Abstract
The DNA damage response (DDR) is critical for maintaining genome stability, and abnormal DDR—resulting from mutations in DNA damage-sensing and repair proteins—is a hallmark of cancer. Here, we aimed to investigate the predictive power of DDR gene mutations and the tumor mutational load (TML) for survival outcomes in a cohort of 22 rectal cancer patients who received pre-operative neoadjuvant therapy. Univariate analysis revealed that TML-high and TP53 mutations were significantly associated with worse overall survival (OS) with TML-high retaining significance in multivariate analyses. Kaplan–Meier survival analyses further showed TML-high was associated with worse disease-free (p = 0.036) and OS (p = 0.024) results in our patient cohort. A total of 53 somatic mutations were identified in 22 samples with eight (36%) containing mutations in DDR genes, including ATM, ATR, CHEK2, MRE11A, RAD50, NBN, ERCC2 and TP53. TP53 was the most frequently mutated gene, and TP53 mutations were significantly associated with worse OS (p = 0.023) in Kaplan–Meier survival analyses. Thus, our data indicate that TML and TP53 mutations have prognostic value for rectal cancer patients and may be important independent biomarkers for patient management. This suggests that prognostic determination for rectal cancer patients receiving pre-operative neoadjuvant therapy should include consideration of the initial TML and tumor genetic status.
1. Introduction
Colorectal cancer (CRC) is the third-most common malignancy in both men and women and the second leading cause of cancer-related death worldwide [1], with over 1.8 million new cases and 900,000 deaths in 2020 [2]. Although the overall rates of CRC have decreased in the past several years, both incidence and mortality are rising in adults under 50 years of age [1,2].
Surgical resection remains the most effective treatment for CRC; however, full resection with clear margins can be difficult due to limited tumor accessibility, particularly within the rectum, resulting in high rates of recurrence for rectal cancer patients [3,4]. Consequently, although colon cancer patients generally receive only postoperative chemotherapy, as needed, based on the risk level and disease severity, for many years, the standard of care for rectal cancer patients involved neoadjuvant chemo/radiotherapy prior to surgery followed by postoperative chemotherapy [3,5,6].
Critically, this treatment paradigm has reduced the recurrence rates and improved the survival outcomes for rectal cancer patients [7]. However, individual patient response is highly variable, with a complete response observed in less than 30% of patients and 40% displaying no significant clinical response [8,9,10]. A more recent approach in which both neoadjuvant chemo/radiotherapy and adjuvant chemotherapy are administered prior to surgery, known as total neoadjuvant chemotherapy, has shown promise and is becoming increasingly common [11,12].
In addition, nonoperative management of rectal cancer for patients who display a clinical complete response after neoadjuvant therapy, referred to as the Watch and Wait approach, is gaining in popularity [13,14]. However, many unanswered questions remain, and there is an urgent need for biomarkers that can be leveraged to identify the individuals most likely to respond well to preoperative neoadjuvant chemo/radiotherapy, while avoiding potential for treatment-associated toxicity in those unlikely to benefit from these therapies. To this end, various molecular biomarkers for predicting CRC prognosis and response to treatment have been proposed; however, none are currently in clinical use [15,16,17,18,19,20,21].
One potential group of molecular biomarkers for CRC are proteins that function in the DNA damage response (DDR). This system detects DNA double-strand breaks (DSBs) and other forms of DNA injury, and in response, it induces both DNA repair pathways and expression of checkpoint proteins, which halt cell cycle progression until the damage can be repaired or apoptosis initiated, thereby, maintaining genome integrity [22]. Thus, mutations in key DDR proteins, such as p53, a mediator of this pathway, often play key roles in cancer development [23].
DDR functionality can further affect the sensitivity to radiotherapy-based cancer treatment, which utilizes ionizing radiation (IR) to generate a wide range of DNA lesions, including DSBs, single-strand breaks (SSBs) and base modifications, leading to cancer cell death. Similarly, chemotherapeutic drugs administered in neoadjuvant therapy, such as 5-fluorurocil (5-FU), induce DNA damage and activate cellular checkpoint proteins, and thus, the drug response is affected by DDR function and DNA repair capabilities [24].
DSBs—the most severe form of DNA damage—are detected by poly(ADP-ribose) polymerase 1 (PARP1) and the ku70–ku80 heterodimer and are repaired via two main pathways: non-homologous end joining (NHEJ) and homologous recombination (HR) [22,25,26]. NHEJ, which is initiated by ku70–ku80, does not require sequence homology and is an error-prone process. In contrast, HR is mediated by the RAD51 recombinase and is homology based, utilizing the undamaged sister chromatid as a template and, thus, is relatively accurate.
These repair processes largely proceed through distinct protein subsets. However, a heterotrimeric complex composed of meiotic recombination 11 (MRE11), DNA repair protein Rad50 (RAD50) and Nijmegen breakage syndrome 1 (NBS1), referred to as the MRN complex, is involved in both NHEJ and HR repair, and mutations in these proteins are associated with numerous cancers, including CRC [27].
A third pathway, known as mismatch repair (MMR), identifies and corrects mismatched base pairs resulting from errors in replication or repair. Mismatch detection is primarily mediated by the MutS homolog (MSH)2–MSH6 heterodimer, known as MutSα, as well as by the MSH2–MSH3 heterodimer (MutSβ) [28]. Other MMR proteins, including MutL homolog (MLH)1, MLH3 and post-meiotic segregation (PMS)2, increased and then form higher order complexes with replication factors and function to remove the lesion and repair the DNA [28]. Critically, as with other DDR pathways, disruptions in MMR lead to genomic instability and increased mutation rates, which can both contribute to cancer development and affect therapeutic outcomes [28].
A number of recent studies have utilized targeted next-generation sequencing (NGS) to assess the prognostic value of specific mutations, as well as the total tumor mutational load (TML), an indicator of genomic instability, for predicting survival and therapeutic outcomes in cancer patients [29,30,31,32,33]. However, to our knowledge, none have used this technology to assess the value of DDR mutations for predicting therapeutic outcomes in CRC. Here, we used the Oncomine™ Tumor Mutation Load Assay to determine whether mutations in DDR/MMR proteins and/or TML status may represent possible biomarkers for predicting the response to chemo/radiotherapy treatment in rectal cancer patients who received pre-operative neoadjuvant therapy.
2. Results
2.1. Patient Characteristics
A total of 22 patients (15 males (66.5%) and 7 females (33.5%)) were included in this study. The patient demographic characteristics are listed in Table 1. The median age was 63 years (range: 35–82 years). All 22 patients received preoperative therapy. Patients were followed for a median period of 3.1 years (range: 0−10.6 years), and the median time to death was 2.1 years after surgery (range: 0−9.2 years).

Table 1.
Patient characteristics.
We first performed TML calculations for all patients using the default Ion Reporter™ TML analysis workflow, in which the initial variant caller considers not only non-synonymous variants but also synonymous mutations, for TML calculation. To further reduce noise in TML estimations, variants were then filtered to eliminate germline variants and select for the highest quality somatic variants using a specifically designed filter (Oncomine™ Variants v5.16).
We found that, in our cohort of 22 samples, the median TML was 24 mutations (Mut)/Mb, with TML values ranging from 2 to 241. Based on previous publications, TML can be categorized as low (<11 Mut/Mb), intermediate (12–48 Mut/Mb) and high (>49 Mut/Mb) TML [30,34]. Here, due to our small sample size (n = 22), we combined the low and intermediate groups into a low/med-TML low group to achieve statistically meaningful data analysis.
Investigation of the association between the TML level and clinicopathological characteristics (Table 1) revealed no significant differences in age, sex, tumor stage, lymph node involvement, metastasis, vascular invasion, perineural invasion, or tumor regression grade (TRG) in patients with low vs. high TML. We further assessed the mutational frequency in known DDR and MMR genes in patients with low/med and high TML status (Table 2).

Table 2.
DDR/MMR mutations in CRC patients.
From this analysis, we found that TP53 was more frequently mutated in TML-high tumors compared to TML-low/med tumors (71.4% vs. 66. 7%), although the difference was not significant, likely due to small sample size (p = 0.83). Overall, mutations in TP53, KRAS and RAD50 were detected in 68.2% (15/22), 27.3% (6/22) and 18.2% (4/22), respectively, of patient samples. A total of nine mutations in four MMR genes, MLH1 (n = 3), MSH2 (n = 3), MSH6 (n = 2), PMS2 (n = 1), were detected in 27.3% (6/22) of samples with TML-low/med and 3/22 (13.6%) samples with TML-high.
2.2. TML Is Significantly Associated with Worse Prognosis for Rectal Cancer Patients
We next performed univariate Cox regression analysis to compare OS in CRC patients with differing clinical features and gene mutations, as well as low/med- vs. high-TML. Among all variables tested, including sex, age at diagnosis, tumor stage, lymph node involvement and DDR gene mutations, we found that TML-high was significantly associated with reduced OS (HR = 0.219, 95% confidence interval [CI]: 0.052–0.924, p = 0.039; Table 3).

Table 3.
Cox regression analyses of TML and DDR gene mutants with the overall survival. Significant values are in bold.
Of the different gene mutations assessed, mutant TP53 was also significantly associated with a worse OS. However, in a multivariate Cox regression model, only TML-high (HR = 0.167, 95% CI: 1.030–0.936, p = 0.042) remained significantly associated with a worse OS, suggesting that TML may be an independent predictive variable for OS of CRC patients. In contrast, MMR gene mutations were not associated with the patient outcome.
We further evaluated TML as a potential prognostic biomarker for poor outcomes in CRC using Kaplan–Meier survival analysis. We found that, in our CRC patient cohort, TML-high was significantly associated with worse DFS (p = 0.036, Figure 1A) and OS (p = 0.024; Figure 1B). This further suggests that TML may be a positive predictive factor for determining the prognosis of patients with CRC.
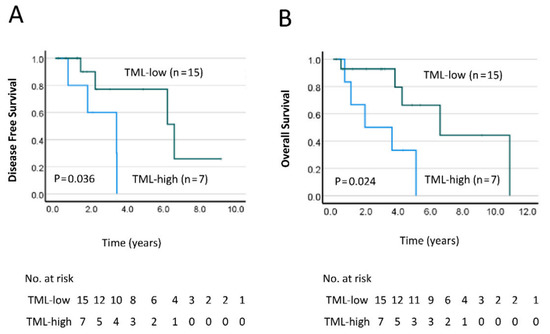
Figure 1.
Kaplan–Meier curves comparing the (A) disease-free survival (DFS) and (B) overall survival (OS) in patients with a low/med- vs. high-tumor mutation load (TML). Differences between the groups were analyzed by the log–rank test, and p-values are shown.
2.3. Mutational Profile of Rectal Cancer
Overall, targeted NGS analysis of tumor tissue from the 22 patients in our CRC cohort revealed 242 unique variants in 53 genes (Table 4). The most commonly mutated gene in our cohort was found to be TP53 (15/22; 68.2%), followed by HNF1A (12/22; 54.5%), ARID1A (10/22; 45.5%) and KRAS (5/22; 22.7%). Eight samples (36.3%) contained mutations in HR-DDR genes, including ATM, ATR, CHEK2, CDK12, FANCA, MRE11A, RAD50, NBN, ERCC2 and TP53, and six samples (27%) harbored mutations in the MMR genes MLH1, MSH2, MSH6 and PMS2.

Table 4.
Somatic mutations in CRC patients treated with neoadjuvant therapy.
As TP53 was the most commonly mutated gene in our cohort, we further examined the p53 mutation to determine whether the genetic changes affected a specific protein function. TP53 mutations were found to be predominantly localized in the DNA-binding domain (amino acid residues 94–290), effectively blocking wild-type (WT) p53 from binding to its transcription response elements and transactivating downstream target genes.
In total, we found the following seven amino acid substitutions in p53 mutational hotspots: R175H (p.Arg175His), H179Y (p.His179Tyr), R248W (p.Arg248Trp), R248Q (p.Arg248Gln), R273L (p.Arg273Leu), P278S (p.Pro278Ser) and R282W (p.Arg282Trp), all of which are localized in the DNA-binding domain. Notably, using Kaplan–Meier survival analysis, we found that patients with mutant p53 displayed worse OS (p = 0.023; Figure 2A) than those with WT p53, suggesting that the TP53 mutational status may also have prognostic value for CRC patients.
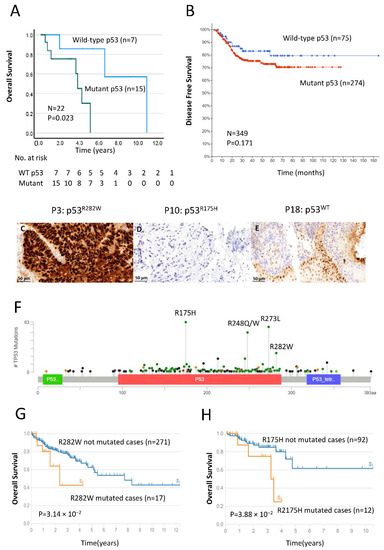
Figure 2.
Survival analysis of patients with TP53 mutations, as determined by next-generation sequencing (NGS), validation of p53 prognostic impact via Cancer Genomics database (http://www.cBioPortol.org, accessed on 15 July 2022) and immunohistochemical staining of the p53 protein in patient samples. (A) Kaplan–Meier curves comparing OS in patients with wild-type (WT) and mutant p53. Differences between the groups were analyzed by the log–rank test, and p-values are shown. (B) Kaplan–Meier survival analysis of DFS in rectal cancer patient groups with wild-type (n = 75, blue line) and mutant p53 (n = 274, red line). Total cases and p-values are shown. (C–E) Representative examples of typical nuclear staining for p53 in CRC patients with (C) TP53 mutant–overexpression, (D) TP53 mutant–reduced expression and (E) TP53 WT–normal expression (40× magnification, scale bar = 50 µm). (F) Diagram represents the protein domains of p53 encoded by TP53 genes. Plot was generated via cBioPortol tools based on a recent rectal cancer study (MSKCC, Nature Medicine 2022), and the p53 DNA-binding domain is shown in a red box. The presence of a mutation is shown on the x-axis, and the frequency of mutations is shown on the y-axis. Missense mutations are shown in green circles, and deleterious mutations (truncating, in frame and splice) are shown in black and brown circles. (G,H) Kaplan–Meier curves comparing p53R282W (G) and p53R175H (H) mutational statuses in patients with not-mutated cases and mutated cases provided by The Cancer Genome Atlas (TCGA-READ and TCGA-COAD).
2.4. Validation by IHC and Confirmation of TP53 Status by Cancer Genomics Databases
Mutation of the p53 gene is a common phenomenon in numerous human tumors, including CRC [35]. This often leads to an accumulation of nonfunctional p53 protein in the cell nuclei or loss of p53 protein, which can be detected by immunohistochemical techniques. Previous studies have further shown that p53 staining in tissue by IHC accurately reflects TP53 mutational status with high specificity, and the patterns of TP53 nucleotide variants correlate with the expression of p53 in cancers [36,37].
Here, to validate our findings by NGS, we performed immunohistochemical analysis to examine the expression of p53 protein in CRC tissue (Figure 2C–E). The results show a typical nuclear staining pattern in CRC tissue with WT TP53 (Figure 2E; patient ID, P18) and aberrant staining patterns in samples with mutant TP53, consistent with accumulation of non-functional p53 (Figure 2C; patient ID, P3) or loss of expression (Figure 2D; patient ID, P10), indicating that abnormal p53 levels are present in CRC tissue containing mutations in the TP53 gene.
Both patients with mutant TP53 (P3 and P10) were treated with neoadjuvant radiotherapy, followed by additional 5-FU-based adjuvant chemotherapy. By NGS, we found that the p53R282W mutation was present in P3, who displayed p53 overexpression (Figure 2C), whereas P10, which showed reduced p53 expression (Figure 2D), contained the p53R175H mutation, which harbors other mutations, including ATM, HNF1A, RB1, TP53 and MRE11A. This suggests that these mutations have distinct effects on the expression and possibly function of the p53 protein.
To confirm our findings in this study, we also performed the validation experiments using Cancer Genomics database (http://www.cBioPortol.org, accessed on 15 July 2022) to validate our results and we compared them with a recent rectal cancer study (MSKCC, Nature Medicine 2022). The Kaplan–Meier curve revealed that TP53 mutational status correlates with worse DFS (n = 349, Figure 2B), indicating potential prognostic role of p53 for CRC patients. In addition, a lollipop plot was generated using the same dataset via cBioPortol. Figure 2F represents the protein domains of p53 encoded by TP53 genes and shows the DNA-binding domain (DBD) of p53 covering from 94 to 290 amino acids.
Mutations in the DBD either interrupt the DNA binding directly or bring local/global change in the p53 structure. Both p53R282W and p53R175H mutations are classified as part of the structure mutations. To explore potential prognostic features for p53R282W and p53R175H mutations, Kaplan–Meier survival analysis was performed using The Cancer Genome Atlas (TCGA) database (n = 300, TCGA-READ and TCGA-COAD). The outcome results revealed both p53R282W and p53R175H mutational statuses are significantly associated with poor OS (p = 3.14 × 10−2, Figure 2G) and (p = 3.88 × 10−2, Figure 2H), respectively.
3. Discussion
Despite declining incidence rates in recent years, CRC remains among the top three most prevalent cancers and a leading cause of cancer-related death worldwide [1,2]. Challenges associated with complete tumor removal, particularly for rectal cancer patients, contribute to high recurrence and mortality rates and necessitate the use of more aggressive preoperative neoadjuvant therapies, which can improve outcomes for those with this CRC subtype [3,4,5,6,7]. However, therapeutic efficacy is highly variable, producing only partial or no response in most patients treated and highlighting the need for molecular biomarkers to identify patients most likely to benefit from such therapies [8,9,10].
Here, to address this need, we used targeted NGS technology to assess the prognostic value of specific DDR/MMR mutations, as well as total TML, for predicting survival and therapeutic outcomes in a cohort of 22 rectal cancer patients who received pre-operative neoadjuvant therapy. We found that both TML-high (>49 Mut/Mb) and mutations in TP53 were significantly associated with worse survival outcomes in our patient cohort, suggesting these molecular features may have prognostic value as biomarkers to determine the optimal therapeutic course for rectal cancer patients.
In total, 53 somatic mutations were identified in 22 patient samples; seven (32%) samples contained mutations in DDR genes, and TP53 was the most frequently mutated gene. We note, however, that our mutation counts likely represent underestimates, as the Oncomine™ NGS panel used in this study does not cover the full length of all DNA damage genes assessed but rather targets known mutational hotspots to estimate mutational burden. Regardless of this limitation, the results from univariate and Kaplan–Meier survival analyses indicate that both TML-high (>49 Mut/Mb) and mutations in TP53 were significantly associated with worse survival outcomes in our patient cohort. Thus, our findings suggest that TML and TP53 mutation have prognostic value as molecular biomarkers for determining the therapeutic course of rectal cancer patients.
Numerous studies have highlighted the prognostic value of tumor TML for predicting response to immune checkpoint inhibitor therapies, with a high TML associated with improved outcomes in patients with a diverse array of malignancies, including melanoma [38], non-small cell lung cancer [30] and CRC [34]. It is hypothesized that an elevated TML leads to production of a broader array of neoantigens for induction of immunogenicity [39,40]. Similarly, higher levels of microsatellite instability (MSI), resulting from MMR deficiency (features present in approximately 15% of CRC tumors [41,42]) are associated with increased neoantigens and better response to immunotherapy in CRC [43,44].
Less is known, however, about the value of TML for predicting prognosis of cancer patients receiving standard chemotherapy or radiotherapy. One study found that TML did not have prognostic value in a large cohort of patients with a diverse array of solid tumors who did not receive immunotherapy [45]. In contrast, findings from a meta-analysis suggested that standard platinum-based chemotherapy may produce better outcomes relative to immunotherapy for patients with low TML [46]. Notably, studies have shown that defective MMR and high MSI are associated with decreased efficacy of 5-FU therapy for CRC patients, although the prognostic value of TML was not assessed [47,48].
Here, we found that for our cohort of rectal cancer patients who underwent preoperative chemo/radiotherapy, TML-high was associated with worse OS in univariate and multivariate analyses. Further, subjects with a higher TML displayed worse DFS and OS in Kaplan–Meier survival analyses. These findings suggest that elevated genomic instability associated with a high TML may promote increased levels of cancer growth and metastasis and resistance to DNA-damaging treatments, possibly via the overexpression of DNA repair proteins and leading to worse outcomes with such therapies.
Thus, TML may represent a valuable molecular biomarker for predicting response to standard chemo/radiotherapy, with low TML patients most likely to benefit from such treatments. As high TML is associated with improved response to immunotherapy, this feature may be leveraged to help to determine the optimal therapeutic course for patients with CRC.
Chemo/radiotherapy kills cancer cells by producing high levels of DNA damage that cannot be repaired, ultimately triggering apoptosis. Treatment efficacy is therefore dependent on the DNA repair capacity of cancer cells, such that malignancies proficient in DNA repair will be more likely to display resistance, whereas those defective for repair are more sensitive [24,49,50]. Moreover, enhanced DNA repair may inhibit anti-tumor immunity, which is induced in response to DNA damage via the production of type I interferon [51].
Accordingly, numerous studies have reported that DDR defects promote enhanced sensitivity to chemo/radiotherapy [50,52], and mutations in DDR/MMR repair genes and/or aberrant expression of these proteins have been proposed as possible biomarkers for various malignancies, including CRC [17,18,19,20,21]. Consistent with this possibility, in a previous study, we found that elevated expression of the DNA repair proteins MRE11 and ATM was predictive for worse outcomes in CRC patients treated with neoadjuvant radiotherapy [16].
Similarly, overexpression of the HR repair protein RAD51 was found to be associated with worse outcomes in a large cohort of CRC patients, including the subset of rectal cancer patients treated with radiotherapy [15]. Qin et al. further reported that expression of the DNA repair protein X-ray repair complementing defective repair in Chinese hamster cells 2 (XRCC2) is linked to enhanced radioresistance and worse survival outcomes of rectal cancer patients, consistent with findings from other cancer types [53].
Paradoxically, however, for other DNA repair proteins, including RAD52 and RAD50, lower levels of expression are associated with worse outcomes for rectal cancer patients treated with neoadjuvant chemo/radiotherapy [54,55], implying that these proteins may have unique functions that contribute to resistance via unknown mechanisms. This is supported by our finding that, unlike RAD50 alone, elevated expression of the MRN complex, which contains RAD50 and plays a role in both NHEJ and HR, is associated with poorer prognosis of chemo/radiotherapy-treated rectal cancer patients [56].
Here, of the 53 somatic mutations identified in our 22-patient cohort, 13 mutations in seven patient samples were located in DDR/MMR genes. Of these, TP53 was the most frequently mutated gene (68.2%; 15/22). This is consistent with findings from previous studies and the central role of p53 in CRC development [35,50,57]. Most TP53 mutations were found within the DNA-binding domain and would thus be expected to disrupt the activation of p53 target genes.
IHC analyses further revealed aberrant patterns of p53 staining in representative TP53 mutant CRC tissue, confirming that the observed mutations also disrupt p53 protein expression. The results from the Kaplan–Meier survival analyses and univariate Cox regression further indicate that TP53 mutation was associated with worse survival outcomes in our rectal cancer patient cohort, although in this case, a significant association was not present in multivariate analyses.
We did not, however, observe significant association between other individual DDR/MMR mutations and patient outcomes in our univariate analysis. Previous studies have shown that the relationship between MMR gene mutations and patient outcome in CRC is complex and largely affected by features such as tumor stage and concurrent BRAF mutations [58,59,60,61]. Here, it is likely that we were unable to detect a positive relationship between MMR gene mutations and patient outcome due to our small patient cohort (n = 22 subjects)—a limitation of this study.
Thus, although our current findings suggest TP53 mutation may have prognostic value for rectal cancer patients who undergo neoadjuvant chemo/radiotherapy, future studies with larger patient populations will be needed to determine the clinical significance of other individual DDR/MMR gene mutations for this subset of CRC patients. Our small sample size may also explain why mutations in KRAS were detected at a slightly lower frequency than what has been reported in previous studies (27.3% vs. 30–50%, respectively) [57,62,63].
As the study focused on known MRN complex and MMR mutations in the DDR/MMR mutation panel we did not assess the mutational frequency of ERCC2/XPD. Lastly, we note that this study was limited by our lack of access to biopsy samples; as a result, all samples were obtained post-treatment, and we were unable to include complete responders in our patient cohort.
4. Materials and Methods
4.1. Patients
This study was approved by the South Western Sydney Local Health District Human Research Ethics Committee (HREC Reference: HREC/14/LPOOL/186; project number 14/103). Specimens were collected from 22 patients who underwent surgery and chemoradiotherapy for CRC between 2000 and 2011. All patients were treated with neoadjuvant chemo/radiotherapy prior to total mesorectal excision and anterior or abdominoperineal resection. Patients were treated with either a 25-Gy dose of radiotherapy administered in five treatment fractions or a 50.4-Gy dose administered in 28 fractions; both treatment groups received 5-FU-based chemotherapy. Follow-up included clinic visits, blood tests, colonoscopy and imaging based on the recommendations of the treating specialist.
4.2. FFPE Tissue Processing and DNA Extraction
Each formalin-fixed paraffin-embedded (FFPE) tissue section ranging in size from 5 to 10 µm in thickness, with no more than 2.25 cm2 of tissue area, was deparaffinized by submerging in xylene and vortexing vigorously 10 sec, followed by centrifugation at 20,000× g for 2 min at room temperature. DNA was then extracted from deparaffinized tissue sections using the QIAamp DNA FFPE Tissue Kit (QIAGEN, Hilden, Germany). All procedures were performed in accordance with the manufacturer’s instructions.
4.3. Qubit Assay
DNA derived from FFPE tissue was quantified using the Qubit dsDNA HS (High Sensitivity) Assay Kit (Thermo Fisher Scientific, Waltham, MA, USA). To prepare samples for analysis, 2 µL of purified DNA was added to 198 µL of Qubit working solution, for final volume of 200 µL. This mixture was then incubated at room temperature for at least 2 min before quantitation was performed with the Qubit 3.0 fluorometer (Thermo Fisher Scientific).
4.4. NGS Library Preparation and TaqMan qPCR
Library preparation was performed for the Oncomine™ Tumor Mutation Load Assay, according to the manufacturer’s protocol (Thermo Fisher Scientific). This targeted NGS panel covers portions of 409 genes that are known to be mutational hotspots in cancer and detects variants within their coding regions. Genomic DNA libraries were prepared using the Ion AmpliSeq™ Library Kit Plus (Thermo Fisher Scientific), starting with 20 ng DNA for each sample.
After combining the target amplification reactions and performing digestion of amplicons, ligation and purification, the purified libraries were quantified using the Ion Library TaqMan® Quantitation Kit (Thermo Fisher Scientific) and run on the QuantStudio 12K Flex Real-Time PCR System (Thermo Fisher Scientific), according to the manufacturer’s instructions. The final concentration of each library after dilution was determined (50 pM).
4.5. Sequencing and Data Analysis
The quantified libraries were clonally amplified by emulsion PCR and enriched on the Ion Chef instrument for automated template preparation using the Ion 540™ Kit-Chef, according to the manufacturer’s protocol (Thermo Fisher Scientific). Templates were loaded onto an Ion 540™ chip (four samples per chip, two chips per run) and sequenced with the Ion GeneStudio™ S5 sequencer system (Thermo Fisher Scientific), according to the manufacturer’s protocols.
NGS results were analyzed using the default settings of the integrated Oncomine™ Tumor Mutation Load-w3.1-DNA-Single Sample analysis workflow pipeline, available on the Ion Reporter software v.5.18 (Thermo Fisher Scientific). In our analysis workflow, the sequenced reads were aligned to the human reference genome hg19, and the resulting BAM files were transferred to Ion Reporter software for variant calling. Variants were identified by filtering with the Oncomine™ Variant Annotator plugin in Ion Reporter v.5.18, with default workflow settings.
4.6. Immunohistochemistry (IHC)
Immunohistochemical staining was performed according to the standard protocol used by the Anatomical Pathology Laboratory, Liverpool Hospital. Prior to staining, the tissue sections were incubated at 60 °C for at least 1 h. Deparaffinization was performed by immersing samples in xylene three times, followed by rehydration through graded, decreasing concentrations of ethanol, ending in running water. The sections were then incubated in pre-heated 98 °C citrate buffer, pH 6.0, in a hot water bath for 20 min, followed by cooling at room temperature for 20 min.
Endogenous peroxidase was blocked with hydrogen peroxidase for 20 min, and then slides were incubated with primary antibody for 20 min at room temperature in a moist chamber (p53, 1:800 dilution; Invitrogen, Thermo Fisher Scientific). Goat secondary antibody (Invitrogen, Thermo Fisher Scientific) for detection of the primary antibody–antigen complex, and high-sensitivity 3, 3′-diaminobenzidine tetrahydrochloride was then added for 15 min, followed by hematoxylin counterstaining and mounting.
4.7. Statical Analysis
Statistical analysis was performed with SPSS for Windows 27.0 (IBM Corporation, Armonk, NY, USA). The overall survival (OS) and disease-free survival (DFS) were assessed by performing Kaplan–Meier analysis, and the significance of the differences between groups was determined by the log–rank test, with p < 0.05 considered to be statistically significant. Univariate and multivariate analyses to compare survival in CRC patients with different clinical features and DDR/MMR gene mutational status were performed using Kaplan–Meier curves and Cox’s proportional hazard ratio (HR) survival modeling. Covariates included sex; age; Tumor, Node and Metastasis (TNM) stage; histological grade; vascular invasion; perineural invasion; chemotherapy; and radiotherapy. Significance for univariate and multivariate analyses was assessed using the Mann–Whitney U test, and in all cases, p < 0.05 was considered statistically significant.
5. Conclusions
In summary, our results indicate that both TML-high (>49 Mut/Mb) and TP53 mutation were associated with worse survival outcomes for rectal cancer patients who received neoadjuvant therapy prior to surgery. Overall, the targeted Oncomine™ Tumor Mutation Load Assay NGS panel uncovered 53 somatic mutations in our 22-patient cohort, with seven (32%) patients containing mutations in 13 DDR/MMR genes and TP53 representing the most frequently mutated gene. Collectively, our findings indicate that TML and TP53 mutation may have prognostic value for CRC patients, suggesting that these features can potentially be leveraged as independent biomarkers for patient management and therapeutic course determination.
Author Contributions
V.H., L.C. and C.S.L. conceived and designed the research; L.C., Y.M., B.W., V.L., A.A. and V.H. collected and did preliminary analysis on the data; V.H., L.C., S.H.L., Y.M., B.W., W.N., M.L., T.L.R., W.C. and C.S.L. further analyzed and interpreted the data; V.H. and L.C. wrote the draft manuscript; S.H.L., Y.M., B.W., W.N., M.L., T.L.R., W.C. and C.S.L. provided critical material. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by the Ainsworth Medical Research Innovation Fund, School of Medicine, Western Sydney University, Australia, grant number 64927.
Institutional Review Board Statement
The study was conducted in accordance with the Declaration of Helsinki and approved by the Human Research Ethics Committee of the South Western Sydney Local Health District (HREC Reference: HREC/14/LPOOL/186; project number 14/103; 25 May 2017).
Informed Consent Statement
Informed consent was obtained from all subjects involved in the study.
Data Availability Statement
The data presented in this study are available on request from the corresponding author. The data are not publicly available due to data size and privacy.
Acknowledgments
We acknowledge the support of the Ainsworth Medical Research Innovation Fund, School of Medicine, Western Sydney University, Australia. We kindly thank Laura Marinelli for her editorial assistance.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Siegel, R.L.; Miller, K.D.; Fuchs, H.E.; Jemal, A. Cancer Statistics, 2022. CA Cancer J. Clin. 2022, 72, 7–33. [Google Scholar] [CrossRef] [PubMed]
- Colorectal Cancer—Statistics. Available online: https://www.cancer.net/cancer-types/colorectal-cancer/statistics (accessed on 5 April 2022).
- Hong, T.S.; Clark, J.W.; Haigis, K.M. Cancers of the Colon and Rectum: Identical or Fraternal Twins? Cancer Discov. 2012, 2, 117–121. [Google Scholar] [CrossRef] [PubMed]
- Manfredi, S.; Bouvier, A.M.; Lepage, C.; Hatem, C.; Dancourt, V.; Faivre, J. Incidence and Patterns of Recurrence after Resection for Cure of Colonic Cancer in a Well Defined Population. Br. J. Surg. 2006, 93, 1115–1122. [Google Scholar] [CrossRef] [PubMed]
- Brenner, H.; Kloor, M.; Pox, C.P. Colorectal Cancer. Lancet 2014, 383, 1490–1502. [Google Scholar] [CrossRef]
- Tamas, K.; Walenkamp, A.M.E.; de Vries, E.G.E.; van Vugt, M.A.T.M.; Beets-Tan, R.G.; van Etten, B.; de Groot, D.J.A.; Hospers, G.A.P. Rectal and Colon Cancer: Not Just a Different Anatomic Site. Cancer Treat. Rev. 2015, 41, 671–679. [Google Scholar] [CrossRef]
- Wang, S.-J.; Hathout, L.; Malhotra, U.; Maloney-Patel, N.; Kilic, S.; Poplin, E.; Jabbour, S.K. Decision-Making Strategy for Rectal Cancer Management Using Radiation Therapy for Elderly or Comorbid Patients. Int. J. Radiat. Oncol. Biol. Phys. 2018, 100, 926–944. [Google Scholar] [CrossRef]
- Ruo, L.; Tickoo, S.; Klimstra, D.S.; Minsky, B.D.; Saltz, L.; Mazumdar, M.; Paty, P.B.; Wong, W.D.; Larson, S.M.; Cohen, A.M.; et al. Long-Term Prognostic Significance of Extent of Rectal Cancer Response to Preoperative Radiation and Chemotherapy. Ann. Surg. 2002, 236, 75–81. [Google Scholar] [CrossRef]
- Das, P.; Skibber, J.M.; Rodriguez-Bigas, M.A.; Feig, B.W.; Chang, G.J.; Wolff, R.A.; Eng, C.; Krishnan, S.; Janjan, N.A.; Crane, C.H. Predictors of Tumor Response and Downstaging in Patients Who Receive Preoperative Chemoradiation for Rectal Cancer. Cancer 2007, 109, 1750–1755. [Google Scholar] [CrossRef]
- Geng, L.; Wang, J. Molecular Effectors of Radiation Resistance in Colorectal Cancer. Precis. Radiat. Oncol. 2017, 1, 27–33. [Google Scholar] [CrossRef]
- Kang, S.; Wu, C. Total Neoadjuvant Therapy Approach in Rectal Adenocarcinoma. Clin. Adv. Hematol. Oncol. 2021, 19, 711–718. [Google Scholar]
- Sclafani, F.; Corrò, C.; Koessler, T. Debating Pros and Cons of Total Neoadjuvant Therapy in Rectal Cancer. Cancers 2021, 13, 6361. [Google Scholar] [CrossRef]
- Cerdán-Santacruz, C.; Vailati, B.B.; São Julião, G.P.; Habr-Gama, A.; Perez, R.O. Watch and Wait: Why, to Whom and How. Surg. Oncol. 2022, 101774. [Google Scholar] [CrossRef]
- Quezada-Diaz, F.F.; Smith, J.J. Nonoperative Management for Rectal Cancer. Hematol. Oncol. Clin. N. Am. 2022, 36, 539–551. [Google Scholar] [CrossRef]
- Tennstedt, P.; Fresow, R.; Simon, R.; Marx, A.; Terracciano, L.; Petersen, C.; Sauter, G.; Dikomey, E.; Borgmann, K. RAD51 Overexpression Is a Negative Prognostic Marker for Colorectal Adenocarcinoma. Int. J. Cancer 2013, 132, 2118–2126. [Google Scholar] [CrossRef]
- Ho, V.; Chung, L.; Revoltar, M.; Lim, S.H.; Tut, T.-G.; Abubakar, A.; Henderson, C.J.; Chua, W.; Ng, W.; Lee, M.; et al. MRE11 and ATM Expression Levels Predict Rectal Cancer Survival and Their Association with Radiotherapy Response. PLoS ONE 2016, 11, e0167675. [Google Scholar] [CrossRef]
- Dayde, D.; Tanaka, I.; Jain, R.; Tai, M.C.; Taguchi, A. Predictive and Prognostic Molecular Biomarkers for Response to Neoadjuvant Chemoradiation in Rectal Cancer. Int. J. Mol. Sci. 2017, 18, 573. [Google Scholar] [CrossRef]
- Peluso, G.; Incollingo, P.; Calogero, A.; Tammaro, V.; Rupealta, N.; Chiacchio, G.; Sandoval Sotelo, M.L.; Minieri, G.; Pisani, A.; Riccio, E.; et al. Current Tissue Molecular Markers in Colorectal Cancer: A Literature Review. Biomed. Res. Int. 2017, 2017, 2605628. [Google Scholar] [CrossRef]
- Yoo, B.C.; Yeo, S.-G. Clinical Utility of Pretreatment Prediction of Chemoradiotherapy Response in Rectal Cancer: A Review. EPMA J. 2017, 8, 61–67. [Google Scholar] [CrossRef]
- Bottarelli, L.; De’Angelis, G.L.; Azzoni, C.; Di Mario, F.; De’Angelis, N.; Leandro, G.; Fornaroli, F.; Gaiani, F.; Negri, F. Potential Predictive Biomarkers in Locally Advanced Rectal Cancer Treated with Preoperative Chemo-Radiotherapy. Acta Biomed. 2018, 89, 102–106. [Google Scholar] [CrossRef]
- Vacante, M.; Borzì, A.M.; Basile, F.; Biondi, A. Biomarkers in Colorectal Cancer: Current Clinical Utility and Future Perspectives. World J. Clin. Cases 2018, 6, 869–881. [Google Scholar] [CrossRef]
- Giglia-Mari, G.; Zotter, A.; Vermeulen, W. DNA Damage Response. Cold Spring Harb. Perspect. Biol. 2011, 3, a000745. [Google Scholar] [CrossRef]
- Vaddavalli, P.L.; Schumacher, B. The P53 Network: Cellular and Systemic DNA Damage Responses in Cancer and Aging. Trends Genet. 2022, 38, 598–612. [Google Scholar] [CrossRef]
- Adamsen, B.L.; Kravik, K.L.; De Angelis, P.M. DNA Damage Signaling in Response to 5-Fluorouracil in Three Colorectal Cancer Cell Lines with Different Mismatch Repair and TP53 Status. Int. J. Oncol. 2011, 39, 673–682. [Google Scholar] [CrossRef][Green Version]
- Yang, G.; Liu, C.; Chen, S.-H.; Kassab, M.A.; Hoff, J.D.; Walter, N.G.; Yu, X. Super-Resolution Imaging Identifies PARP1 and the Ku Complex Acting as DNA Double-Strand Break Sensors. Nucleic Acids Res. 2018, 46, 3446–3457. [Google Scholar] [CrossRef]
- Scully, R.; Panday, A.; Elango, R.; Willis, N.A. DNA Double-Strand Break Repair-Pathway Choice in Somatic Mammalian Cells. Nat. Rev. Mol. Cell Biol. 2019, 20, 698–714. [Google Scholar] [CrossRef]
- Situ, Y.; Chung, L.; Lee, C.S.; Ho, V. MRN (MRE11-RAD50-NBS1) Complex in Human Cancer and Prognostic Implications in Colorectal Cancer. Int. J. Mol. Sci. 2019, 20, 816. [Google Scholar] [CrossRef]
- Pećina-Šlaus, N.; Kafka, A.; Salamon, I.; Bukovac, A. Mismatch Repair Pathway, Genome Stability and Cancer. Front. Mol. Biosci. 2020, 7, 122. [Google Scholar] [CrossRef] [PubMed]
- Yeh, Y.-M.; Lee, C.-H.; Chen, S.-H.; Lee, C.-T.; Chen, Y.-L.; Lin, B.-W.; Lin, S.-C.; Chan, R.-H.; Lee, J.-C.; Shen, M.-R.; et al. Comprehensive Assessment of HER2 Alteration in a Colorectal Cancer Cohort: From next-Generation Sequencing to Clinical Significance. Cancer Manag. Res. 2019, 11, 7867–7875. [Google Scholar] [CrossRef]
- Alborelli, I.; Leonards, K.; Rothschild, S.I.; Leuenberger, L.P.; Savic Prince, S.; Mertz, K.D.; Poechtrager, S.; Buess, M.; Zippelius, A.; Läubli, H.; et al. Tumor Mutational Burden Assessed by Targeted NGS Predicts Clinical Benefit from Immune Checkpoint Inhibitors in Non-Small Cell Lung Cancer. J. Pathol. 2020, 250, 19–29. [Google Scholar] [CrossRef]
- Harpaz, N.; Gatt, Y.E.; Granit, R.Z.; Fruchtman, H.; Hubert, A.; Grinshpun, A. Mucinous Histology, BRCA1/2 Mutations, and Elevated Tumor Mutational Burden in Colorectal Cancer. J. Oncol. 2020, 2020, e6421205. [Google Scholar] [CrossRef]
- Park, J.; Lee, S.-I.; Shin, S.; Hong, J.H.; Yoo, H.M.; Kim, J.G. Genetic Profiling of Somatic Alterations by Oncomine Focus Assay in Korean Patients with Advanced Gastric Cancer. Oncol. Lett. 2020, 20, 1. [Google Scholar] [CrossRef] [PubMed]
- Özdoğan, M.; Papadopoulou, E.; Tsoulos, N.; Tsantikidi, A.; Mariatou, V.-M.; Tsaousis, G.; Kapeni, E.; Bourkoula, E.; Fotiou, D.; Kapetsis, G.; et al. Comprehensive Tumor Molecular Profile Analysis in Clinical Practice. BMC Med. Genom. 2021, 14, 105. [Google Scholar] [CrossRef] [PubMed]
- Schrock, A.B.; Ouyang, C.; Sandhu, J.; Sokol, E.; Jin, D.; Ross, J.S.; Miller, V.A.; Lim, D.; Amanam, I.; Chao, J.; et al. Tumor Mutational Burden Is Predictive of Response to Immune Checkpoint Inhibitors in MSI-High Metastatic Colorectal Cancer. Ann. Oncol. 2019, 30, 1096–1103. [Google Scholar] [CrossRef] [PubMed]
- Nakayama, M.; Oshima, M. Mutant P53 in Colon Cancer. J. Mol. Cell Biol. 2019, 11, 267–276. [Google Scholar] [CrossRef]
- Manoharan, V.; Karunanayake, E.H.; Tennekoon, K.H.; De Silva, S.; Imthikab, A.I.A.; De Silva, K.; Angunawela, P.; Vishwakula, S.; Lunec, J. Pattern of Nucleotide Variants of TP53 and Their Correlation with the Expression of P53 and Its Downstream Proteins in a Sri Lankan Cohort of Breast and Colorectal Cancer Patients. BMC Cancer 2020, 20, 72. [Google Scholar] [CrossRef]
- Singh, N.; Piskorz, A.M.; Bosse, T.; Jimenez-Linan, M.; Rous, B.; Brenton, J.D.; Gilks, C.B.; Köbel, M. P53 Immunohistochemistry Is an Accurate Surrogate for TP53 Mutational Analysis in Endometrial Carcinoma Biopsies. J. Pathol. 2020, 250, 336–345. [Google Scholar] [CrossRef]
- Snyder, A.; Makarov, V.; Merghoub, T.; Yuan, J.; Zaretsky, J.M.; Desrichard, A.; Walsh, L.A.; Postow, M.A.; Wong, P.; Ho, T.S.; et al. Genetic Basis for Clinical Response to CTLA-4 Blockade in Melanoma. N. Engl. J. Med. 2014, 371, 2189–2199. [Google Scholar] [CrossRef]
- Goodman, A.M.; Kato, S.; Bazhenova, L.; Patel, S.P.; Frampton, G.M.; Miller, V.; Stephens, P.J.; Daniels, G.A.; Kurzrock, R. Tumor Mutational Burden as an Independent Predictor of Response to Immunotherapy in Diverse Cancers. Mol. Cancer Ther. 2017, 16, 2598–2608. [Google Scholar] [CrossRef]
- Okada, M.; Shimizu, K.; Fujii, S. Identification of Neoantigens in Cancer Cells as Targets for Immunotherapy. Int. J. Mol. Sci. 2022, 23, 2594. [Google Scholar] [CrossRef]
- Ionov, Y.; Peinado, M.A.; Malkhosyan, S.; Shibata, D.; Perucho, M. Ubiquitous Somatic Mutations in Simple Repeated Sequences Reveal a New Mechanism for Colonic Carcinogenesis. Nature 1993, 363, 558–561. [Google Scholar] [CrossRef]
- Cancer Genome Atlas Network. Comprehensive Molecular Characterization of Human Colon and Rectal Cancer. Nature 2012, 487, 330–337. [Google Scholar] [CrossRef]
- Du, F.; Liu, Y. Predictive Molecular Markers for the Treatment with Immune Checkpoint Inhibitors in Colorectal Cancer. J. Clin. Lab. Anal. 2021, 36, e24141. [Google Scholar] [CrossRef]
- Zheng, Y.; Fu, Y.; Wang, P.-P.; Ding, Z.-Y. Neoantigen: A Promising Target for the Immunotherapy of Colorectal Cancer. Dis. Markers 2022, 2022, 8270305. [Google Scholar] [CrossRef]
- Shao, C.; Li, G.; Huang, L.; Pruitt, S.; Castellanos, E.; Frampton, G.; Carson, K.R.; Snow, T.; Singal, G.; Fabrizio, D.; et al. Prevalence of High Tumor Mutational Burden and Association with Survival in Patients with Less Common Solid Tumors. JAMA Netw. Open 2020, 3, e2025109. [Google Scholar] [CrossRef]
- Galvano, A.; Gristina, V.; Malapelle, U.; Pisapia, P.; Pepe, F.; Barraco, N.; Castiglia, M.; Perez, A.; Rolfo, C.; Troncone, G.; et al. The Prognostic Impact of Tumor Mutational Burden (TMB) in the First-Line Management of Advanced Non-Oncogene Addicted Non-Small-Cell Lung Cancer (NSCLC): A Systematic Review and Meta-Analysis of Randomized Controlled Trials. ESMO Open 2021, 6, 100124. [Google Scholar] [CrossRef]
- Ribic, C.M.; Sargent, D.J.; Moore, M.J.; Thibodeau, S.N.; French, A.J.; Goldberg, R.M.; Hamilton, S.R.; Laurent-Puig, P.; Gryfe, R.; Shepherd, L.E.; et al. Tumor Microsatellite-Instability Status as a Predictor of Benefit from Fluorouracil-Based Adjuvant Chemotherapy for Colon Cancer. N. Engl. J. Med. 2003, 349, 247–257. [Google Scholar] [CrossRef]
- Sargent, D.J.; Marsoni, S.; Monges, G.; Thibodeau, S.N.; Labianca, R.; Hamilton, S.R.; French, A.J.; Kabat, B.; Foster, N.R.; Torri, V.; et al. Defective Mismatch Repair As a Predictive Marker for Lack of Efficacy of Fluorouracil-Based Adjuvant Therapy in Colon Cancer. J. Clin. Oncol. 2010, 28, 3219–3226. [Google Scholar] [CrossRef]
- Mladenov, E.; Magin, S.; Soni, A.; Iliakis, G. DNA Double-Strand Break Repair as Determinant of Cellular Radiosensitivity to Killing and Target in Radiation Therapy. Front. Oncol. 2013, 3, 113. [Google Scholar] [CrossRef]
- Reilly, N.M.; Novara, L.; Di Nicolantonio, F.; Bardelli, A. Exploiting DNA Repair Defects in Colorectal Cancer. Mol. Oncol. 2019, 13, 681–700. [Google Scholar] [CrossRef]
- Härtlova, A.; Erttmann, S.F.; Raffi, F.A.; Schmalz, A.M.; Resch, U.; Anugula, S.; Lienenklaus, S.; Nilsson, L.M.; Kröger, A.; Nilsson, J.A.; et al. DNA Damage Primes the Type I Interferon System via the Cytosolic DNA Sensor STING to Promote Anti-Microbial Innate Immunity. Immunity 2015, 42, 332–343. [Google Scholar] [CrossRef]
- Goldstein, M.; Kastan, M.B. The DNA Damage Response: Implications for Tumor Responses to Radiation and Chemotherapy. Annu. Rev. Med. 2015, 66, 129–143. [Google Scholar] [CrossRef]
- Qin, C.-J.; Song, X.-M.; Chen, Z.-H.; Ren, X.-Q.; Xu, K.-W.; Jing, H.; He, Y.-L. XRCC2 as a Predictive Biomarker for Radioresistance in Locally Advanced Rectal Cancer Patients Undergoing Preoperative Radiotherapy. Oncotarget 2015, 6, 32193–32204. [Google Scholar] [CrossRef]
- Ho, V.; Chung, L.; Singh, A.; Lea, V.; Revoltar, M.; Lim, S.H.; Tut, T.-G.; Ng, W.; Lee, M.; de Souza, P.; et al. Early Postoperative Low Expression of RAD50 in Rectal Cancer Patients Associates with Disease-Free Survival. Cancers 2017, 9, 163. [Google Scholar] [CrossRef]
- Ho, V.; Chung, L.; Singh, A.; Lea, V.; Abubakar, A.; Lim, S.H.; Chua, W.; Ng, W.; Lee, M.; Roberts, T.L.; et al. Aberrant Expression of RAD52, Its Prognostic Impact in Rectal Cancer and Association with Poor Survival of Patients. Int. J. Mol. Sci. 2020, 21, 1768. [Google Scholar] [CrossRef]
- Ho, V.; Chung, L.; Singh, A.; Lea, V.; Abubakar, A.; Lim, S.H.; Ng, W.; Lee, M.; de Souza, P.; Shin, J.-S.; et al. Overexpression of the MRE11-RAD50-NBS1 (MRN) Complex in Rectal Cancer Correlates with Poor Response to Neoadjuvant Radiotherapy and Prognosis. BMC Cancer 2018, 18, 869. [Google Scholar] [CrossRef]
- Zhou, Z.; Xie, X.; Wang, X.; Zhang, X.; Li, W.; Sun, T.; Cai, Y.; Wu, J.; Dang, C.; Zhang, H. Correlations Between Tumor Mutation Burden and Immunocyte Infiltration and Their Prognostic Value in Colon Cancer. Front. Genet. 2021, 12, 623424. [Google Scholar] [CrossRef]
- French, A.J.; Sargent, D.J.; Burgart, L.J.; Foster, N.R.; Kabat, B.F.; Goldberg, R.; Shepherd, L.; Windschitl, H.E.; Thibodeau, S.N. Prognostic Significance of Defective Mismatch Repair and BRAF V600E in Patients with Colon Cancer. Clin. Cancer Res. 2008, 14, 3408–3415. [Google Scholar] [CrossRef] [PubMed]
- Sinicrope, F.A.; Shi, Q.; Allegra, C.J.; Smyrk, T.C.; Thibodeau, S.N.; Goldberg, R.M.; Meyers, J.P.; Pogue-Geile, K.L.; Yothers, G.; Sargent, D.J.; et al. Association of DNA Mismatch Repair and Mutations in BRAF and KRAS With Survival After Recurrence in Stage III Colon Cancers: A Secondary Analysis of 2 Randomized Clinical Trials. JAMA Oncol. 2017, 3, 472–480. [Google Scholar] [CrossRef] [PubMed]
- Hestetun, K.E.; Aasebø, K.; Rosenlund, N.B.; Müller, Y.; Dahl, O.; Myklebust, M.P. Mismatch Repair Phenotype Determines the Implications of Tumor Grade and CDX2 Expression in Stage II-III Colon Cancer. Mod. Pathol. 2021, 34, 161–170. [Google Scholar] [CrossRef] [PubMed]
- Tan, E.; Whiting, J.; Xie, H.; Imanirad, I.; Carballido, E.; Felder, S.; Frakes, J.; Mo, Q.; Walko, C.; Permuth, J.B.; et al. BRAF Mutations Are Associated with Poor Survival Outcomes in Advanced-Stage Mismatch Repair-Deficient/Microsatellite High Colorectal Cancer. Oncologist 2022, 27, 191–197. [Google Scholar] [CrossRef] [PubMed]
- Wolff, R.K.; Hoffman, M.D.; Wolff, E.C.; Herrick, J.S.; Sakoda, L.C.; Samowitz, W.S.; Slattery, M.L. Mutation Analysis of Adenomas and Carcinomas of the Colon: Early and Late Drivers. Genes Chromosomes Cancer 2018, 57, 366–376. [Google Scholar] [CrossRef]
- Yaeger, R.; Chatila, W.K.; Lipsyc, M.D.; Hechtman, J.F.; Cercek, A.; Sanchez-Vega, F.; Jayakumaran, G.; Middha, S.; Zehir, A.; Donoghue, M.T.A.; et al. Clinical Sequencing Defines the Genomic Landscape of Metastatic Colorectal Cancer. Cancer Cell 2018, 33, 125–136.e3. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).