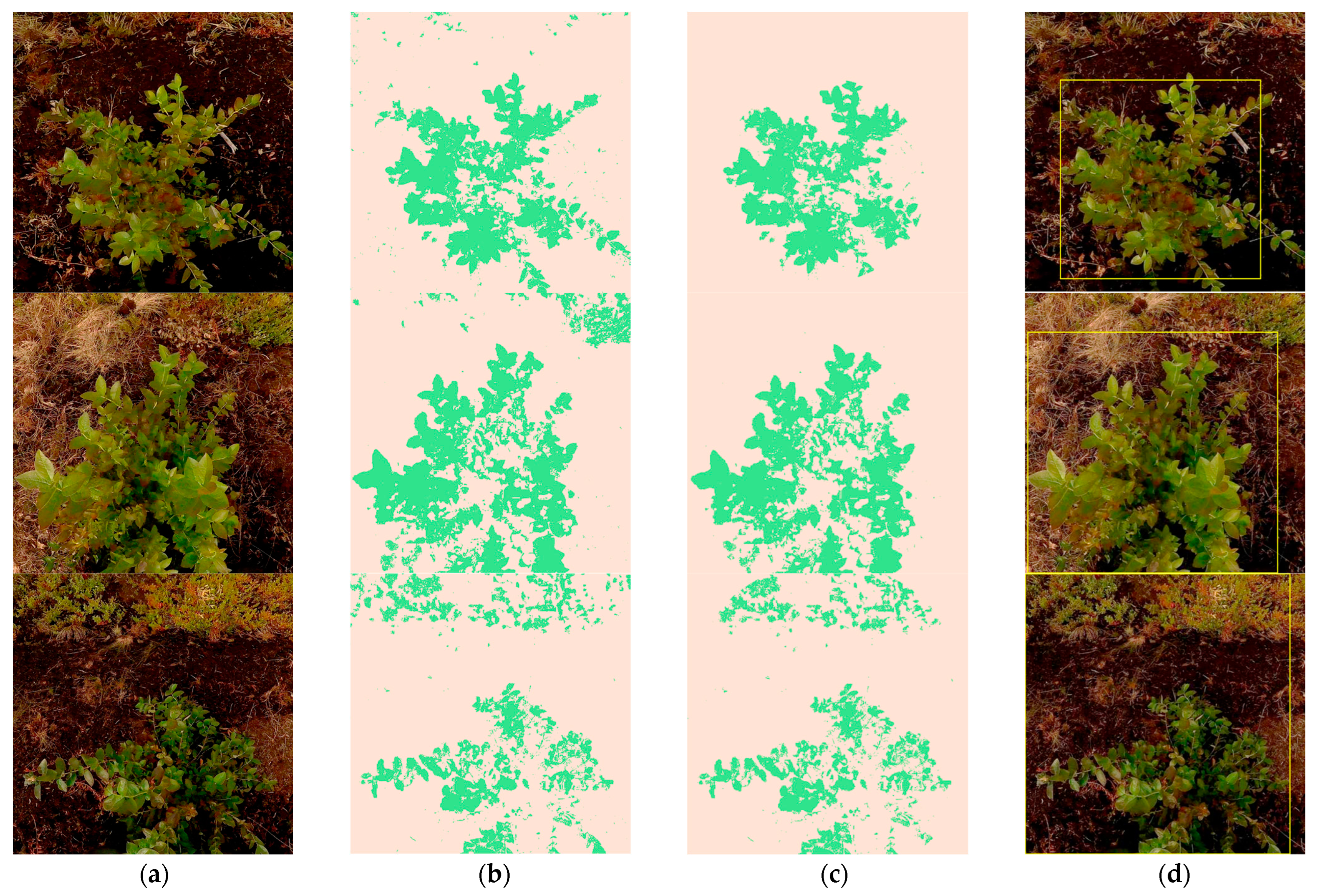
| Plant Custer 1 | Bounding Box Coordinates | NDVI | |
|---|---|---|---|
| Min (x, y) | Max (x, y) | ||
| P009 (Figure 7g) | (11, 31) | (491, 474) | 0.9218 |
| P006 (Figure 7h) | (92, 123) | (468, 479) | 0.7515 |
| P050 (Figure 7i) | (217, 162) | (428, 357) | 0.7743 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zaman, A.G.M.; Roy, K.; Olt, J. Normalized Difference Vegetation Index Prediction for Blueberry Plant Health from RGB Images: A Clustering and Deep Learning Approach. AgriEngineering 2024, 6, 4831-4850. https://doi.org/10.3390/agriengineering6040276
Zaman AGM, Roy K, Olt J. Normalized Difference Vegetation Index Prediction for Blueberry Plant Health from RGB Images: A Clustering and Deep Learning Approach. AgriEngineering. 2024; 6(4):4831-4850. https://doi.org/10.3390/agriengineering6040276
Chicago/Turabian StyleZaman, A. G. M., Kallol Roy, and Jüri Olt. 2024. "Normalized Difference Vegetation Index Prediction for Blueberry Plant Health from RGB Images: A Clustering and Deep Learning Approach" AgriEngineering 6, no. 4: 4831-4850. https://doi.org/10.3390/agriengineering6040276
APA StyleZaman, A. G. M., Roy, K., & Olt, J. (2024). Normalized Difference Vegetation Index Prediction for Blueberry Plant Health from RGB Images: A Clustering and Deep Learning Approach. AgriEngineering, 6(4), 4831-4850. https://doi.org/10.3390/agriengineering6040276







