Power–Cadence Relationships in Cycling: Building Models from a Limited Number of Data Points
Abstract
1. Introduction
2. Materials and Methods
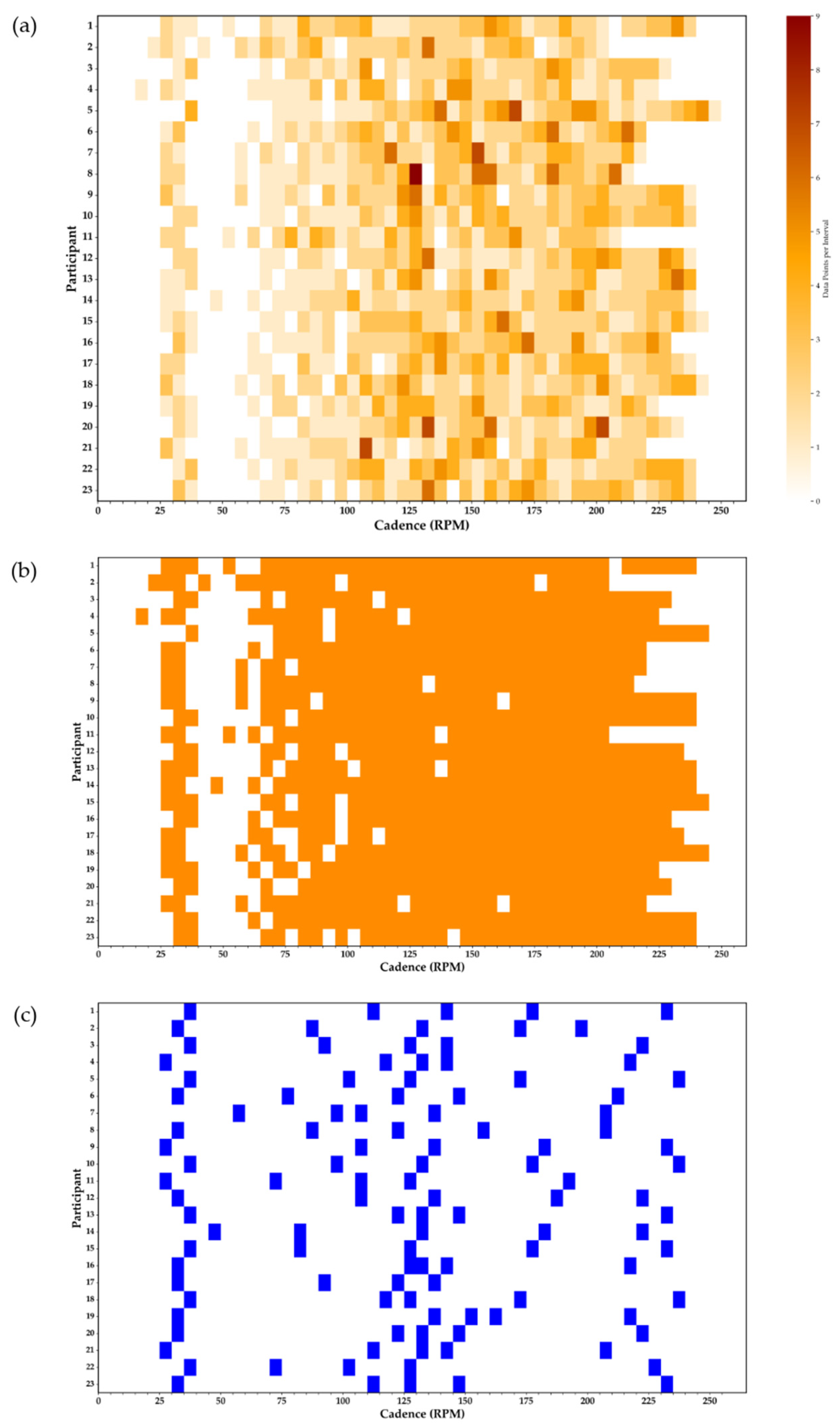
- Moderate-Number (MN) model: Included only the point with the highest positive residual (i.e., furthest above the fitted curve) in each 5-RPM cadence interval. This method, previously described [5], is intended to eliminate submaximal points and retain the most likely maximal point per interval (Figure 1b and Figure 2c, Table 1). On average, MN models included 34 ± 2 data points per participant (about 40% of the HN dataset).
- Low-Number (LN) model: Included only five data points selected from strategic portions of the power–cadence relationship to ensure proper representation of three critical regions: (i) low cadences to inform the linear coefficient (ax), (ii) near the apex to capture peak curvature (bx2), and (iii) high cadences to define the descending portion of the curve (cx3). These data points were selected as (i) the point with the highest residual in the low-cadence range (0–60 rpm), (ii) the point of peak power output (Ppeak), the point with the highest residual 5–50 rpm below Ppeak cadence (left side of the apex), and the point with the highest residual 5–50 rpm above Ppeak cadence (right side of the apex), and (iii) the point with the highest residual within 10 rpm of the maximal cadence recorded during the flywheel-only sprint. This method was designed to capture strategically located maximal values for optimal estimation of Equation (1) coefficients (Figure 1c and Figure 2d, Table 1).
3. Results
3.1. Goodness of Fit of the Models
3.2. Optimal Cadence (Copt): X-Coordinate of the Curve Apex
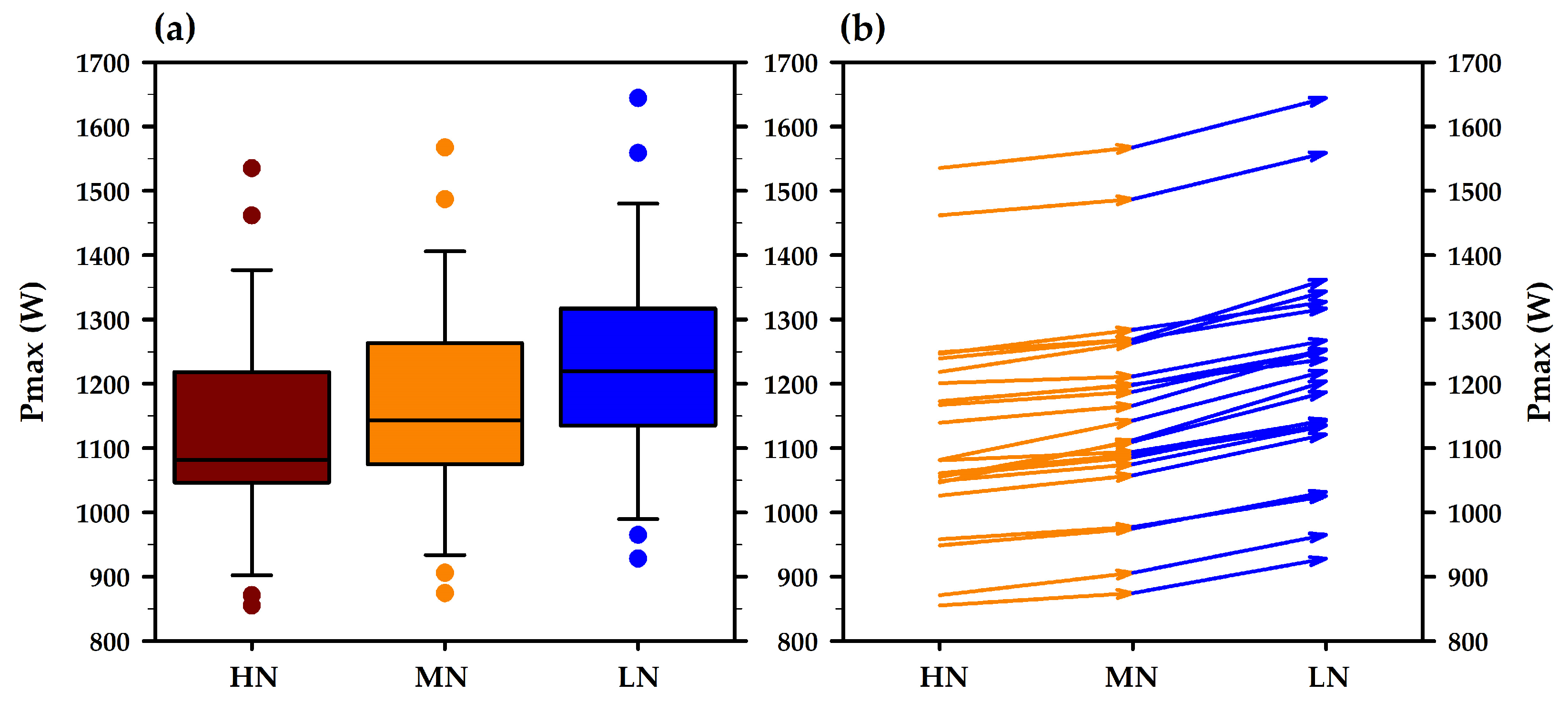
3.3. Modeled Maximal Power Output (Pmax): Y-Coordinate of the Curve Apex
3.4. Accuracy of the Models for Predicting Ppeak
3.5. Sensitivity Analysis of the LN Model
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| Cppeak | Cadence at the highest level of power measured experimentally |
| Copt | x-coordinate of the apex of the modeled power vs. cadence relationship |
| HN | High-number of data points model |
| LN | Low-number of data points model |
| LLO | Leave-One-Out sensitivity analysis |
| MAE | Mean Absolute Error |
| MAPE | Mean Absolute Percentage Error |
| MN | Moderate-number of data points model |
| Pmax | y-coordinate of the apex of the modeled power vs. cadence relationship |
| Ppeak | Highest level of power measured experimentally |
| RPM | Rotation per minute |
| SEE | Standard Error of the Estimate |
| W | Watt |
References
- Dotan, R.; Bar-Or, O. Load optimization for the Wingate Anaerobic Test. Eur. J. Appl. Physiol. Occup. Physiol. 1983, 51, 409–417. [Google Scholar] [CrossRef] [PubMed]
- Driss, T.; Vandewalle, H. The measurement of maximal (anaerobic) power output on a cycle ergometer: A critical review. Biomed. Res. Int. 2013, 2013, 589361. [Google Scholar] [CrossRef]
- Sargeant, A.J.; Dolan, P.; Young, A. Optimal Velocity for Maximal Short-term (Anaerobic) Power Output in Cycling. Int. J. Sports Med. 1984, 5, 124–125. [Google Scholar] [CrossRef]
- Vandewalle, H.; Peres, G.; Heller, J.; Monod, H. All out anaerobic capacity tests on cycle ergometers. A comparative study on men and women. Eur. J. Appl. Physiol. Occup. Physiol. 1985, 54, 222–229. [Google Scholar] [CrossRef]
- Rudsits, B.L.; Hopkins, W.G.; Hautier, C.A.; Rouffet, D.M. Force-velocity test on a stationary cycle ergometer: Methodological recommendations. J. Appl. Physiol. 2018, 124, 831–839. [Google Scholar] [CrossRef]
- Bertucci, W.; Taiar, R.; Grappe, F. Differences between sprint tests under laboratory and actual cycling conditions. J. Sports Med. Phys. Fit. 2005, 45, 277–283. [Google Scholar]
- Dorel, S.; Hautier, C.A.; Rambaud, O.; Rouffet, D.; Van Praagh, E.; Lacour, J.R.; Bourdin, M. Torque and power-velocity relationships in cycling: Relevance to track sprint performance in world-class cyclists. Int. J. Sports Med. 2005, 26, 739–746. [Google Scholar] [CrossRef]
- Gardner, A.S.; Martin, J.C.; Martin, D.T.; Barras, M.; Jenkins, D.G. Maximal torque- and power-pedaling rate relationships for elite sprint cyclists in laboratory and field tests. Eur. J. Appl. Physiol. 2007, 101, 287–292. [Google Scholar] [CrossRef]
- Vandewalle, H.; Peres, G.; Heller, J.; Panel, J.; Monod, H. Force-velocity relationship and maximal power on a cycle ergometer. Correlation with the height of a vertical jump. Eur. J. Appl. Physiol. Occup. Physiol. 1987, 56, 650–656. [Google Scholar] [CrossRef]
- Wackwitz, T.A.; Minahan, C.L.; King, T.; Du Plessis, C.; Andrews, M.H.; Bellinger, P.M. Quantification of maximal power output in well-trained cyclists. J. Sports Sci. 2021, 39, 84–90. [Google Scholar] [CrossRef]
- Yeo, B.K.; Rouffet, D.M.; Bonanno, D.R. Foot orthoses do not affect crank power output during maximal exercise on a cycle-ergometer. J. Sci. Med. Sport. 2016, 19, 368–372. [Google Scholar] [CrossRef] [PubMed]
- Dwyer, D.B.; Molaro, C.; Rouffet, D.M. Force-velocity profiles of track cyclists differ between seated and non-seated positions. Sports Biomech. 2023, 22, 621–632. [Google Scholar] [CrossRef]
- Taylor, K.B.; Deckert, S.; Sanders, D. Field-testing to determine power—and torque—cadence profiles in professional road cyclists. Eur. J. Sport. Sci. 2023, 23, 1085–1093. [Google Scholar] [CrossRef] [PubMed]
- Crielaard, J.M.; Pirnay, F. Anaerobic and aerobic power of top athletes. Eur. J. Appl. Physiol. Occup. Physiol. 1981, 47, 295–300. [Google Scholar] [CrossRef]
- Kordi, M.; Folland, J.; Goodall, S.; Barratt, P.; Howatson, G. Isovelocity vs. Isoinertial Sprint Cycling Tests for Power- and Torque-cadence Relationships. Int. J. Sports Med. 2019, 40, 897–902. [Google Scholar] [CrossRef]
- Buttelli, O.; Seck, D.; Vandewalle, H.; Jouanin, J.C.; Monod, H. Effect of fatigue on maximal velocity and maximal torque during short exhausting cycling. Eur. J. Appl. Physiol. Occup. Physiol. 1996, 73, 175–179. [Google Scholar] [CrossRef]
- Gardner, A.S.; Martin, D.T.; Jenkins, D.G.; Dyer, I.; Van Eiden, J.; Barras, M.; Martin, J.C. Velocity-specific fatigue: Quantifying fatigue during variable velocity cycling. Med. Sci. Sports Exerc. 2009, 41, 904–911. [Google Scholar] [CrossRef]
- McCartney, N.; Heigenhauser, G.J.; Jones, N.L. Power output and fatigue of human muscle in maximal cycling exercise. J. Appl. Physiol. Respir. Environ. Exerc. Physiol. 1983, 55, 218–224. [Google Scholar] [CrossRef]
- O’Bryan, S.J.; Taylor, J.L.; D’Amico, J.M.; Rouffet, D.M. Quadriceps Muscle Fatigue Reduces Extension and Flexion Power During Maximal Cycling. Front. Sports Act. Living 2021, 3, 797288. [Google Scholar] [CrossRef]
- Davies, C.T.; Young, K. Effects of external loading on short term power output in children and young male adults. Eur. J. Appl. Physiol. Occup. Physiol. 1984, 52, 351–354. [Google Scholar] [CrossRef]
- Doré, E.; Duché, P.; Rouffet, D.; Ratel, S.; Bedu, M.; Van Praagh, E. Measurement error in short-term power testing in young people. J. Sports Sci. 2003, 21, 135–142. [Google Scholar] [CrossRef]
- Arsac, L.M.; Belli, A.; Lacour, J.R. Muscle function during brief maximal exercise: Accurate measurements on a friction-loaded cycle ergometer. Eur. J. Appl. Physiol. Occup. Physiol. 1996, 74, 100–106. [Google Scholar] [CrossRef] [PubMed]
- Capriotti, P.V.; Sherman, W.M.; Lamb, D.R. Reliability of power output during intermittent high-intensity cycling. Med. Sci. Sports Exerc. 1999, 31, 913–915. [Google Scholar] [CrossRef]
- Martin, J.C.; Diedrich, D.; Coyle, E.F. Time course of learning to produce maximum cycling power. Int. J. Sports Med. 2000, 21, 485–487. [Google Scholar] [CrossRef]
- Davies, C.T.; Sandstrom, E.R. Maximal mechanical power output and capacity of cyclists and young adults. Eur. J. Appl. Physiol. Occup. Physiol. 1989, 58, 838–844. [Google Scholar] [CrossRef]
- Driss, T.; Vandewalle, H.; Chevalier, J.M.L.; Monod, H. Force-velocity relationship on a cycle ergometer and knee-extensor strength indices. Can. J. Appl. Physiol. 2002, 27, 250–262. [Google Scholar] [CrossRef]
- Driss, T.; Vandewalle, H.; Monod, H. Maximal power and force-velocity relationships during cycling and cranking exercises in volleyball players. Correlation with the vertical jump test. J. Sports Med. Phys. Fit. 1998, 38, 286–293. [Google Scholar]
- Seck, D.; Vandewalle, H.; Decrops, N.; Monod, H. Maximal power and torque-velocity relationship on a cycle ergometer during the acceleration phase of a single all-out exercise. Eur. J. Appl. Physiol. Occup. Physiol. 1995, 70, 161–168. [Google Scholar] [CrossRef]
- Dorel, S.; Couturier, A.; Lacour, J.R.; Vandewalle, H.; Hautier, C.; Hug, F. Force-velocity relationship in cycling revisited: Benefit of two-dimensional pedal forces analysis. Med. Sci. Sports Exerc. 2010, 42, 1174–1183. [Google Scholar] [CrossRef]
- Kruger, R.L.; Peyrard, A.; di Domenico, H.; Rupp, T.; Millet, G.Y.; Samozino, P. Optimal load for a torque-velocity relationship test during cycling. Eur. J. Appl. Physiol. 2020, 120, 2455–2466. [Google Scholar] [CrossRef]
- Nadeau, M.; Brassard, A.; Cuerrier, J.P. The bicycle ergometer for muscle power testing. Can. J. Appl. Sport. Sci. 1983, 8, 41–46. [Google Scholar]
- Leo, P.; Martinez-Gonzalez, B.; Mujika, I.; Giorgi, A. Mechanistic influence of the torque cadence relationship on power output during exhaustive all-out field tests in professional cyclists. J. Sports Sci. 2025, 43, 887–894. [Google Scholar] [CrossRef] [PubMed]
- Bogdanis, G.C.; Papaspyrou, A.; Theos, A.; Maridaki, M. Influence of resistive load on power output and fatigue during intermittent sprint cycling exercise in children. Eur. J. Appl. Physiol. 2007, 101, 313–320. [Google Scholar] [CrossRef] [PubMed]
- Capmal, S.; Vandewalle, H. Torque-velocity relationship during cycle ergometer sprints with and without toe clips. Eur. J. Appl. Physiol. Occup. Physiol. 1997, 76, 375–379. [Google Scholar] [CrossRef]
- Linossier, M.T.; Denis, C.; Dormois, D.; Geyssant, A.; Lacour, J.R. Ergometric and metabolic adaptation to a 5-s sprint training programme. Eur. J. Appl. Physiol. Occup. Physiol. 1993, 67, 408–414. [Google Scholar] [CrossRef]
- Martin, J.C.; Wagner, B.M.; Coyle, E.F. Inertial-load method determines maximal cycling power in a single exercise bout. Med. Sci. Sports Exerc. 1997, 29, 1505–1512. [Google Scholar] [CrossRef]
- McCartney, N.; Obminski, G.; Heigenhauser, G.J. Torque-velocity relationship in isokinetic cycling exercise. J. Appl. Physiol. 1985, 58, 1459–1462. [Google Scholar] [CrossRef]
- Sargeant, A.J.; Hoinville, E.; Young, A. Maximum leg force and power output during short-term dynamic exercise. J. Appl. Physiol. Respir. Environ. Exerc. Physiol. 1981, 51, 1175–1182. [Google Scholar] [CrossRef]
- Hautier, C.A.; Linossier, M.T.; Belli, A.; Lacour, J.R.; Arsac, L.M. Optimal velocity for maximal power production in non-isokinetic cycling is related to muscle fibre type composition. Eur. J. Appl. Physiol. Occup. Physiol. 1996, 74, 114–118. [Google Scholar] [CrossRef]
- Rouffet, D.M. Étude de L’activation Musculaire À L’aide de L’électromyographie de Surface—Aspects Méthodologiques et Apports Dans L’analyse du Mouvement Humain. Ph.D. Thesis, Université Claude Bernard Lyon 1, Villeurbanne, France, 2007. [Google Scholar]
- Lakomy, H.K. Measurement of work and power output using friction-loaded cycle ergometers. Ergonomics 1986, 29, 509–517. [Google Scholar] [CrossRef]
- Morin, J.B.; Belli, A. A simple method for measurement of maximal downstroke power on friction-loaded cycle ergometer. J. Biomech. 2004, 37, 141–145. [Google Scholar] [CrossRef] [PubMed]




| HN | MN | LN | |
| Data points (n) | 81 ± 7 | 34 ± 2 | 5 ± 0 |
| Min cadence (RPM) | 31 ± 3 | 33 ± 2 | 35 ± 6 |
| Max cadence (RPM) | 229 ± 12 | 229 ± 12 | 216 ± 23 |
| Exp. Data | HN | MN | LN | |
| R2 | 0.938 ± 0.027 | 0.962 ± 0.014 | 0.995 ± 0.008 | |
| SEE (W) | 72 ± 17 | 53 ± 14 | 29 ± 15 | |
| Ppeak/Pmax (W) | 1218 ± 178 | 1126 ± 163 | 1157 ± 162 | 1220 ± 168 |
| Cppeak/Copt (RPM) | 127 ± 11 | 125 ± 8 | 127 ± 8 | 126 ± 8 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Rouffet, D.M.; Rudsits, B.L.; Daniels, M.W.; Ariyo, T.; Hautier, C.A. Power–Cadence Relationships in Cycling: Building Models from a Limited Number of Data Points. Signals 2025, 6, 32. https://doi.org/10.3390/signals6030032
Rouffet DM, Rudsits BL, Daniels MW, Ariyo T, Hautier CA. Power–Cadence Relationships in Cycling: Building Models from a Limited Number of Data Points. Signals. 2025; 6(3):32. https://doi.org/10.3390/signals6030032
Chicago/Turabian StyleRouffet, David M., Briar L. Rudsits, Michael W. Daniels, Temi Ariyo, and Christophe A. Hautier. 2025. "Power–Cadence Relationships in Cycling: Building Models from a Limited Number of Data Points" Signals 6, no. 3: 32. https://doi.org/10.3390/signals6030032
APA StyleRouffet, D. M., Rudsits, B. L., Daniels, M. W., Ariyo, T., & Hautier, C. A. (2025). Power–Cadence Relationships in Cycling: Building Models from a Limited Number of Data Points. Signals, 6(3), 32. https://doi.org/10.3390/signals6030032







