Hyperspectral Response of the Soybean Crop as a Function of Target Spot (Corynespora cassiicola) Using Machine Learning to Classify Severity Levels
Abstract
1. Introduction
2. Materials and Methods
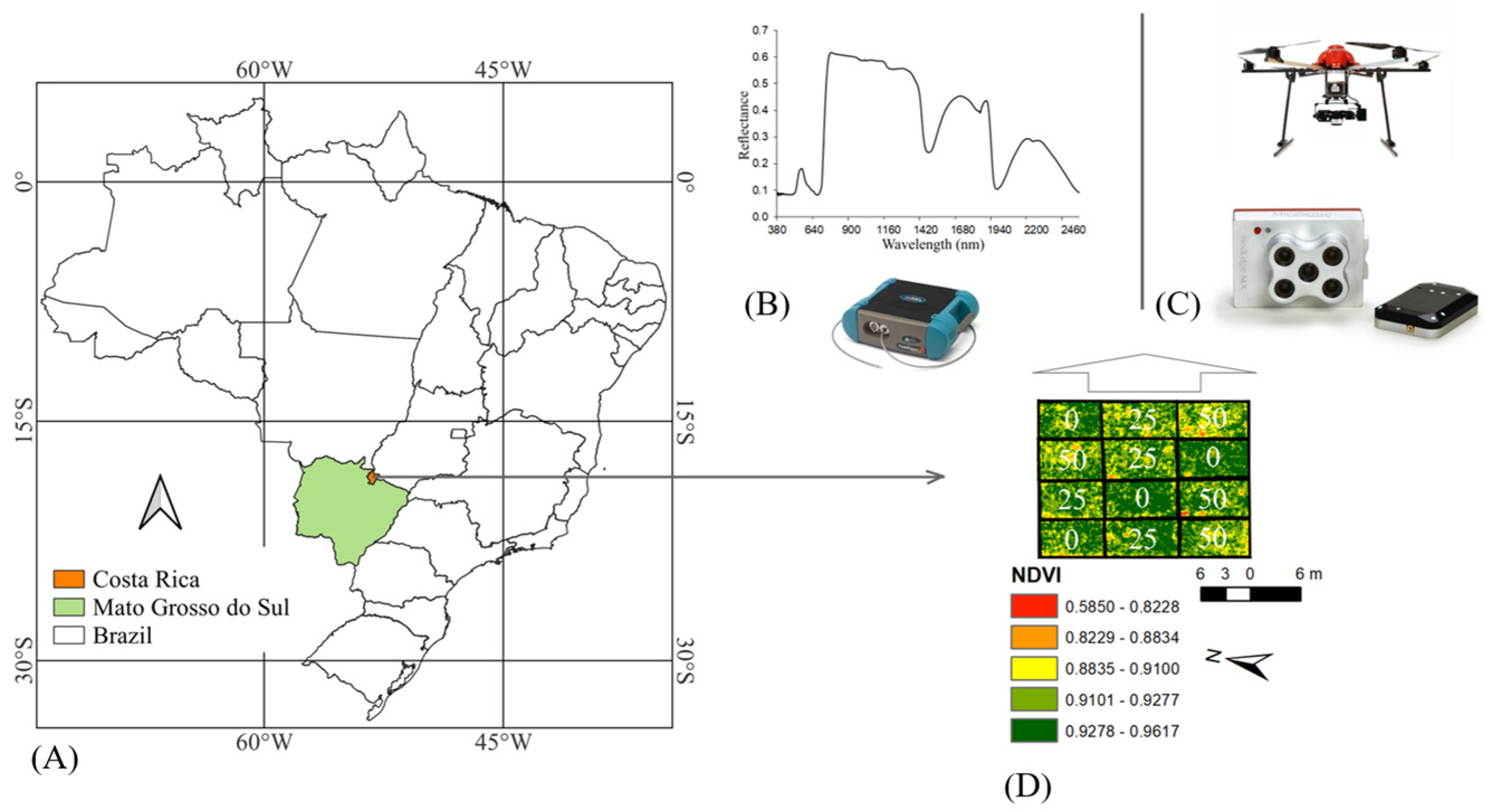
2.1. Study Area
2.2. Experimental Design
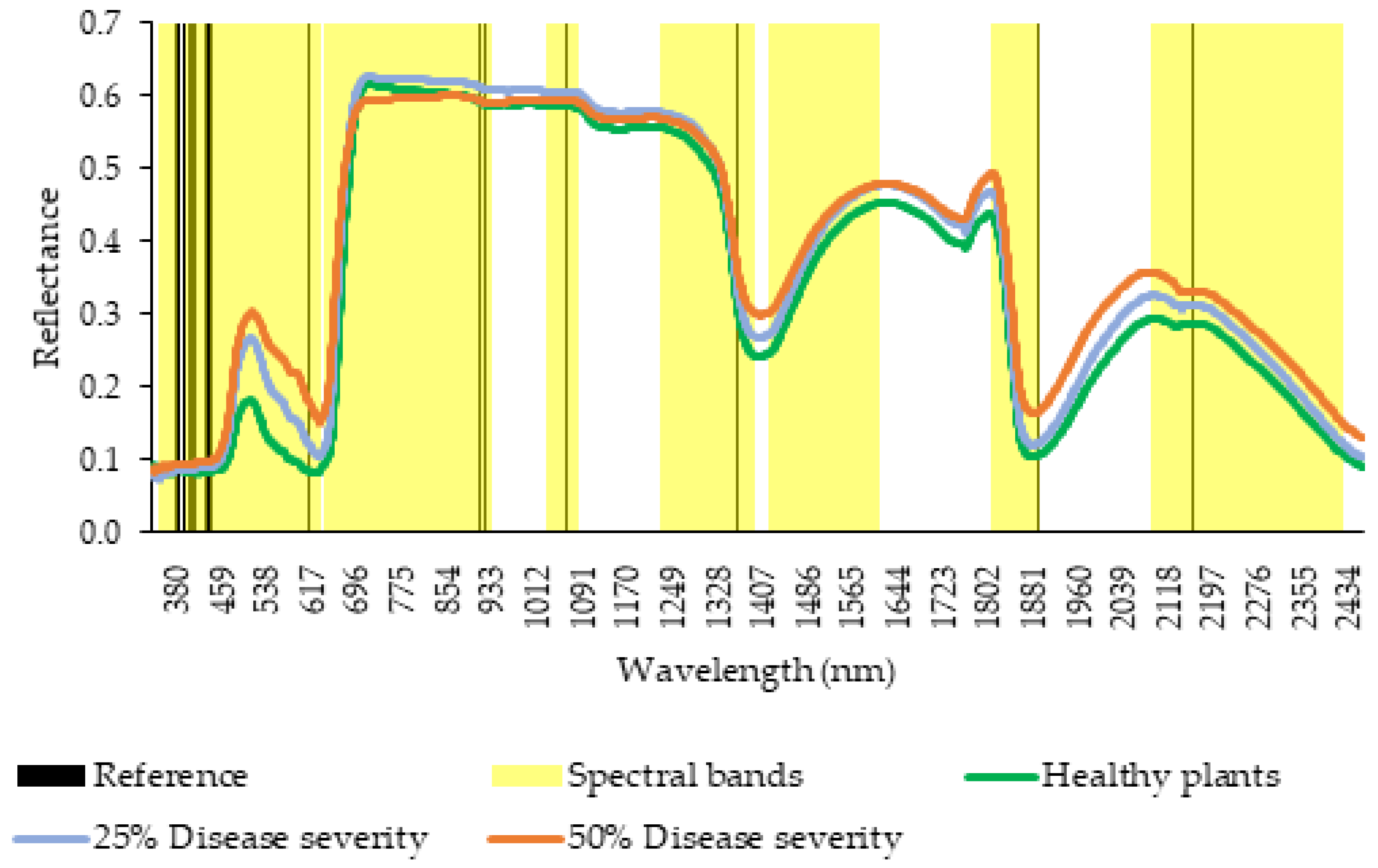
2.3. Spectral Analysis
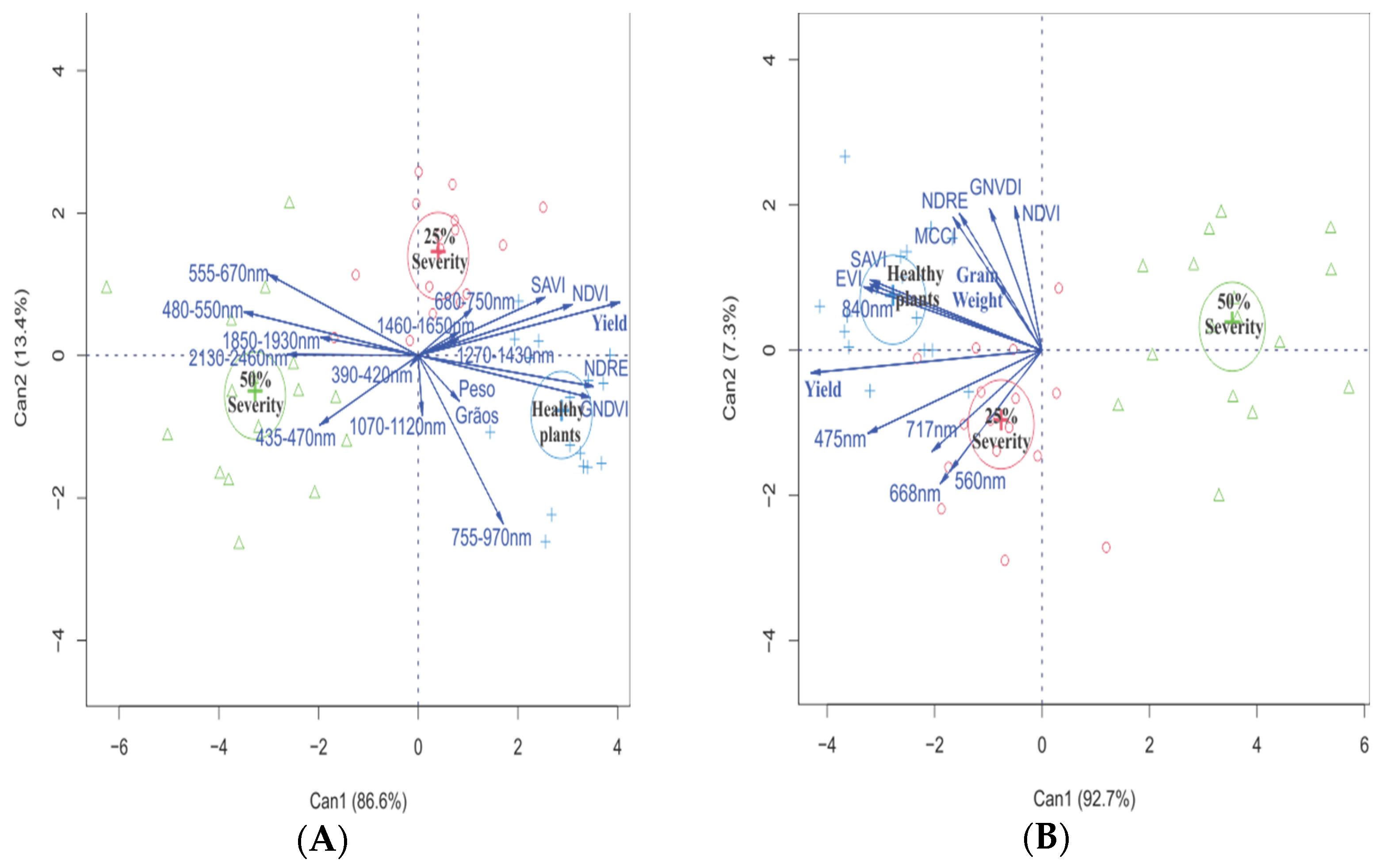
2.4. Data Analyses
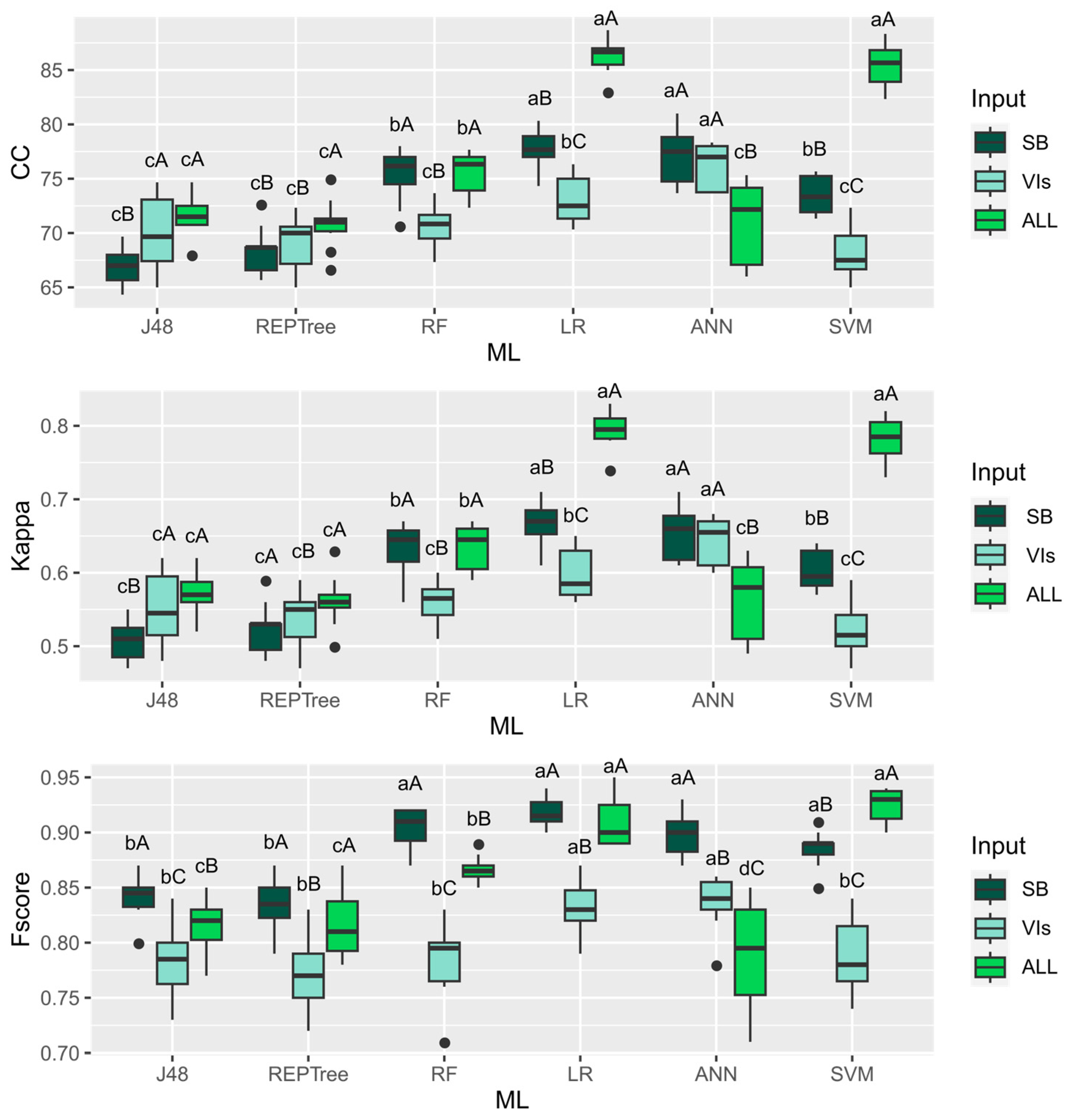
3. Results
4. Discussion
5. Conclusions
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Zhao, M.; Dong, Y.; Huang, W.; Ruan, C.; Guo, J. Regional-Scale Monitoring of Wheat Stripe Rust Using Remote Sensing and Geographical Detectors. Remote Sens. 2023, 15, 4631. [Google Scholar] [CrossRef]
- Chen, H.; Li, H.; Liu, Z.; Zhang, C.; Zhang, S.; Atkinson, P.M. A Novel Greenness and Water Content Composite Index (GWCCI) for Soybean Mapping from Single Remotely Sensed Multispectral Images. Remote Sens. Environ. 2023, 295, 113679. [Google Scholar] [CrossRef]
- Arantes, B.H.T.; Martins, G.D.; Carvalho, E.R.; Nogueira, L.C.A. Identificação de Ferrugem Na Soja Por Meio de Imagens de Alta Resolução Espacial. Rev. Bras. Geogr. Física 2019, 12, 1003–1016. [Google Scholar] [CrossRef]
- Zhang, S.-L.; Sun, Q.; Cao, Y.; Ji, Y.-P.; Zhang, Y.-J.; Herrera-Balandrano, D.D.; Chen, X.; Shi, X.-C.; Wang, S.-Y.; Laborda, P. Biocontrol of Corynespora Cassiicola in Soybean Using a New Phenethyl Alcohol-Producing Meyerozyma Caribbica Strain. Biol. Control 2023, 184, 105287. [Google Scholar] [CrossRef]
- Edwards Molina, J.P.; Paul, P.A.; Amorim, L.; Da Silva, L.; Siqueri, F.V.; Borges, E.P.; Campos, H.D.; Venancio, W.S.; Meyer, M.C.; Martins, M.C. Effect of Target Spot on Soybean Yield and Factors Affecting This Relationship. Plant Pathol. 2019, 68, 107–115. [Google Scholar] [CrossRef]
- Dixon, L.J.; Schlub, R.L.; Pernezny, K.; Datnoff, L.E. Host Specialization and Phylogenetic Diversity of Corynespora Cassiicola. Phytopathology 2009, 99, 1015–1027. [Google Scholar] [CrossRef]
- Sumabat, L.G.; Kemerait, R.C., Jr.; Brewer, M.T. Phylogenetic Diversity and Host Specialization of Corynespora Cassiicola Responsible for Emerging Target Spot Disease of Cotton and Other Crops in the Southeastern United States. Phytopathology 2018, 108, 892–901. [Google Scholar] [CrossRef]
- Aguiar, F.M.; Vallad, G.E.; Timilsina, S.; Veloso, J.S.; Fonseca, M.E.N.; Boiteux, L.S.; Reis, A. Phylogenetic Network Analysis of South and North American Corynespora Cassiicola Isolates from Tomato, Cucumber, and Novel Hosts. Eur. J. Plant Pathol. 2022, 163, 657–671. [Google Scholar] [CrossRef]
- Yamamoto, S.; Nomoto, S.; Hashimoto, N.; Maki, M.; Hongo, C.; Shiraiwa, T.; Homma, K. Monitoring Spatial and Time-Series Variations in Red Crown Rot Damage of Soybean in Farmer Fields Based on UAV Remote Sensing. Plant Prod. Sci. 2023, 26, 36–47. [Google Scholar] [CrossRef]
- Bajwa, S.G.; Rupe, J.C.; Mason, J. Soybean Disease Monitoring with Leaf Reflectance. Remote Sens. 2017, 9, 127. [Google Scholar] [CrossRef]
- Shahi, T.B.; Xu, C.-Y.; Neupane, A.; Guo, W. Recent Advances in Crop Disease Detection Using UAV and Deep Learning Techniques. Remote Sens. 2023, 15, 2450. [Google Scholar] [CrossRef]
- Anderson, K.; Gaston, K.J. Lightweight Unmanned Aerial Vehicles Will Revolutionize Spatial Ecology. Front. Ecol. Env. 2013, 11, 138–146. [Google Scholar] [CrossRef]
- Colomina, I.; Molina, P. Unmanned Aerial Systems for Photogrammetry and Remote Sensing: A Review. ISPRS J. Photogramm. Remote Sens. 2014, 92, 79–97. [Google Scholar] [CrossRef]
- Farber, C.; Mahnke, M.; Sanchez, L.; Kurouski, D. Advanced Spectroscopic Techniques for Plant Disease Diagnostics. A Review. Trends Anal. Chem. 2019, 118, 43–49. [Google Scholar] [CrossRef]
- Koc, A.; Odilbekov, F.; Alamrani, M.; Henriksson, T.; Chawade, A. Predicting Yellow Rust in Wheat Breeding Trials by Proximal Phenotyping and Machine Learning. Plant Methods 2022, 18, 30. [Google Scholar] [CrossRef] [PubMed]
- dos Santos, H.G.; Jacomine, P.K.T.; Dos Anjos, L.H.C.; De Oliveira, V.A.; Lumbreras, J.F.; Coelho, M.R.; De Almeida, J.A.; de Araujo Filho, J.C.; De Oliveira, J.B.; Cunha, T.J.F. Sistema Brasileiro de Classificação de Solos; Embrapa: Brasília, Brazil, 2018; ISBN 8570358172. [Google Scholar]
- Soja, E.; Dinali, C.; Seixas, S.; Alvadi, N.N.; Balbinot, A.; Francisco, J.; Krzyzanowski, C.; Villas, R.M.; De, B.; Leite, C.; et al. Sistemas de Produção 17 Tecnologias de Produção de Soja; Embrapa Soja: Londrina, Brazil, 2020. [Google Scholar]
- Soares, R.M.; Godoy, C.V.; Oliveira, M.C.N.D. Escala Diagramática Para Avaliação Da Severidade Da Mancha Alvo Da Soja. Trop. Plant Pathol. 2009, 34, 333–338. [Google Scholar] [CrossRef][Green Version]
- da Silva Junior, C.A.; Nanni, M.R.; Shakir, M.; Teodoro, P.E.; de Oliveira-Júnior, J.F.; Cezar, E.; de Gois, G.; Lima, M.; Wojciechowski, J.C.; Shiratsuchi, L.S. Soybean Varieties Discrimination Using Non-Imaging Hyperspectral Sensor. Infrared Phys. Technol. 2018, 89, 338–350. [Google Scholar] [CrossRef]
- Rouse, J.W.; Haas, R.H.; Schell, J.A.; Deering, D.W. Monitoring Vegetation Systems in the Great Plains with ERTS. NASA Spec. Publ. 1974, 351, 309. [Google Scholar]
- Gitelson, A.; Merzlyak, M.N. Spectral Reflectance Changes Associated with Autumn Senescence of Aesculus Hippocastanum L. and Acer Platanoides L. Leaves. Spectral Features and Relation to Chlorophyll Estimation. J. Plant Physiol. 1994, 143, 286–292. [Google Scholar] [CrossRef]
- Huete, A.R. A Soil-Adjusted Vegetation Index (SAVI). Remote Sens. Environ. 1988, 25, 295–309. [Google Scholar] [CrossRef]
- Gitelson, A.A.; Merzlyak, M.N. Signature Analysis of Leaf Reflectance Spectra: Algorithm Development for Remote Sensing of Chlorophyll. J. Plant Physiol. 1996, 148, 494–500. [Google Scholar] [CrossRef]
- Perry, E.M.; Goodwin, I.; Cornwall, D. Remote Sensing Using Canopy and Leaf Reflectance for Estimating Nitrogen Status in Red-Blush Pears. HortScience Horts. 2018, 53, 78–83. [Google Scholar] [CrossRef]
- Egmont-Petersen, M.; de Ridder, D.; Handels, H. Image Processing with Neural Networks—A Review. Pattern Recognit. 2002, 35, 2279–2301. [Google Scholar] [CrossRef]
- Quinlan, J.R. C4. 5: Programming for Machine Learning. Morgan Kauffmann 1993, 38, 49. [Google Scholar]
- Štepanovský, M.; Ibrová, A.; Buk, Z.; Velemínská, J. Novel Age Estimation Model Based on Development of Permanent Teeth Compared with Classical Approach and Other Modern Data Mining Methods. Forensic Sci. Int. 2017, 279, 72–82. [Google Scholar] [CrossRef] [PubMed]
- Al Snousy, M.B.; El-Deeb, H.M.; Badran, K.; Al Khlil, I.A. Suite of Decision Tree-Based Classification Algorithms on Cancer Gene Expression Data. Egypt. Inform. J. 2011, 12, 73–82. [Google Scholar] [CrossRef]
- Belgiu, M.; Drăguţ, L. Random Forest in Remote Sensing: A Review of Applications and Future Directions. ISPRS J. Photogramm. Remote Sens. 2016, 114, 24–31. [Google Scholar] [CrossRef]
- Kalmegh, S. Analysis of WEKA Data Mining Algorithm REPTree, Simple Cart and RandomTree for Classification of Indian News; Academia Inc.: San Francisco, CA, USA, 2015; Volume 2. [Google Scholar]
- Nalepa, J.; Kawulok, M. Selecting Training Sets for Support Vector Machines: A Review. Artif. Intell. Rev. 2019, 52, 857–900. [Google Scholar] [CrossRef]
- Bhering, L.L. Rbio: A Tool for Biometric and Statistical Analysis Using the R Platform. Crop Breed. Appl. Biotechnol. 2017, 17, 187–190. [Google Scholar] [CrossRef]
- West, J.S.; Bravo, C.; Oberti, R.; Moshou, D.; Ramon, H.; McCartney, H.A. Detection of Fungal Diseases Optically and Pathogen Inoculum by Air Sampling. In Precision Crop Protection—The Challenge and Use of Heterogeneity; Springer Dordrecht: Dordrecht, The Netherlands, 2010; pp. 135–149. [Google Scholar]
- Mahlein, A.-K. Plant Disease Detection by Imaging Sensors–Parallels and Specific Demands for Precision Agriculture and Plant Phenotyping. Plant Dis. 2016, 100, 241–251. [Google Scholar] [CrossRef]
- Christenson, B.S.; Schapaugh, W.T., Jr.; An, N.; Price, K.P.; Fritz, A.K. Characterizing Changes in Soybean Spectral Response Curves with Breeding Advancements. Crop Sci. 2014, 54, 1585–1597. [Google Scholar] [CrossRef]
- Zahir, S.A.D.M.; Omar, A.F.; Jamlos, M.F.; Azmi, M.A.M.; Muncan, J. A Review of Visible and Near-Infrared (Vis-NIR) Spectroscopy Application in Plant Stress Detection. Sens. Actuators A Phys. 2022, 338, 113468. [Google Scholar] [CrossRef]
- Moreira, M.A. Fundamentos Do Sensoriamento Remoto e Metodologias de Aplicação; 3° Edição; UFV: Chilliwack, BC, Canada, 2005. [Google Scholar]
- Negrisoli, M.M.; Negrisoli, R.; da Silva, F.; Lopes, L.S.; de Souza Júnior, F.S.; Velini, E.D.; Carbonari, C.A.; Rodrigues, S.A.; Raetano, C.G. Soybean Rust Detection and Disease Severity Classification by Remote Sensing. Agron. J. 2022, 114, 3246–3262. [Google Scholar] [CrossRef]
- Ahmed, M.; Seraj, R.; Islam, S.M.S. The K-Means Algorithm: A Comprehensive Survey and Performance Evaluation. Electronics 2020, 9, 1295. [Google Scholar] [CrossRef]
- Saravia, D.; Salazar, W.; Valqui-Valqui, L.; Quille-Mamani, J.; Porras-Jorge, R.; Corredor, F.-A.; Barboza, E.; Vásquez, H.V.; Casas Diaz, A.V.; Arbizu, C.I. Yield Predictions of Four Hybrids of Maize (Zea mays) Using Multispectral Images Obtained from UAV in the Coast of Peru. Agronomy 2022, 12, 2630. [Google Scholar] [CrossRef]
- Carter, G.A.; Knapp, A.K. Leaf Optical Properties in Higher Plants: Linking Spectral Characteristics to Stress and Chlorophyll Concentration. Am. J. Bot. 2001, 88, 677–684. [Google Scholar] [CrossRef] [PubMed]
- Junges, A.H.; Almança, M.A.K.; Fajardo, T.V.M.; Ducati, J.R. Leaf Hyperspectral Reflectance as a Potential Tool to Detect Diseases Associated with Vineyard Decline. Trop. Plant Pathol. 2020, 45, 522–533. [Google Scholar] [CrossRef]
- Knauer, U.; Matros, A.; Petrovic, T.; Zanker, T.; Scott, E.S.; Seiffert, U. Improved Classification Accuracy of Powdery Mildew Infection Levels of Wine Grapes by Spatial-Spectral Analysis of Hyperspectral Images. Plant Methods 2017, 13, 47. [Google Scholar] [CrossRef] [PubMed]
- Heim, R.H.J.; Wright, I.J.; Chang, H.; Carnegie, A.J.; Pegg, G.S.; Lancaster, E.K.; Falster, D.S.; Oldeland, J. Detecting Myrtle Rust (Austropuccinia psidii) on Lemon Myrtle Trees Using Spectral Signatures and Machine Learning. Plant Pathol. 2018, 67, 1114–1121. [Google Scholar] [CrossRef]
- Mahlein, A.-K.; Oerke, E.-C.; Steiner, U.; Dehne, H.-W. Recent Advances in Sensing Plant Diseases for Precision Crop Protection. Eur. J. Plant Pathol. 2012, 133, 197–209. [Google Scholar] [CrossRef]
- de Oliveira Pires, M.S.; de Carvalho Alves, M.; Pozza, E.A. Multispectral Radiometric Characterization of Coffee Rust Epidemic in Different Irrigation Management Systems. Int. J. Appl. Earth Obs. Geoinf. 2020, 86, 102016. [Google Scholar] [CrossRef]
- Bose, P.; Dutta, S.; Goyal, V.; Bandyopadhyay, S.K. Leaf Diseases Detection of Medicinal Plants Based on Support Vector Machine Classification Algorithm. J. Pharm. Res. Int. 2021, 33, 111–119. [Google Scholar] [CrossRef]
- Santana, D.C.; Teixeira Filho, M.C.M.; da Silva, M.R.; das Chagas, P.H.M.; de Oliveira, J.L.G.; Baio, F.H.R.; Campos, C.N.S.; Teodoro, L.P.R.; da Silva Junior, C.A.; Teodoro, P.E. Machine Learning in the Classification of Soybean Genotypes for Primary Macronutrients’ Content Using UAV–Multispectral Sensor. Remote Sens. 2023, 15, 1457. [Google Scholar] [CrossRef]
- da Silva Barros, P.P.; Rosalen, D.L.; Iost Filho, F.H.; Martins, G.D.; Di Leo, N. Monitoramento Fitossanitário Utilizando Sensoriamento Remoto: Avanços e Desafios. Rev. Bras. Cart. 2021, 73, 489. [Google Scholar] [CrossRef]
- Gewali, U.B.; Monteiro, S.T.; Saber, E. Machine Learning Based Hyperspectral Image Analysis: A Survey. arXiv 2018, arXiv:1802.08701. [Google Scholar]


| pH | H + Al | Ca | Mg | Al | CEC | B | Cu | Fe | Mn | Zn | K | P | OM | Clay | V | m |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| cmolc dm−3 | mg dm−3 | g dm−3 | % | |||||||||||||
| 5.3 | 4.6 | 5.5 | 1.7 | 0.05 | 12.0 | 0.33 | 1.4 | 45.0 | 16.6 | 5.2 | 97.0 | 37.4 | 36.1 | 67 | 60.2 | 0.8 |
| Spectral Band Center (nm) | Plant Physiological Characteristics |
|---|---|
| 370 | Phototropism |
| 420 | a-carotene |
| 425 | b-carotene |
| 430 | Chlorophyll absorption |
| 440 | a-carotene |
| 445 | xanthophyll |
| 445 | Chlorophyll synthesis |
| 450 | b-carotene |
| 453 | Chlorophyll b |
| 470 | a-carotene |
| 475 | Chlorophyll b |
| 480 | a-carotene |
| 650 | Chlorophyll synthesis |
| 960 | Chlorophyll absorption |
| 1100 | Chlorophyll absorption |
| 1400 | Water absorption |
| 1930 | Water absorption |
| 2200 | Al-OH, Mg-OH and CO3 peak |
| Band | Spectral Range (nm) |
|---|---|
| B1 | 390–420 |
| B2 | 435–470 |
| B3 | 480–550 |
| B4 | 555–670 |
| B5 | 680–750 |
| B6 | 755–970 |
| B7 | 1070–1120 |
| B8 | 1270–1430 |
| B9 | 1460–1650 |
| B10 | 1850–1930 |
| B11 | 2130–2460 |
| Abbreviation | Vegetation Index | Equation | Reference |
|---|---|---|---|
| NDVI | Normalized difference Vegetation index | [20] | |
| NDRE | Normalized difference Red edge index | [21] | |
| SAVI | Soil-adjusted Vegetation index | [22] | |
| GNDVI | Green normalized Difference vegetation | [23] | |
| EVI | Enhanced vegetation Index | [22] | |
| MCCI | Modified canopy Chlorophyll content index | [24] |
| ML | Machine Learning Model | Reference |
|---|---|---|
| ANN | Multilayer perceptron artificial neural network | [25] |
| J48 | J48 decision tree | [26] |
| RL | Logistic regression | [27] |
| DT | REPTree decision tree | [28] |
| RF | Random forest | [29] |
| Rt | Random tree decision tree | [30] |
| SVM | Support vector machine | [31] |
| Abbreviation | Accuracy | Equation |
|---|---|---|
| CC | Correct classifications | |
| F-score | F-score | |
| Kappa | Kappa coefficient |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
de Queiroz Otone, J.D.; Theodoro, G.d.F.; Santana, D.C.; Teodoro, L.P.R.; de Oliveira, J.T.; de Oliveira, I.C.; da Silva Junior, C.A.; Teodoro, P.E.; Baio, F.H.R. Hyperspectral Response of the Soybean Crop as a Function of Target Spot (Corynespora cassiicola) Using Machine Learning to Classify Severity Levels. AgriEngineering 2024, 6, 330-343. https://doi.org/10.3390/agriengineering6010020
de Queiroz Otone JD, Theodoro GdF, Santana DC, Teodoro LPR, de Oliveira JT, de Oliveira IC, da Silva Junior CA, Teodoro PE, Baio FHR. Hyperspectral Response of the Soybean Crop as a Function of Target Spot (Corynespora cassiicola) Using Machine Learning to Classify Severity Levels. AgriEngineering. 2024; 6(1):330-343. https://doi.org/10.3390/agriengineering6010020
Chicago/Turabian Stylede Queiroz Otone, José Donizete, Gustavo de Faria Theodoro, Dthenifer Cordeiro Santana, Larissa Pereira Ribeiro Teodoro, Job Teixeira de Oliveira, Izabela Cristina de Oliveira, Carlos Antonio da Silva Junior, Paulo Eduardo Teodoro, and Fabio Henrique Rojo Baio. 2024. "Hyperspectral Response of the Soybean Crop as a Function of Target Spot (Corynespora cassiicola) Using Machine Learning to Classify Severity Levels" AgriEngineering 6, no. 1: 330-343. https://doi.org/10.3390/agriengineering6010020
APA Stylede Queiroz Otone, J. D., Theodoro, G. d. F., Santana, D. C., Teodoro, L. P. R., de Oliveira, J. T., de Oliveira, I. C., da Silva Junior, C. A., Teodoro, P. E., & Baio, F. H. R. (2024). Hyperspectral Response of the Soybean Crop as a Function of Target Spot (Corynespora cassiicola) Using Machine Learning to Classify Severity Levels. AgriEngineering, 6(1), 330-343. https://doi.org/10.3390/agriengineering6010020












