1. Introduction
Cancer is one of the most common causes of death worldwide with millions of cases being diagnosed every year. Therapy and identification of cancer constitute one of the most prominent research fields in the international community, with numerous innovative approaches being developed for early diagnosis and efficient management of the disease. Surgical removal of the primary cancer with or without adjuvant therapy constitutes the standard of care for the disease. Upon tumor resection, the medical expert who is performing the operation can form an abstract visual picture of the major area of malignant tissue but not the “edges” (points formulating its boundary) at a microscopic level, which might be penetrating into the surrounding region of non-malignant tissue area [
1]. To ensure the adequate removal of the cancerous tissue, the protocol requires removing enough normal tissue to achieve a “clean margin” while maintaining the functionality and formation of the organ. In addition, tumor margins are routinely assessed post-operatively for pathological conditions to ensure that the tumor was effectively eliminated. A major reason is that incompletely resected margins require re-excision, causing additional corporal and psychological encumbrance to the patient. Moreover, the medical expert also takes into consideration the small, yet aggravating possibility of local recurrence in treated patients triggered by miss-detected marginal regions of the tumor during pathological sampling [
2]. Therefore, except for tumor detection, it is of paramount importance to trace and further examine the accurate position and structural composition of the cancerous tissue segments so as to facilitate efficient excision, while, in parallel, making certain that, firstly, healthy tissue is not damaged and, secondly, the possibility of cancer recurrence is sufficiently decreased (at least in terms of unsuccessful surgery or surgical failures). Existing imaging modalities, such as positron emission tomography (PET) and magnetic resonance imaging (MRI), experience difficulties in precisely discriminating cancer from normal tissue at the tumor margins. However, it is essential that this specific limitation is resolved promptly, as guided surgery requires the proper detection of tumor margins and identification of its morphological characteristics intraoperatively. Hence, for resection of malignant tissue, developing a competent framework for demarcating tumor margins non-invasively could prove extremely beneficial in clinical practice. It may both facilitate efficient removal of cancer via surgical procedures and prevent the disease from occurring again [
2,
3].
Brain tumors are considered rather rare yet fatal cancers with intrinsic difficulties in identifying health risks and determinant conditions in the overall community. Several of these kinds of malignancy are by nature refractive to treat due to their position in the brain. Surgery solutions can be adopted for low-grade tumors, while chemotherapy and radiotherapy could serve as possible choices upon visible malignant tissue remaining after surgical procedures. For higher-grade disease, a combination of these approaches could be utilized in therapy plans. Yet, all three treatment options sustain increased risk for long-term morbidity of patients and insufficient cure of the disease. Methodologies for early identification and diagnosis of brain tumors are fundamental to prevent destruction or deterioration of brain cells due to disease evolution or therapy side effects. The key step for developing such methods lies in studying and deeply understanding the mechanisms beyond brain tumor angiogenesis and growth. Nervous system tumors (including brain and other types) are the 10th leading cause of death for both genders. Based on epidemiological predictions, it is estimated that 18,020 adults, namely, 10,190 male and 7830 female, lost their lives from malignant brain and central nervous system (CNS) tumors in 2020. Another important statistical finding is that the 5-year survival rate for patients who suffer from these types of cancer is nearly 36%, while the 10-year survival rate is almost 31%, which decreases with increasing age. At younger ages, i.e., for populations between fifteen to thirty-nine years old, the 5-year survival rate is almost 71%. The 5-year survival rate for people over forty years old drops to 21%. However, survival rates are dependent on several factors, such as genetic, environmental, immunological, biochemical, and biological factors, including the location and kind of spinal cord or brain tumor. For instance, environmental conditions are considered as potential risk factors for carcinogenesis of brain tumors by means of radiation exposure, poisonous agents (N-nitroso composites, plant protection products), atmospheric pollution, and electromagnetic radiation through radio waves. However, for certain contributing factors, there is no definite scientific evidence to advocate their correlation. On the other hand, 5% of cerebral tumors are linked to various hereditary cancer predisposition syndromes. People with these disorders inherit a germline mutation in a tumor suppressor gene. Although the relationship between hereditary genetic factors and brain tumors has been established, greater efforts should be focused on identifying the underlying genes. Finally, immunological, biological, and biochemical factors such as exposure to infections, viruses, and allergens, may be among the risk factors for developing brain tumors, although more studies validating such hypotheses are imperative [
3]. Glioblastoma multiforme (GBM) is the most frequent and aggressive malignant primary brain tumor in people over 18 years old and is classified, together with other types of tumors, in the general class of gliomas. Several studies demonstrate that more complete resection of these tumors leads to improved outcomes for patients [
4,
5,
6,
7], and, thus, many approaches are being implemented to improve the extent of tumor resection. In clinical trials, Stummer and coworkers [
8] have demonstrated that image-guided resection of GBM results in more accurate removal of tumorous tissue, as well as higher survival rates and improved living standards for patients. The first in vivo trials for image-guided surgery in GBM using an administered probe were performed by the group of V. Ntzichristos [
9,
10], indicating that multispectral optical methodologies achieve high-resolution quantitative imaging of tissue and cancer biomarkers. Similarly, the work of van Dam [
11] supports the potential benefits of intra-operative optical imaging, showing that the use of fluorescence imaging achieves certain and case-sensitive detection of tumor tissue in real-time mode during surgery, enabling the detection of remarkably increased malignant lesions compared to those that could have been detected by visual observation. Cutter et al. [
12] used animal models of GBM, which demonstrated that imaging probes can be added topically to the resection cavity and elaborate the tumor margin. The majority of the non-invasive near-infrared fluorescent (NIRF) imaging probes constructed so far are based on a consensus sequence of peptides to operate/behave as an enzyme substrate that enables probe activation. Cutter and her team [
12] utilized fluorescently quenched activity-based probes (ABP), built on tiny molecule suicide inhibitors of tumor-associated proteases. Several studies emphasized the efficiency and utilization of NIRF probes. They proposed this particular topology for the detection of overexpressed tumor-associated markers [
13] and the imaging of tumor-expressed proteases in tissue culture, as well as in tumors that were hypodermically implanted in mice [
14].
The main contribution of this study is the application of tumor-margin identification for enhanced and user-friendly visual inspection of brain tissue along with efficient guidance of tumor removal. Thus, we do not aim to adopt or develop complex and advanced image processing concepts but mainly focus on the potential of the proposed system (hardware and software) through preliminary laboratory assessments in this area, due to its low cost, simplicity, and time-effective extraction of results. Specifically, this paper addresses the potential of a digital fluorescence imaging system (custom-made design, not a commercially utilized product) supported by an adaptive segmentation pipeline, with the aim of enhancing the imaging and contrast of tumor brain regions. Brain cancer cells were irradiated by an Argon laser operating at 488 nm and connected to the microscope. Output optical data (auto-fluorescence images) were acquired and treated utilizing unsupervised clustering in conjunction with adaptive thresholding algorithms, without any prior knowledge of the sample data. By utilizing the image intensity distribution in three complementary ways and fusing the processing results, the designed system can be viewed as an efficient, user-independent tool for assisting image-guided surgery, providing increased penetration depth and efficiency in locating the tumor margins and area. In this mode, enabling the surgical resection of invasive tumor tissues without causing significant damage to healthy tissue (proposed system follows a non-invasive approach since no straightforward contact between the illumination source and the tissue sample/patient is required), via the accurate specification and extraction of the tumor margins, would yield more effective supplementary therapy and extend patient survival. Focusing on the efficacy of brain tumor resection, the implemented technical and algorithmic framework could be a compact screening and guidance tool, enabling fast, clear identification and the removal of margin-penetrating cells, which could play a key role in future operational and treatment assessments, especially in combination with molecular imaging probes targeting brain tumor markers. The processing unit of the system addresses the joint incorporation of different forms of structural information hidden within image samples via a direct procedure that can be easily understood and adopted by the medical expert. Indeed, the proposed imaging modality enables the facile and non-detrimental application to tissues, in conjunction with accurate and near-instant activation of fluorescence probe, constituting a cost-effective and powerful tool for near real-time visualization and identification of tumor-associated markers during surgery.
After the introduction of the aims, novelty, and contribution of the current study, a detailed review of the relevant scientific research is presented in
Section 2 in support of the adoption of the proposed optical system design.
Section 3 describes the basic concepts of the experimental setup, while
Section 4 presents the brain tumor image processing steps.
Section 5 illustrates the results of the proposed tumor identification procedure, emphasizing the basic conclusions and prospects. The last section (Conclusions) summarizes the description of the proposed unsupervised learning fluorescence imaging system along with the outcome and potential of the presented work.
2. Literature Review
Optical coherence tomography (OCT) is one the most extensively studied technologies for the identification of tumor margins. This approach facilitates the extraction of information about tumor margins during surgery in real-time mode, enabling inspection/scanning of foci of the cancerous area or tumor cells that have spread to a different region than their origin (metastasized cancer). One of the most notable works in this research field addresses image-guided surgery (IGS) of breast cancer using OCT [
15]. The specific methodology utilizes optical “ranging” of near InfraRed (IR) wavelengths to extract structural information in the spatial domain and increased imaging resolution at the microscopic/cellular level (at the order of microns). This is achieved via constructing cross-sectional maps in the two-dimensional image space based on the physical phenomenon of the interaction of light with an illuminated surface (optical backscattering). Many other studies demonstrate OCT as an efficient framework for the delineation of boundaries between normal and diseased breast tissue, utilizing techniques for the evaluation of human breast tissue through processing and analysis in frequency (Fourier transform) and spatial domain [
16]. Another direction for assessments lies in the 3D volumetric data acquisition of human breast cancer tumor margins and axillary lymph nodes by rotating and retracting an OCT needle-probe below the tissue surface during imaging [
17]. Despite the high resolution and efficiency in discriminating normal from cancerous tissue, the limited image penetration depth achieved (2–3 mm) has restricted the utilization of OCT as a real-time imaging modality. In addition, quantitative and objective assessments are based on the subjective interpretation of the acquired image by a specialized and well-trained medical expert. Similarly, optical radiomic signatures, extracted from corresponding optical data (optical coherence tomography images), led to enhanced identification of melanoma, with unique contrast and specificity [
18].
The concept of elastic scattering spectroscopy (ESS) was used in the ultra-violet (UV)-visible spectral bands [
19] and constitutes one of the earliest studies attempting to evaluate and characterize the boundaries of cancerous breast tissue samples through the application of optical spectroscopy. An analogous attempt was performed in [
20] to examine the extent of cancer removal through surgical operations applying the technology of inelastic scattering of monochromatic light (Raman spectroscopy). Although these techniques are quite prone to morphological alterations at both sub-cellular and cellular plane (microscopic level) and enable efficient discrimination of different tissues states via their light fingerprints, their small effective illumination-source-to-detector separation limits the ability to sense the presence of malignant tissue up to 1–2 mm from the surface. A group of researchers from MIT, in particular, a team from the GRH Spectroscopy Laboratory, have developed a mobile scanning system (spectroscopic device incorporating a fiber-optic probe) and properly formulated to evaluate tumor margins intraoperatively; the portable scanner is based on two principles, namely intrinsic fluorescence spectroscopy (IFS) and diffuse reflectance spectroscopy (DRS) [
21].
Fluorescence-based image acquisition and analysis methods have facilitated both the accurate demarcation of the infiltrating contour of tumors in real-time and in vivo mode and the evaluation of their histological characteristics. For instance, confocal laser endomicroscopy (CLE) enables the in vivo extraction of brain tissue optical data (fluorescent-based images) with highly increased resolution (at the cellular level), as accomplished in optical biopsies. On the other hand, the principle of fluorescence-guided surgery (FGS) is adopted in applications where improved visualization quality of tumor margins is required so that the extent of cancerous tissue removal in surgical operation increases. A systematic study of several means for fluorescence image-guided glioma surgery, based on preclinical and clinical assessments, is reported in [
22].
In other works, photo-acoustic imaging was also used for potential intraoperative tumor margin detection [
23]. Photo-acoustic tomography (PAT) has been utilized for breast cancer imaging, brain imaging, and other applications. Even though the limited penetration depth of the technique can be overcome by utilizing microwaves or radiowaves, ultrasounds sustain intense artifact effects upon the reflection of signals from gas to liquid structures and gas to solid ones due to differentiations in acoustic impedances. In addition, ultrasounds suffer from considerable signal attenuation and corruption (phase distortion) in dense/compact structures (thick bones properties), such as the human skull. As a further observation, such acoustic signals are characterized by a decreased ability to efficiently penetrate through gas hollows (such as lung tissues and/or brain tumors). Finally, in order to efficiently detect ultrasonic waves, direct communication/connection of the sensor module (transducer) and the biological structure (tissue sample) is required.
In [
24], the authors investigated the feasibility of the multispectral dye-enhanced polarized light imaging method to delineate non-melanoma tumor margins, a tissue type where regions of increased hemoglobin concentration and dye absorption need to be isolated. This work successfully distinguished and identified the tumor margins using the specific imaging modality.
Apart from the imaging modalities mentioned above, nanoscale structures are also used to enhance margin detection. For example, nanotechnology-based contrast agents enhance optical imaging methods and aid in margin detection. Gold nanoparticles (GNPs) are particularly suited for this purpose as they absorb and scatter light strongly and the presence of gold metal intensifies the signal greatly. These gold nanoparticles are coupled with a dye and used in Raman spectroscopy and other optical spectroscopic techniques to produce the surface-enhanced Raman spectroscopic (SERS) effect. Near IR lasers and fluorescence detectors are also combined to record the Raman signals and measure fluorescence in the presence of gold nanoparticles, which concentrate in tumor regions. Due to their nano size, they can easily penetrate blood vessels that are leaky due to the tumor and get trapped in the edge of these tumors as such blood vessels are dominant in tumor edges. This effect enhances margin detection enormously. Similar studies introduced the use of gold nanoparticles in NIR narrow-band imaging (NIR NBI) [
25], utilizing two lighting emitting diodes (LEDs) with wavelengths at the green and NIR range to illuminate the GNP infiltrated blood vessels. A charge-coupled device (CCD) captures the reflected illumination signal from the sample. In the corresponding results, the gold nanoparticles clearly demarcated the tumor margins, demonstrating the potential of this technology in intraoperative detection. Targeted gold nanoparticles that deliver a fluorescent payload to tumors have also been used to demarcate the tumor from normal tissues [
26].
During the last decade, Giakos et al. [
27,
28,
29,
30] have extensively worked on a number of approaches for the differentiation of healthy and cancerous cells, introducing label-free NIR–IR polarimetric diffuse reflectance-based cancer detection methodologies in conjunction with a wavelet and fractal analysis. Visualizing the interaction of IR with healthy and malignant (cancerous) lung cells through polarimetry under diffuse reflectance geometry, in connection with polarimetric exploratory data analysis (pEDA) [
29] enables the development of robust and competent diagnostic tools. Image and signal generation through the determination of the polarization states of light proves quite effective and sustains specific benefits for a wide variety of identification and classification tasks and applications, mostly based on the intrinsic nature of optical backscattering to provide increased contrast in varying polarization conditions. As a result, under backscattered geometry, multiple kinds of early-stage malignancies (cancer) could be discriminated, considering and quantifying their unique diffuse reflectance polarimetric signatures. Another related method uses polarimetric discrimination of the wavelet-fractal domain for histological analysis of monolayer lung cancer cells [
31,
32]. Wavelet polarimetric evaluation of healthy, squamous carcinoma, and adenocarcinoma lung tissue cell lines proves quite promising and reliable in accurately and robustly classifying cells as healthy or cancerous ones, in conjunction with proper discrimination between malignant cells originating from different types of lung cancer [
32].
In order to provide efficient interfaces to medical doctors and guide them to easily focus on affected regions, imaging modalities must be supported by digital image processing and segmentation schemes, especially in the case of biomedical applications, as medical images suffer from low contrast and noise. Thus it is of paramount importance to select appropriate and application adaptive approaches in order to increase contrast, decrease information loss and artifacts from captured image data. Histogram-based techniques constitute a common yet quite effective tool for medical image enhancement, finding numerous applications and algorithmic modifications in international literature for many years [
33,
34]. The latest advances in magnetic resonance (MR) image enhancement include metaheuristics and particle swarm optimization (PSO). Rundo et al. [
35,
36] proposed an automatic global thresholding and segmentation framework based on genetic algorithms (GAs), confirming through MRI data that there is an underlying bimodal distribution of pixel intensities. The principal idea lies in calculating the optimal threshold (best solution) under an evolutionary computational approach that better discriminates the two Gaussian distributions, formulating a bimodal pixel intensity representation function (histogram) in a medical image sub-region. Acharya and Kumar introduced an efficient particle swarm optimized histogram equalization scheme to automatically extract optimal threshold for texture identification of regions through an iterative approach under a fitness function that combines different performance metrics [
37]. The main limitation of such approaches is the increased computational and algorithmic design complexity along with increased overall processing time. Machine learning and computational intelligence approaches are outside the scope of the present study, as we focus on developing a framework that is easy for medical experts to use and set up and can be adopted in real- or near real-time image analysis applications.
Clustering constitutes a widely-established methodology for statistical image analysis, based on assigning groups of pixels into classes (clusters) based on a distance index/metric (intensity similarity and topological proximity), so that the topological inter-cluster similarity and the extra-cluster separability are increased. The selection of the optimal clustering scheme and algorithmic setup (namely, the feature definition, the initial cluster centers or seed points, the distance function to be adopted, or the number of dominant classes) depends on the input dataset origin and type along with the application requirements. The classic k-means and the mean shift algorithms constitute two fundamental frameworks for image classification and decomposition. The first one (known as “k-means clustering”) [
38] is an iterative approach that aims to divide a set of data samples (pixel intensities in the case of images) into K groups (clusters) by exploiting the data-driven probability density function in the topology of the feature space. The algorithm converges to a preset number K of clusters in the data distribution, but the quality of the extracted outcome and classification performance are strongly affected and controlled by the initialization condition and the number of clusters a priori defined. The mean shift (MSH) technique (known as “mean shift clustering”) [
39,
40] is considered an identical solution to overcome the previously mentioned key drawback of k-means clustering. MSH is a robust, statistical, nonparametric detection methodology that works in the density distribution space. Its basic ideas lie in determining and controlling the mode (the highest density of data points) adaptation in terms of a kernel-weighted mean value (average) estimate of data points (observations) within a region of movement (sliding smoothing window). The computational procedure is performed iteratively until convergence to a global or local solution (local density mode) according to a well-defined and carefully selected stopping criterion.
Another type of segmentation technique that directly utilizes the intensity distribution of data (histogram) defines thresholds for classes based on certain discrimination criteria. Otsu thresholding has been one of the standard thresholding techniques used for segmenting an image into two key principal regions (object and background classes) [
41]. It is based on the Gaussian modeling of each histogram mode as a natural model of the probability distribution function of a physical object. The margin of the Bayesian detection error of a bi-modal joint distribution defines the appropriate threshold, as a balance point between the misdetection probabilities of the two classes. Notice that the approach can be easily extended to the multiclass case by modeling a multi-modal data distribution and defining the appropriate threshold between consecutive pairs of Gaussian models. On similar grounds, entropy-based thresholding exploits the Shannon entropy [
42] of distribution in each histogram mode and interprets the maximization of the thresholded image entropy as indicative of maximum information transfer [
43].
These techniques were used in this study to threshold the fluorescence images and discriminate the cancerous regions from the healthy area of the brain tissue; Otsu’s approach is selected because it calculates histograms in a probabilistic manner and produces instant segmentation results under the assumption of the normality of illumination without any prior knowledge of the image properties. In addition, we employ the mean shift procedure in order to refine the specification of image modes within the data distribution and regulate the derivation of empirical probability density functions. As a complementary function, entropy thresholding is also selected to address the information content and interaction of classes. The latter scheme has proven effective in locating sharp discontinuities (edges) within an image in a robust and flexible way, overcoming the sensitivity to noise and time complexity of gradient-based edge detectors utilizing first and second-order derivatives [
44]. We expect that this efficiency will be beneficial for the accurate separation of healthy and tumor tissue regions, with well-defined borders, for real-time image-guided surgery. For the efficient segmentation of fluorescence tissue images addressing the sensitivity close to the class borders, we implement the fusion of the previous approaches by means of the union of their results. In particular, the entropy-based result is more closely examined for the discrimination of small cancerous formations in the healthy tissue, whereas the combination of individual results is enhanced by mathematical morphology into closed sections for deriving the gross total area of cancer. These established segmentation approaches were adopted for the proposed fused algorithmic scheme, considering the following conditions:
- (a)
We propose a technique to be directly utilized in laboratory assessments, so that the algorithmic procedures become familiar and easily manipulated by medical experts, simulating their own form of processing of microscopy samples.
- (b)
The nature of the input fluorescent images is not appropriate for region-based segmentation, because they do not express tumors as compact regions; as a result, edge-based techniques cannot be favored for the identification of tumor borders, as no clear connected contours can be determined along the tumorous segments of the tissue sample under examination.
- (c)
The proposed approach aims at a time-efficient tumor visual inspection methodology, not a complete, advanced, and “from scratch” image processing framework. In this context, supervised segmentation and machine learning-based schemes are beyond the scope of the present study.
3. Proposed Tumor Visualization and Identification Framework
This study operates on tissue autofluorescence imaging principles, an optical data acquisition framework that has recently shown promise as a diagnostic modality. Optical fluorescence microscopy is a promising technique for high-quality histological imaging, as it does not harm the tissue samples and saves time and labor [
29]. In this experiment, the most common fluorophores within the tissue absorb blue light and re-emit a portion of this radiation at higher wavelengths (green) as a fluorescence signal. Typically, autofluorescence arises from different kinds of fluorophores in the tissue, such as proteins and enzymes; it is sensitive to both morphological, biochemical, and metabolic changes associated with tissue pathologies. Segmentation and enhancement of the obtained images are necessary for the accurate and unsupervised distinction of the tumor margins, even more so, if the background and the object in the image are not very well demarcated due to intensity and texture differences. An unsupervised method aims to minimize errors subjectivity introduced by human intervention/supervision.
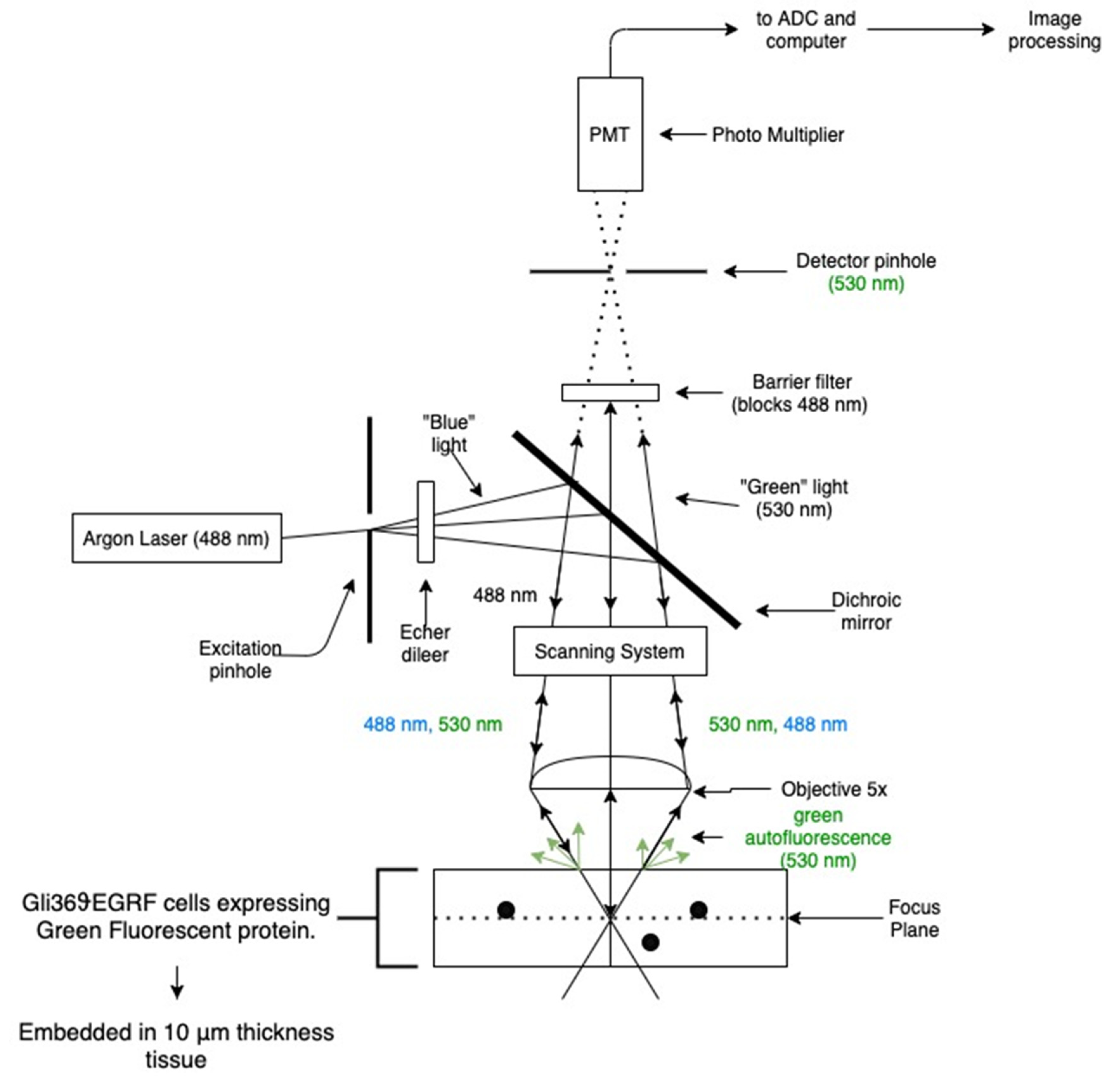
Optical fluorescence optical data (microscopy images) depicting brain tumor slice samples were acquired via an imaging system designed with a Zeiss laser scanning microscope (LSM), setting up magnification at 5×. The overall experimental arrangement is shown in
Figure 1.
Samples of human Gli36ϑEGRF glioblastoma cell lines expressing green fluorescent protein are used throughout this study. Biological material (tumor cells) comes in the form of 10 µm tissue slices, which were cut beginning from the initial 2 mm of the former brain section. The dataset includes five slides (each one with three sections), which correspond to the distance (depth) of the slice with respect to the starting point of the anterior brain section (beginning from 10 µm and ending to 150 µm, in slices of 10 µm size). The tumor is positioned on the top boundary of these slices, containing cells that express a green fluorescent protein (GFP). Fluorescence imaging modality is utilized for cancer identification, based on the fact that specific biomarker fluorophores are expressed at a substantially more intensive level in tumor cells with respect to healthy ones, enhancing imaging quality for discrimination purposes. Specifically, brain tumor samples were examined using 488 nm Argon laser attached to the microscope; blue light, after passing through a dichroic lens, is incident on the sample inducing green-light auto fluorescence. Diffuse reflectance-auto-fluorescent images are acquired in the “green” spectral range (specific wavelength band selected at 530 nm); while the reflected illumination blue light is blocked with a green-pass filter, set in front of the receiver photomultiplier. Captured images were then processed using unsupervised clustering in conjunction with adaptive thresholding algorithms.
The samples were a gift from Dr. EA Chiocca [
45] and prepared from cloning of human Gli36 cells. The cell lines were cultured in Dulbecco’s Modified Eagle Medium (DMEM) (solution contains 4.5 g/L glucose, L-glutamine) supplemented qq2 with 10% concentration in fetal bovine serum (FBS), penicillin levels at 180 U/mL, streptomycin values at 180 mg/mL, and a level of 0.45 mg/mL for amphotericin B (all reagents were developed from Gibco, Invitrogen Corporation). By nature, specific cells overexpress the vIII mutant forms of the estimated glomerular filtration rate (EGFR) gene and have been extensively used for in vivo studies, because they grow rapidly in rat brains. For this purpose, the cell lines were injected into the brain of athymic female nude (nu/nu) mice (6–8 weeks old at the time of surgery), which were grown and preserved at the Animal Resource Center (installations at Case Western Reserve University) in agreement with corresponding institutional policies (CWRU IACUC Animal Experimental Protocol: 2009-0019). Regarding the procedure for brain tumor implants, the animal population was anesthetized and glioblastoma cells were deposited in the right striatum at a slow rate of 1 mL/min and at a depth of 23 mm from the dura. Post imaging brain slices were fixed in a solution medium of 4% paraformaldehyde, 30% sucrose protected, and physically firmed in optimum cutting temperature compound (OCT) for cryostat sectioning (instrument model Leica CM3050S). Tissue sections were acquired sequentially at 10 or 25 mm cut directly onto slides and preserved at −80 °C until manipulation. In order to be visualized via probes, biological material was heated to room temperature for a time interval of 10 min, washed in phosphate buffer solution (PBS) and cover slipped with fluorescent mounting media. As a final step, fluorescent microscopy images were acquired and analyzed performing an unsupervised clustering framework combined with adaptive thresholding techniques.
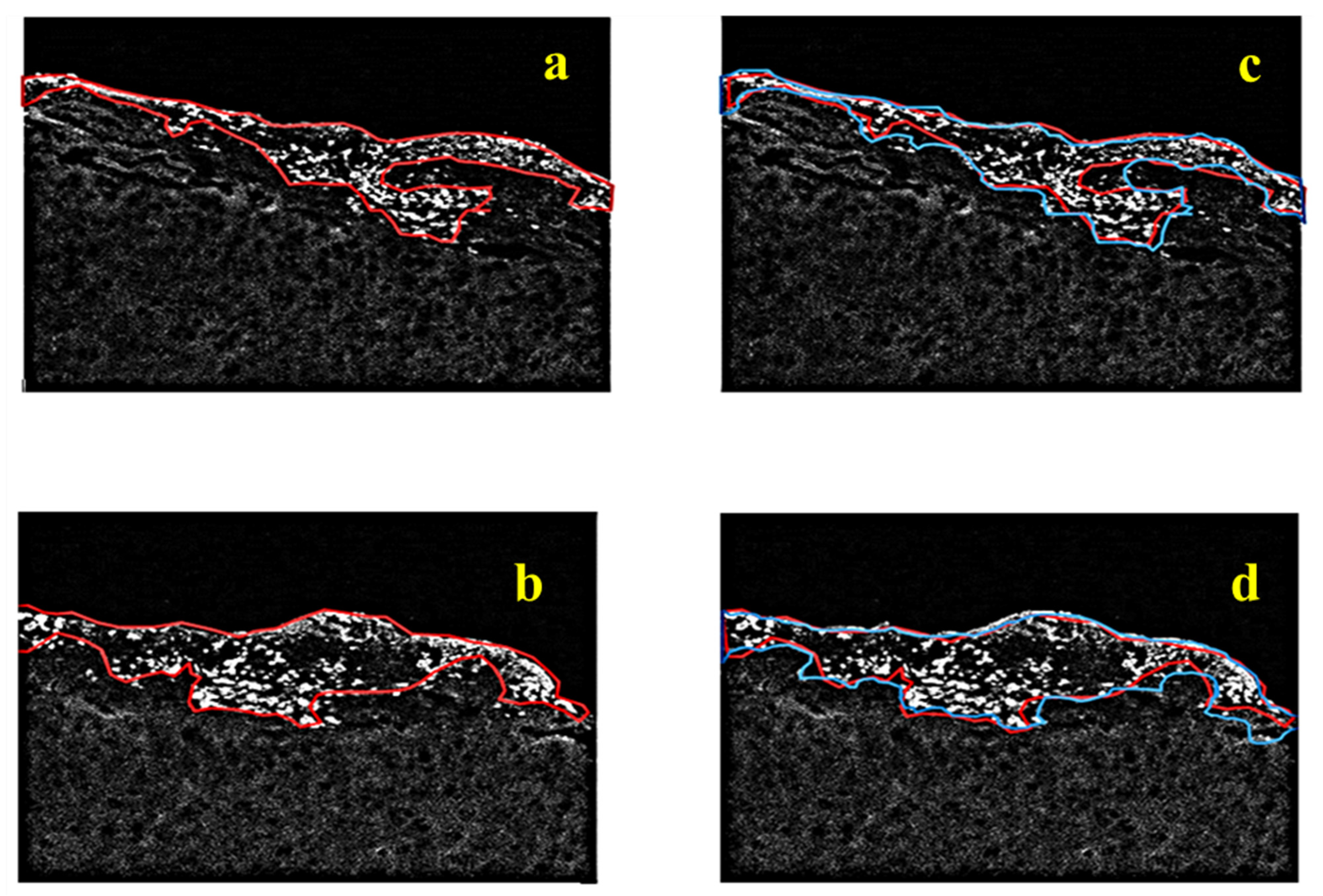
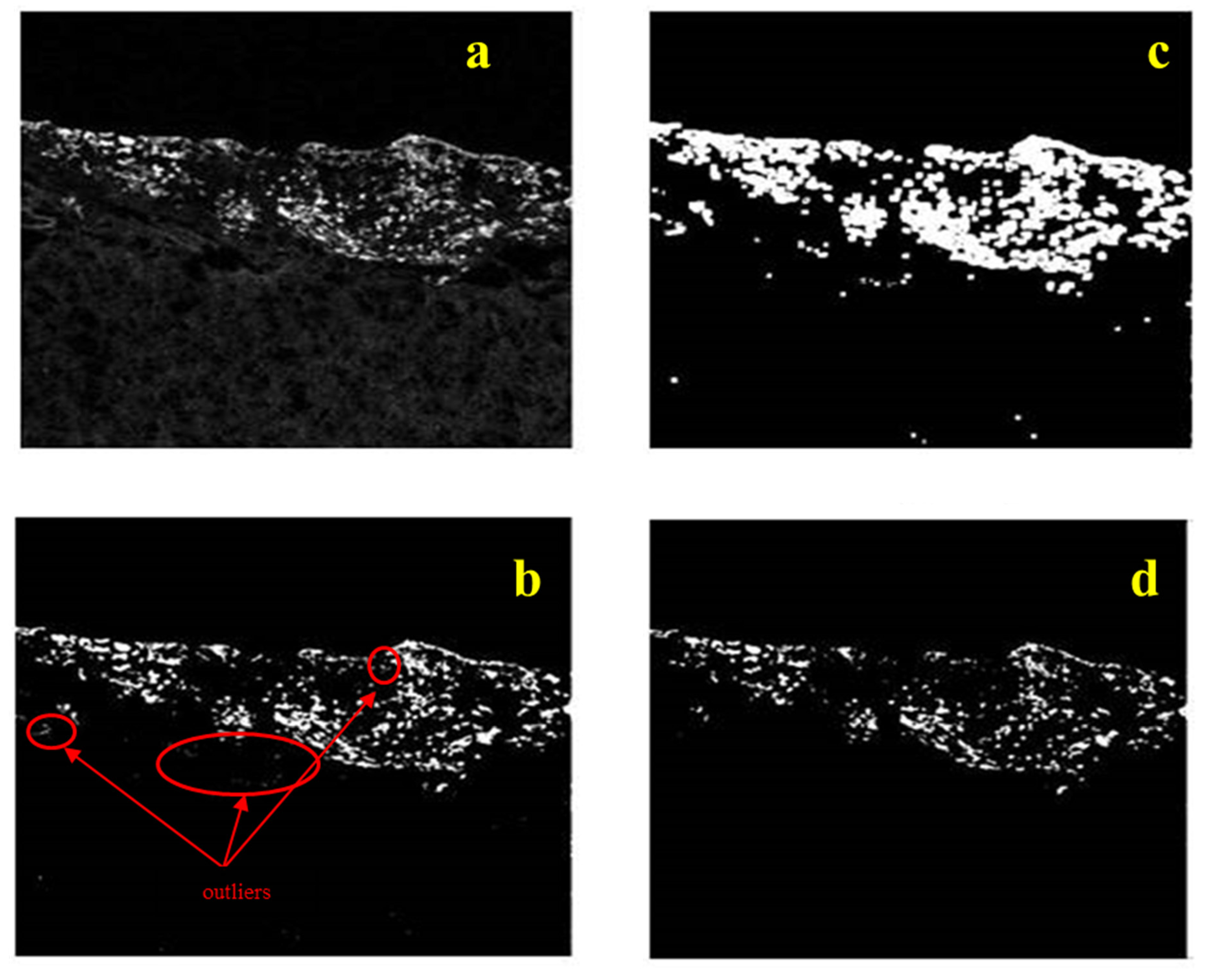
The proposed tumor identification and visualization pipeline of algorithms are shown in
Figure 2. It is based on a three-step framework run in sequential mode, i.e., image processing/segmentation, image fusion, and image refinement modules, each one with a unique contribution to information mining. To quantitatively evaluate the detection accuracy of our fluorescence tumor imaging and identification system, a ground truth image was generated, superimposing the assigned tumor margins marked by a medical expert on the raw fluorescence sample image. Since the accurate detection of tumor margins is impossible, even with dense sampling and biopsy, the validation of the extracted results is performed based on a closed and compact region approximation of the overall tumor surrounding contour produced by the pathologist. Thus, we should note here that the following study is semi-quantitative, as it is affected by the qualitative judgment of the expert. Thus, our evaluation at this stage considers the agreement between the algorithmic and the medical perspectives on the significant tumor area, its extent, and structure. As already mentioned, assurance about the local condition can only be provided by selective biopsy. In our study, the biopsy was performed at three specific small areas in the region of strongest fluorescent response (this biopsy was guided by the proposed imaging modality), which verified the tumorous nature of all tested sites.
The algorithmic identification (segmentation) procedure, the first step of the proposed tumor visualization pipeline, is initiated by performing median filtering (3 × 3 window size) on the original fluorescence image in order to eliminate speckle noise, possibly introduced during the image acquisition procedure. The image processing module is gradually performed in three discrete stages run in parallel: an initial estimation of tumor region is performed by applying the Otsu thresholding technique; meanwhile, an enrichment step takes place via the entropy thresholding approach, which introduces information regarding the background area and refined borders of the cancerous region. At the third branch of the image processing module, the smoothed key segments (those that differentiate most from the background) of the tumor margins are estimated performing the mean shift clustering methodology to the fluorescence sample image. The second step (image fusion module) of the proposed overall scheme proceeds with fusing the results of the three individual image processing techniques applied in the first step and enables us to form the binary mask containing pixels that are active (non-zero intensity value) in more than one segmented images, resulting to a robust estimation of the “strong and consistent components” of tumor margins, i.e., the pixels that are detected as tumor belonging ones by at least two of the three adopted segmentation techniques. The final step of the overall pipeline of algorithms proceeds by applying mathematical morphology (image closing and region filling) to the fused quantized image, acting as a noise/outliers filter, filling out holes and refining borders, and derives the reconstructed geometry of the overall cancerous area as a closed and compact region, illustrating the potential for numerical computations regarding the tumor size, area, shape, and depth.
The notion of “Otsu thresholding” was initially introduced as a non-parametric and unsupervised approach based on the assumption that an image is formulated by pixels from two principal classes (background region and foreground one containing the object/scene of interest), indicating that the image can be represented via a bimodal histogram [
46]. Having established this image model, the method defines an optimal threshold value, under an iterative calculation procedure, in order to binarize the image. The threshold value is adaptively determined so that the intra-class divergence (variability between data points belonging to one class) is minimized and, at the same time, the inter-class deviation (differentiation between individuals of the two distinct classes) is maximized. This condition imposes efficient segmentation of the image, because the variance is considered a reliable index for determining the homogeneity of a region. The intra-class divergence is determined as the weighted sum of the variances within any of the two dominant classes, with “weight” representing the a priori likelihood of that particular class. On the contrary, the inter-class variance reaches its maximum by initializing a “starting point” value (threshold) and repeatedly updating it to attain the maximum differentiation between the two pixel distributions.
Another thresholding technique exploited in this study is based on information theory and Shannon entropy maximization. Entropy can be considered as an index that represents the uncertainty of a random variable, thus it quantifies the information present in a dataset or a message [
47]. The principal objective of this approach is to partition the image histogram (pixel intensity values considered as the probability distribution) into two different (independent) distributions via the determination of an optimal threshold value that maximizes the randomness of both image region classes, and, thus, enriched information content can be extracted [
48,
49]. Let P
1, P
2, ..., P
n be the probability distributions of different pixel intensity values (levels of grey) represented within the image. Let “
s” be the threshold value determined for binarizing the image (binary image segmentation). Thus, two independent classes (probability distributions) are outlined; the first one corresponds to pixel intensities varying from 0 to “
s” and the second one representing grey levels from “
s+1” to “
n”. Let these sets be defined as A and B, respectively, according to the following representation:
The entropy values are defined as follows [
50]:
Note that the term in the parenthesis in equation 2 encodes the entropy of one part {
P1,
P2,
P3, ...,
Ps} of the original distribution {
P1,
P2,
P3, ...,
Ps} and should be represented by
Hs, for simplicity, and, thus, the quantity
H(
A) takes the following form:
In a similar way,
where
Hn refers to the entropy of the original distribution.
The total entropy of the two individual distributions is defined by
ψ(
s) and is given as:
The value of “s” that maximizes entropy value (maximum information retrieved) then accounts for the optimal threshold.
The third segmentation technique employed in our proposed algorithmic scheme, the mean shift clustering, attempts to extract complementary information at a different level of data organization by closely analyzing the probability distribution of the sample fluorescence image. This method has been developed on the grounds of statistical clustering as applied in a feature space of the image itself. These features may reflect the intensity distribution (1D histogram space) alone or in combination with the spatial pixel distribution (defining a 3D space), or even the color distribution (in a 3D space). The principal motivation beyond mean shift clustering is the modeling and approximation of input data points as an empirical probability density function, according to which a large number of neighboring samples in feature space (dense regions) are translated into local maxima or modes (most frequent value met in a set of data points) of the corresponding statistical representation (distribution). For every individual (data point) in the processed population, a gradient ascent process is iteratively applied to the locally estimated density until the algorithm converges to an optimal solution. The data points associated with the same stationary point (mode) formulate dominant regions within the image (clusters that contain pixels close to each other and with similar intensity values).
In our application, the mean shift approach operates in the 1D space of intensities. It initially defines a window around each point in the space of intensities and calculates its mean as the “center of gravity” based on the included intensities. In the second step, it translates (“shifts”) this midpoint (center of the current window) to the mean value and repeats the procedure until the stopping criterion is met (convergence is achieved). Conceptually, in each step of the iterative calculations, the window shifts to a denser region of grayscale intensities, until it approximates a local maximum of the total distribution function. This iterative scheme repeatedly assigns every pixel (intensity point) to a class center (mean point) and intuitively calculates the number of dominant regions (segments) within the input signal/image via the final number of the cluster-centroids determined.
The principal concepts of the mathematical background of the mean shift algorithm are briefly presented below. Kernel density estimation can estimate the density function of a random variable in a non-parametric mode. A kernel φ(x) is a non-negative function of a multidimensional feature vector, with which the vector is integrated over the domain of its definition.
This is usually implemented as a Parzen window with its kernel
K() representing a rectangular window operating in the space of the n data points {
x1,
x2, …,
xn} and a bandwidth parameter
h notation that sustains the physical meaning of the window size indicator. The corresponding kernel density estimator for a given set of d-dimensional points (features) is given as:
where
h (bandwidth parameter) defines the radius of the kernel. The sample mean (distribution density) around a point
x using
K is computed as:
According to the mean shift approach, the vector
x is moved to
m(x) and the process is repeated until convergence. This iterative movement of
x can be interpreted as a gradient ascent on the density contour:
where
K′() denotes the derivative of
K(). Using the specific kernel form
where
C is a constant and setting
g(x) = −
k′(
x) as the derivative of the selected kernel profile, we finally obtain the summarized formulas-definitions:
where, the first quantity is proportional to the density estimate at
x computed with a new kernel (G(
x) =
Cg g(||
x||
2). Equation (10) indicates that the mean shift is proportional to the local gradient estimate at point
x obtained with kernel
K, so that it can represent a path that directs the solution to a stationary point of the estimated density, i.e., a mode of the distribution (or a cluster centroid). It can be observed that the mean shift movement is quick/long for low-density regions (valleys of the corresponding density distribution) but slow/short as term
x draws near a mode (until it finally approximates zero value upon coincidence of the point with the mode). The unique quantity of the MSH algorithm to be initially adjusted by the user is bandwidth parameter h, representing the fixed window size to be examined around each data point. This algorithmic parameter is intuitive and application-dependent [
51]. Another drawback of MSH is the low speed of convergence (especially in cases of high-resolution color images), but substantial research effort has been devoted to speed-ups and improvements.
At this point we can define a contour plot for each algorithmically defined image, denoting the borders of cancer areas detected. These areas essentially form small clusters of tumors affecting the healthy tissue and, as such, they can be used as topological markers for obtaining samples for biopsy. The result of entropy thresholding appears to be most appropriate for such purposes, as explained in the results section. However, to increase our confidence in the definition of cancer areas, we fuse the results of individual segmentation schemes. Our assumption for fusion is that if a pixel is “active” (value 1 in the binary output image, yet the background is set to zero value) in two or more segmentation techniques, then this point is assigned with an increased probability of belonging to the tumor section since corresponding pixel information is examined and validated in more than one segmented image of the fused pipeline. Thus, the proposed segmentation mask is derived from the cancer pixels detected in at least two of the three segmentation masks. At the final stage of processing, morphology operators are applied to the output binary mask to derive a compact region representing our gross estimate of the tumor spread with filled gaps and connected components. For the requirements of this study, we utilize the morphological operators of “image closing” with a disk structuring element of radius 19 and “flood-filling” of 8-neighbor connectivity, in order to refine the overall tumor region and fill in the holes within the extracted region of interest. A detailed analysis of the mathematical morphology and the corresponding operators can be found in [
52]. This solid estimate of the tumor can be further analyzed for shape and size calculations, providing additional topographic information to the medical expert.