Using Deep Neural Networks to Evaluate Leafminer Fly Attacks on Tomato Plants
Abstract
1. Introduction
2. Material and Methods
2.1. Database Creation
2.2. Annotation of Images and Creation of Reference Masks
2.3. Data Preprocessing
2.4. Configuration of the Experiment
2.5. Model Evaluation Metrics
- TP =
- true positive is the number of pixels correctly assigned to the evaluated semantic class (background, healthy leaf, and injured leaf);
- FP =
- false positive is the number of pixels incorrectly classified in the semantic class, although they belong to another class;
- FN =
- false negative is the number of pixels belonging to the semantic class assigned to another class.
2.6. Severity Estimation
3. Results
3.1. Comparison of Models and Backbones
3.2. Severity Estimated by Models
4. Discussion
5. Conclusions
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- FAO, FAOSTAT—Food and Agriculture Organization of the United Nations. Statistical Database 2020. 2020. Available online: http://faostat.fao.org (accessed on 17 December 2020).
- Nalam, V.; Louis, J.; Shah, J. Plant defense against aphids, the pest extraordinaire. Plant Sci. 2019, 279, 96–107. [Google Scholar] [CrossRef] [PubMed]
- Gusmão, M.R.; Picanço, M.; Leite, G.L.; Moura, M.F. Seletividade de inseticidas a predadores de pulgões. Hortic. Bras. 2000, 18, 130–133. [Google Scholar] [CrossRef]
- Picanço, M.C.; Bacci, L.; Crespo, A.L.B.; Miranda, M.M.M.; Martins, J.C. Effect of integrated pest management practices on tomato production and conservation of natural enemies. Agric. For. Entomol. 2007, 9, 327–335. [Google Scholar] [CrossRef]
- Gilbertson, R.L.; Batuman, O. Emerging viral and other diseases of processing tomatoes: Biology, diagnosis and management. Acta Hortic. 2013, 971, 35–48. [Google Scholar] [CrossRef]
- Liu, J.; Wang, X. Tomato diseases and pests detection based on improved YOLO V3 convolutional neural network. Front. Plant Sci. 2020, 11, 898. [Google Scholar] [CrossRef] [PubMed]
- Scheffer, S.J.; Hawthorne, D.J. Molecular evidence of host-associated genetic divergence in the holly leafminer Phytomyza glabricola (Diptera: Agromyzidae): Apparent discordance among marker systems. Mol. Ecol. 2007, 16, 2627–2637. [Google Scholar] [CrossRef] [PubMed]
- Johnson, M.W.; Welter, S.C.; Toscano, N.C.; Ting, P.I.; Trumble, J.T. Reduction of tomato leaf photosynthesis rates by mining activity of Liriomyza sativae (Diptera: Agromyzidae). J. Econ. Entomol. 1983, 76, 1061–1063. [Google Scholar] [CrossRef]
- Zhang, J.; Huang, Y.; Pu, R.; Gonzalez-Moreno, P.; Yuan, L.; Wu, K.; Huang, W. Monitoring plant diseases and pests through remote sensing technology: A review. Comput. Electron. Agric. 2019, 165, 104943. [Google Scholar] [CrossRef]
- Moura, A.P.; Michereff Filho, M.; Guimarães, J.A.; Liz, R.S. Manejo Integrado de Pragas do Tomateiro para Processamento Industrial; Circular Técnica; Embrapa: Brasilia, Brazil, 2014; Volume 129. [Google Scholar]
- Lin, T.L.; Chang, H.Y.; Chen, K.H. The pest and disease identification in the growth of sweet peppers using faster R-CNN and mask R-CNN. J. Internet Technol. 2020, 21, 605–614. [Google Scholar]
- Wang, K.; Zhang, S.; Wang, Z.; Liu, Z.; Yang, F. Mobile smart device-based vegetable disease and insect pest recognition method. Intell. Autom. Soft Comput. 2013, 19, 263–273. [Google Scholar] [CrossRef]
- Barbedo, J.G.A. Detecting and classifying pests in crops using proximal images and machine learning: A review. AI 2020, 1, 312–328. [Google Scholar] [CrossRef]
- Bock, C.H.; Barbedo, J.G.; Del Ponte, E.M.; Bohnenkamp, D.; Mahlein, A.K. From visual estimates to fully automated sensor-based measurements of plant disease severity: Status and challenges for improving accuracy. Phytopathol. Res. 2020, 2, 9. [Google Scholar] [CrossRef]
- Dawei, W.; Limiao, D.; Jiangong, N.; Jiyue, G.; Hongfei, Z.; Zhongzhi, H. Recognition pest by image-based transfer learning. J. Sci. Food Agric. 2019, 99, 4524–4531. [Google Scholar] [CrossRef] [PubMed]
- Liu, T.; Chen, W.; Wu, W.; Sun, C.; Guo, W.; Zhu, X. Detection of aphids in wheat fields using a computer vision technique. Biosyst. Eng. 2016, 141, 82–93. [Google Scholar] [CrossRef]
- Fuentes, A.; Yoon, S.; Kim, S.C.; Park, D.S. A robust deep-learning-based detector for real-time tomato plant diseases and pests recognition. Sensors 2017, 17, 2022. [Google Scholar] [CrossRef]
- Huang, M.; Wan, X.; Zhang, M.; Zhu, Q. Detection of insect-damaged vegetable soybeans using hyperspectral transmittance image. J. Food Eng. 2013, 116, 45–49. [Google Scholar] [CrossRef]
- Ma, Y.; Huang, M.; Yang, B.; Zhu, Q. Automatic threshold method and optimal wavelength selection for insect-damaged vegetable soybean detection using hyperspectral images. Comput. Electron. Agric. 2014, 106, 102–110. [Google Scholar] [CrossRef]
- Barbedo, J.G.A. A review on the main challenges in automatic plant disease identification based on visible range images. Biosyst. Eng. 2016, 144, 52–60. [Google Scholar] [CrossRef]
- Divya, B.; Santhi, M. SVM-based pest classification in agriculture field. Int. J. Recent Technol. Eng. 2019, 7, 150–155. [Google Scholar]
- Mustafa, W.A.; Yazid, H. Illumination and contrast correction strategy using bilateral filtering and binarization comparison. J. Telecommun. Electron. Comput. Eng. 2016, 8, 67–73. [Google Scholar]
- Chen, S.; Zhang, K.; Zhao, Y.; Sun, Y.; Ban, W.; Chen, Y.; Zhuang, H.; Zhang, X.; Liu, J.; Yang, T. An approach for rice bacterial leaf streak disease segmentation and disease severity estimation. Agriculture 2021, 11, 420. [Google Scholar] [CrossRef]
- Cheng, X.; Zhang, Y.; Chen, Y.; Wu, Y.; Yue, Y. Pest identification via deep residual learning in complex background. Comput. Electron. Agric. 2017, 141, 351–356. [Google Scholar] [CrossRef]
- Chowdhury, M.E.; Rahman, T.; Khandakar, A.; Ayari, M.A.; Khan, A.U.; Khan, M.S.; Al-Emadi, N.; Reaz, M.B.I.; Islam, M.T.; Ali, S.H.M. Automatic and reliable leaf disease detection using deep learning techniques. AgriEngineering 2021, 3, 294–312. [Google Scholar] [CrossRef]
- Fuentes, A.F.; Yoon, S.; Lee, J.; Park, D.S. High-performance deep neural network-based tomato plant diseases and pests diagnosis system with refinement filter bank. Front. Plant Sci. 2018, 9, 1–15. [Google Scholar] [CrossRef] [PubMed]
- Hidayatuloh, A.; Nursalman, M.; Nugraha, E. Identification of tomato plant diseases by leaf image using squeezenet model. In Proceedings of the 2018 International Conference on Information Technology Systems and Innovation, ICITSI 2018, Bandung, Indonesia, 22–26 October 2018; pp. 199–204. [Google Scholar]
- Weng, W.; Zhu, X. INet: Convolutional networks for biomedical image segmentation. IEEE Access 2021, 9, 16591–16603. [Google Scholar] [CrossRef]
- Ronneberger, O.; Fischer, P.; Brox, T. U-Net: Convolutional networks for biomedical image segmentation. In Proceedings of the International Conference on Medical Image Computing and Computer-assisted Intervention, Munich, Germany, 5–9 October 2015; Springer: Berlin/Heidelberg, Germany, 2015; pp. 234–241. [Google Scholar]
- Lin, T.Y.; Dollár, P.; Girshick, R.; He, K.; Hariharan, B.; Belongie, S. Feature pyramid networks for object detection. In Proceedings of the 30th IEEE Conference on Computer Vision and Pattern Recognition, CVPR 2017, Honolulu, HI, USA, 21–26 July 2017; pp. 936–944. [Google Scholar]
- Seferbekov, S.; Iglovikov, V.; Buslaev, A.; Shvets, A. Feature pyramid network for multi-class land segmentation. In Proceedings of the IEEE Computer Society Conference on Computer Vision and Pattern Recognition Workshops, Salt Lake City, UT, USA, 18–22 June 2018; pp. 272–275. [Google Scholar]
- Chaurasia, A.; Culurciello, E. LinkNet: Exploiting encoder representations for efficient semantic segmentation. In Proceedings of the 2017 IEEE Visual Communications and Image Processing, IEEE, St. Petersburg, FL, USA, 10–13 December 2017; pp. 1–4. [Google Scholar]
- Dhiman, P.; Kukreja, V.; Manoharan, P.; Kaur, A.; Kamruzzaman, M.M.; Dhaou, I.B.; Iwendi, C. A novel deep learning model for detection of severity level of the disease in citrus fruits. Electronics 2022, 11, 495. [Google Scholar] [CrossRef]
- Wang, J.; Perez, L. The effectiveness of data augmentation in image classification using deep learning. Convolutional Neural Netw. Vis. Recognit. 2017, 11, 1–8. [Google Scholar]
- Takahashi, R.; Matsubara, T.; Uehara, K. Data augmentation using random image cropping and patching for deep CNNs. IEEE Trans. Circuits Syst. Video Technol. 2020, 30, 2917–2931. [Google Scholar] [CrossRef]
- Szegedy, C.; Vanhoucke, V.; Ioffe, S.; Shlens, J.; Wojna, Z. Rethinking the inception architecture for computer vision. In Proceedings of the IEEE Computer Society Conference on Computer Vision and Pattern Recognition, Las Vegas, NV, USA, 27–30 June 2016; pp. 2818–2826. [Google Scholar]
- Russakovsky, O.; Deng, J.; Su, H.; Krause, J.; Satheesh, S.; Ma, S.; Huang, Z.; Karpathy, A.; Khosla, A.; Bernstein, M.; et al. ImageNet large scale visual recognition challenge. Int. J. Comput. Vis. 2015, 115, 211–252. [Google Scholar] [CrossRef]
- Freudenberg, M.; Nölke, N.; Agostini, A.; Urban, K.; Wörgötter, F.; Kleinn, C. Large scale palm tree detection in high resolution satellite images using U-Net. Remote Sens. 2019, 11, 312. [Google Scholar] [CrossRef]
- Wang, C.; Zhao, Z.; Yu, Y. Fine retinal vessel segmentation by combining nest U-Net and patch-learning. Soft Comput. 2021, 25, 5519–5532. [Google Scholar] [CrossRef]
- Jadon, S. A survey of loss functions for semantic segmentation. In Proceedings of the 2020 IEEE Conference on Computational Intelligence in Bioinformatics and Computational Biology, CIBCB 2020, Online. 27–29 October 2020. [Google Scholar]
- Lin, T.Y.; Goyal, P.; Girshick, R.; He, K.; Dollár, P. Focal loss for dense object detection. IEEE Trans. Pattern Anal. Mach. Intell. 2020, 42, 318–327. [Google Scholar] [CrossRef] [PubMed]
- Bock, C.H.; Parker, P.E.; Cook, A.Z.; Gottwald, T.R. Characteristics of the perception of different severity measures of citrus canker and the relationships between the various symptom types. Plant Dis. 2008, 92, 927–939. [Google Scholar] [CrossRef] [PubMed]
- Bock, C.H.; Poole, G.H.; Parker, P.E.; Gottwald, T.R. Plant disease severity estimated visually, by digital photography and image analysis, and by hyperspectral imaging. Crit. Rev. Plant Sci. 2010, 29, 59–107. [Google Scholar] [CrossRef]
- Madden, L.; Hughes, G.; Van Den Bosch, F. Measuring Plant Diseases. In the Study of Plant Disease Epidemics; American Phytopathological Society: St. Paul, MN, USA, 2007; pp. 11–31. [Google Scholar]
- Piao, S.; Liu, J. Accuracy improvement of UNet based on dilated convolution. J. Phys. Conf. Ser. 2019, 1345, 052066. [Google Scholar] [CrossRef]
- Gonçalves, J.P.; Pinto, F.A.C.; Queiroz, D.M.; Villar, F.M.M.; Barbedo, J.G.A.; Del Ponte, E.M. Deep learning architectures for semantic segmentation and automatic estimation of severity of foliar symptoms causedby diseases or pests. Biosyst. Eng. 2021, 210, 129–142. [Google Scholar] [CrossRef]
- Torres, D.L.; Feitosa, R.Q.; Happ, P.N.; La Rosa, L.E.C.; Marcato Junior, J.; Martins, J.; Bressan, P.O.; Gonçalves, W.N.; Liesenberg, V. Applying fully convolutional architectures for semantic segmentation of a single tree species in urban environment on high resolution UAV optical imagery. Sensors 2020, 20, 563. [Google Scholar] [CrossRef]
- Zhu, Q.; Zheng, Y.; Jiang, Y.; Yang, J. Efficient multi-class semantic segmentation of high resolution aerial imagery with dilated linknet. In Proceedings of the International Geoscience and Remote Sensing Symposium (IGARSS), Yokohama, Japan, 28 July–2 August 2019; pp. 1065–1068. [Google Scholar]
- Esgario, J.G.; Krohling, R.A.; Ventura, J.A. Deep learning for classification and severity estimation of coffee leaf biotc stress. Comput. Eletronics Agric. 2020, 169, 105162. [Google Scholar] [CrossRef]
- Cui, B.; Chen, X.; Lu, Y. Semantic segmentation of remote sensing images using transfer learning and deep convolutional neural network with dense connection. IEEE Access 2020, 8, 116744–116755. [Google Scholar] [CrossRef]
- Rahman, M.A.; Wang, Y. Optimizing intersection-over-union in deep neural networks for image segmentation. In Lecture Notes in Computer Science; including subseries Lecture Notes in Artificial Intelligence and Lecture Notes in Bioinformatics; Springer International publishing: Las Vegas, NV, USA, 2016; Volume 10072, pp. 234–244. [Google Scholar]


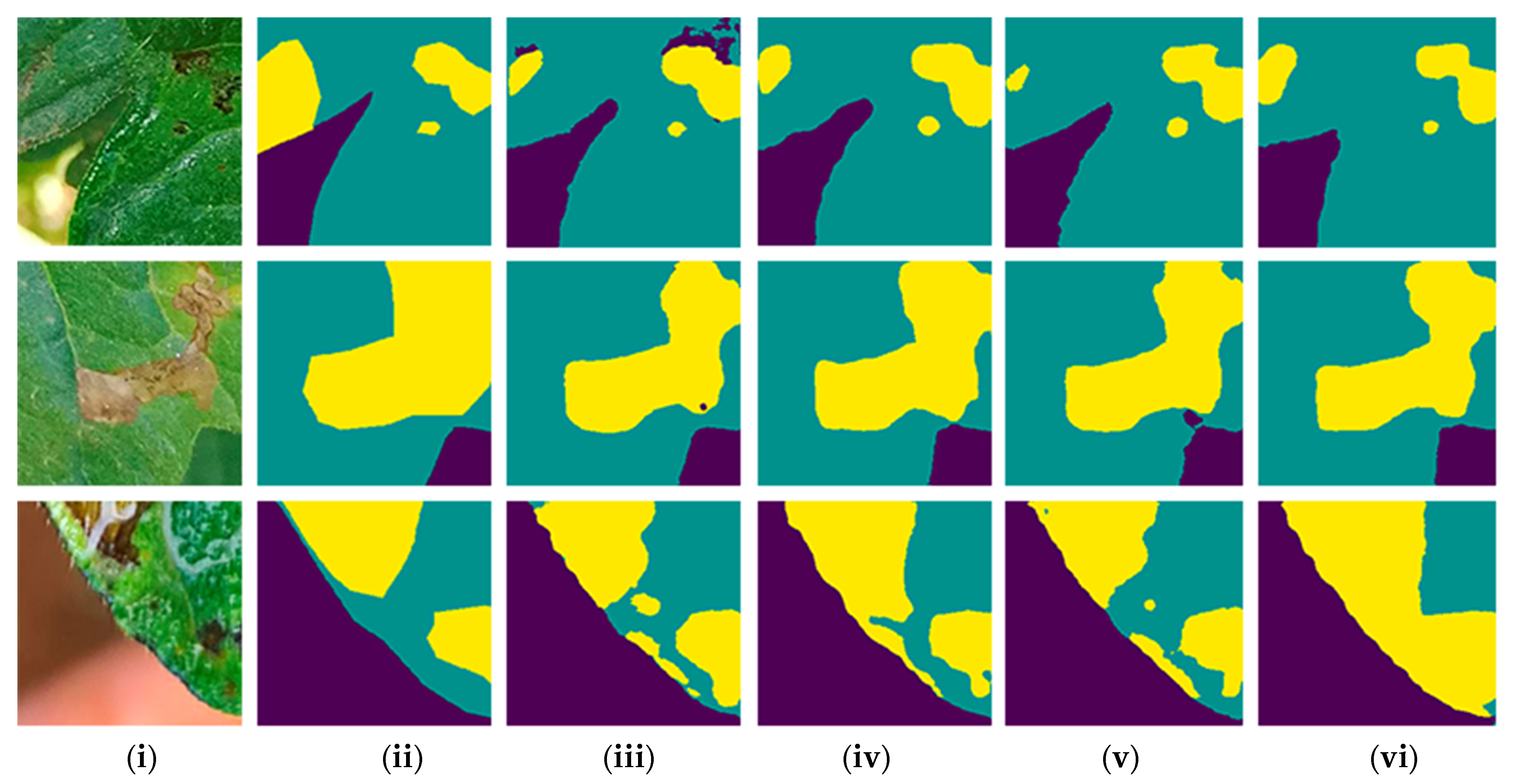
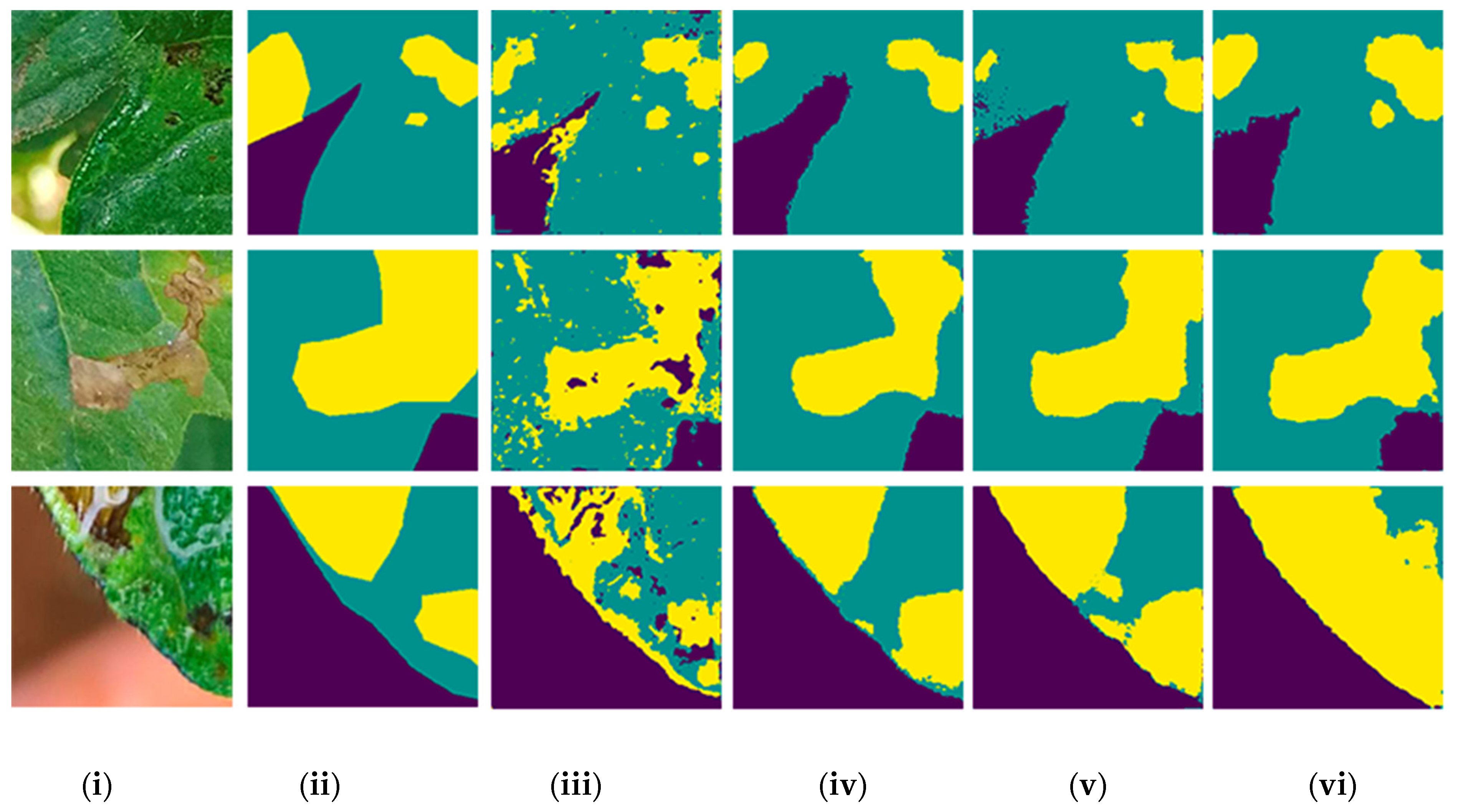
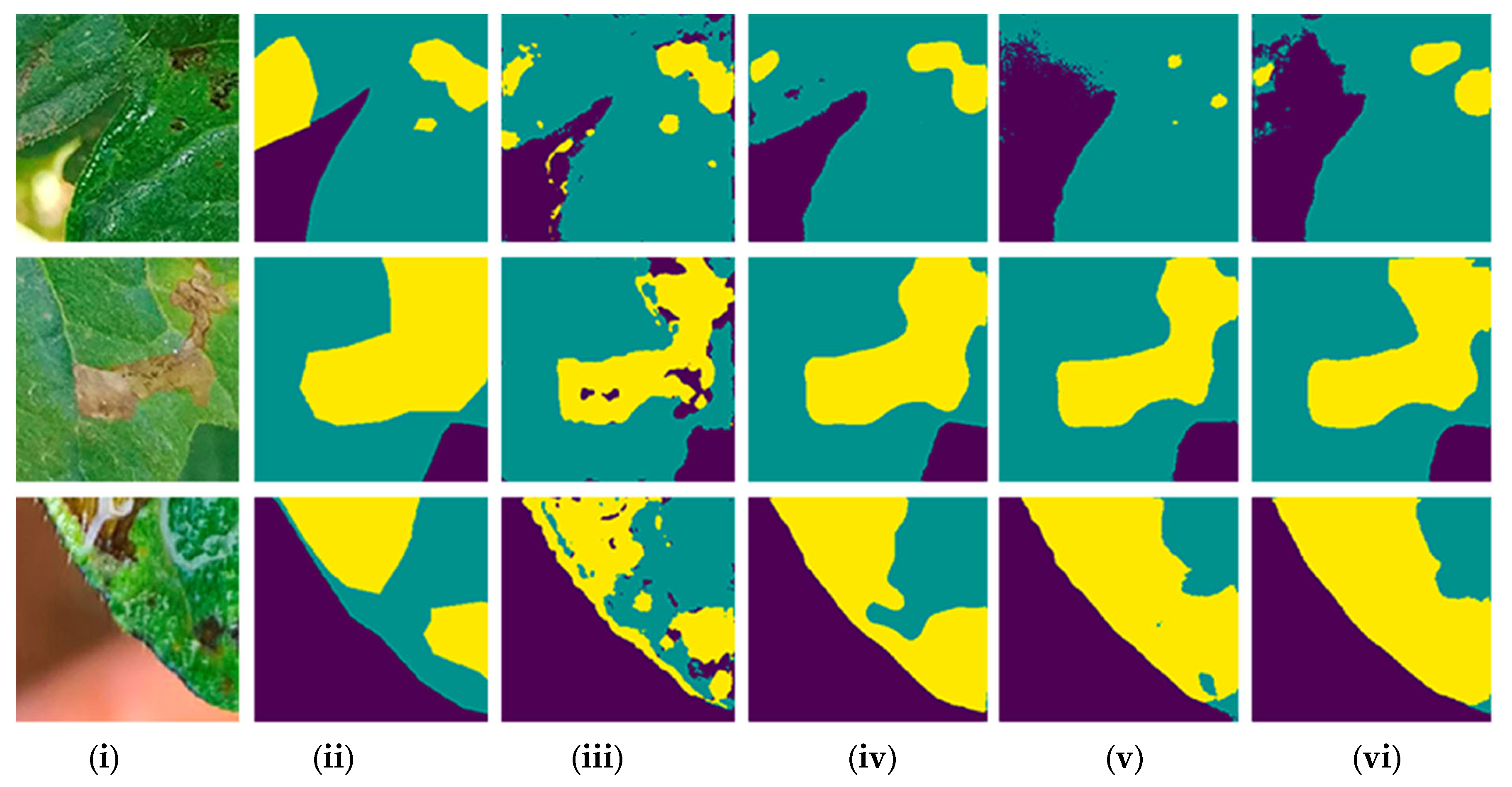



| Category | Deep Learning Model | Backbone | Class | Average IoU (%) |
|---|---|---|---|---|
| Best models | U-Net | Inceptionv3 | Background | 86 |
| U-Net | Inceptionv3 | Leaf | 87 | |
| FPN | DenseNet121 | Symptoms | 61 | |
| Worst models | U-Net | VGG16 | Background | 65 |
| LinkNet | VGG16 | Leaf | 69 | |
| LinkNet | VGG16 | Symptoms | 25 |
| Deep Learning Model | Backbone | Test Accuracy (%) | Average Precision (%) | Average Recall (%) | Average IoU (%) |
|---|---|---|---|---|---|
| U-Net | VGG16 | 83.90 | 80.63 | 78.31 | 61.76 |
| ResNet34 | 90.00 | 86.63 | 85.81 | 72.73 | |
| Inceptionv3 | 91.58 | 87.84 | 87.59 | 77.71 | |
| DenseNet121 | 91.25 | 87.83 | 87.74 | 76.33 | |
| LinkNet | VGG16 | 81.23 | 73.94 | 67.87 | 53.03 |
| ResNet34 | 88.66 | 84.62 | 84.77 | 73.21 | |
| Inceptionv3 | 91.06 | 87.09 | 87.39 | 75.67 | |
| DenseNet121 | 90.72 | 87.00 | 86.79 | 75.99 | |
| FPN | VGG16 | 89.27 | 85.58 | 84.31 | 73.12 |
| ResNet34 | 90.53 | 87.29 | 86.27 | 74.61 | |
| Inceptionv3 | 91.10 | 87.98 | 86.82 | 75.12 | |
| DenseNet121 | 91.56 | 88.63 | 87.71 | 76.62 |
| Backbone | Deep Learning Model | ||
|---|---|---|---|
| U-Net | LinkNet | FPN | |
| Trainable Parameters | Trainable Parameters | Trainable Parameters | |
| VGG16 | 23,748,531 | 20,318,611 | 17,572,547 |
| ResNet34 | 24,439,094 | 21,620,118 | 23,915,590 |
| Inceptionv3 | 29,896,979 | 26,228,243 | 24,994,851 |
| DenseNet121 | 12,059,635 | 8,267,411 | 9,828,099 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Martins Crispi, G.; Valente, D.S.M.; Queiroz, D.M.d.; Momin, A.; Fernandes-Filho, E.I.; Picanço, M.C. Using Deep Neural Networks to Evaluate Leafminer Fly Attacks on Tomato Plants. AgriEngineering 2023, 5, 273-286. https://doi.org/10.3390/agriengineering5010018
Martins Crispi G, Valente DSM, Queiroz DMd, Momin A, Fernandes-Filho EI, Picanço MC. Using Deep Neural Networks to Evaluate Leafminer Fly Attacks on Tomato Plants. AgriEngineering. 2023; 5(1):273-286. https://doi.org/10.3390/agriengineering5010018
Chicago/Turabian StyleMartins Crispi, Guilhermi, Domingos Sárvio Magalhães Valente, Daniel Marçal de Queiroz, Abdul Momin, Elpídio Inácio Fernandes-Filho, and Marcelo Coutinho Picanço. 2023. "Using Deep Neural Networks to Evaluate Leafminer Fly Attacks on Tomato Plants" AgriEngineering 5, no. 1: 273-286. https://doi.org/10.3390/agriengineering5010018
APA StyleMartins Crispi, G., Valente, D. S. M., Queiroz, D. M. d., Momin, A., Fernandes-Filho, E. I., & Picanço, M. C. (2023). Using Deep Neural Networks to Evaluate Leafminer Fly Attacks on Tomato Plants. AgriEngineering, 5(1), 273-286. https://doi.org/10.3390/agriengineering5010018








