Shear-Wave Elastography Using Commercially Available Ultrasound in a Mouse Model of Chronic Liver Disease
Abstract
:1. Introduction
2. Materials and Methods
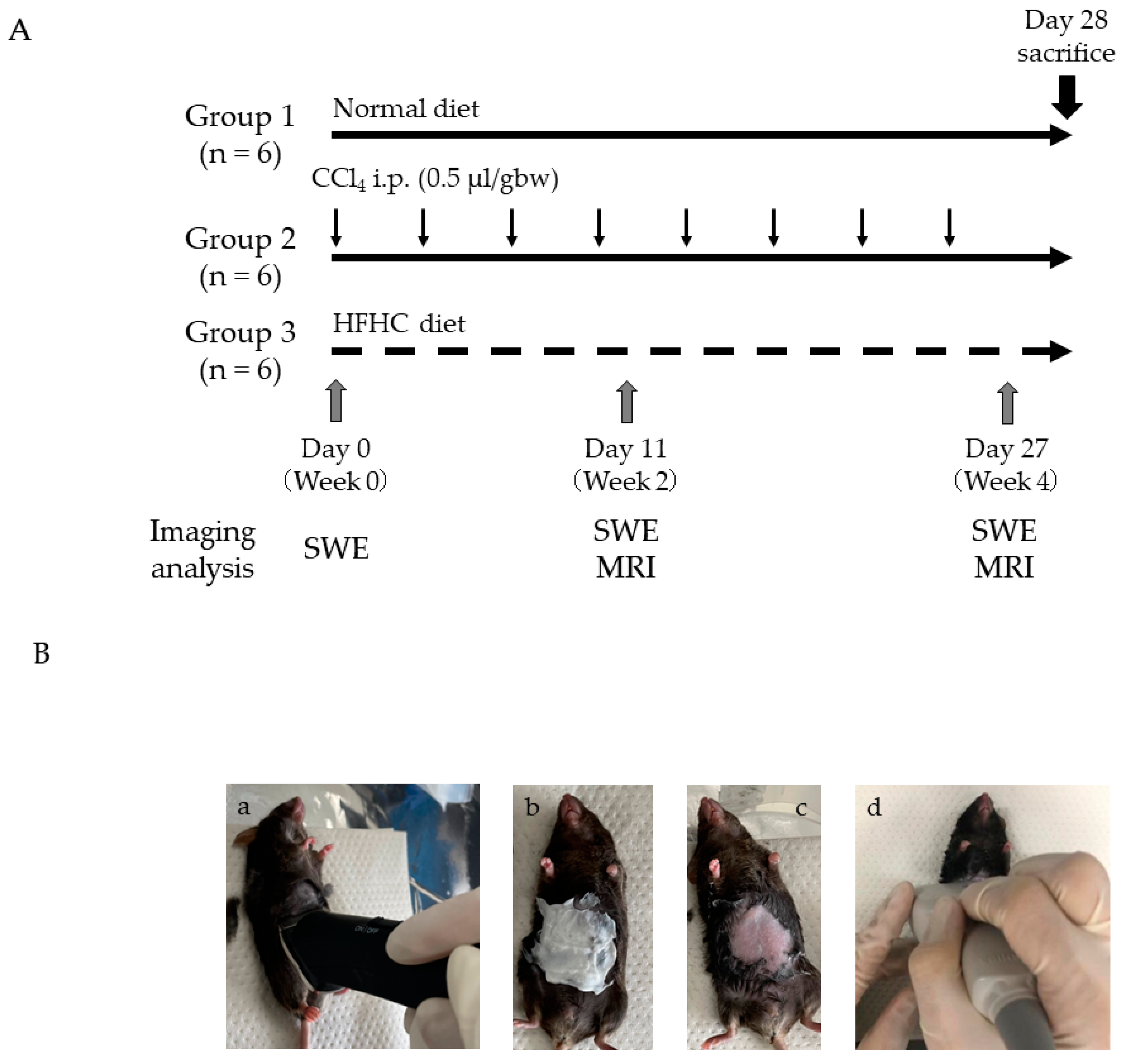
2.1. Experimental Protocol
2.2. Hematoxylin-and-Eosin (H&E) Staining and Picrosirius Red Staining
2.3. SWE Measurements
2.4. MRI
2.5. RNA Isolation and Quantitative Real-Time Polymerase Chain Reaction (RT-PCR)
2.6. M2BP ELISA Measurement
2.7. Statistical Analysis
3. Results
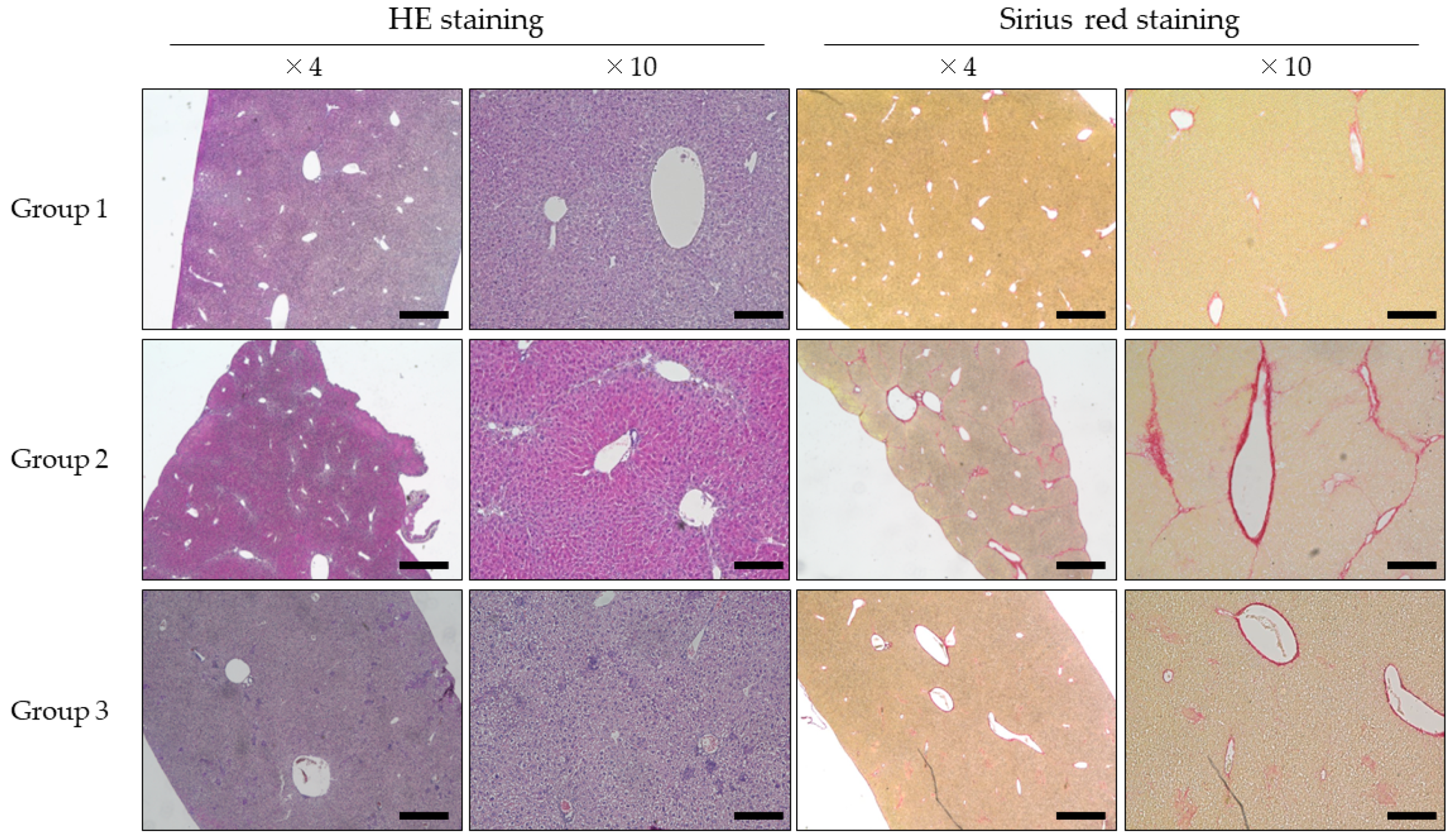
3.1. Evaluation of Mouse Liver Histology
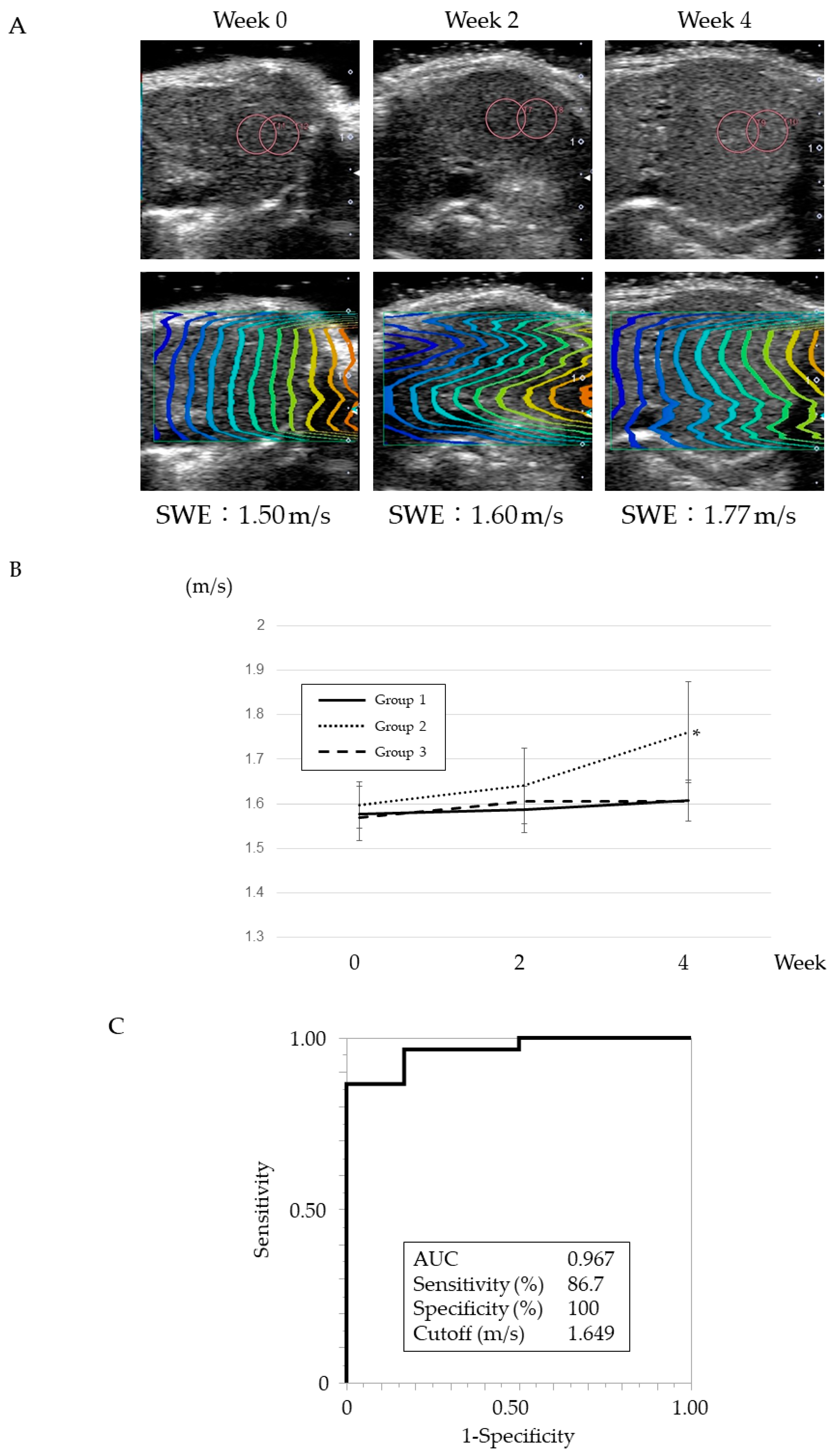
3.2. Changes in SWE Values
3.3. MRI (T1rho, T2) Maps
3.4. Relationship between SWE Values and MRI Parameters
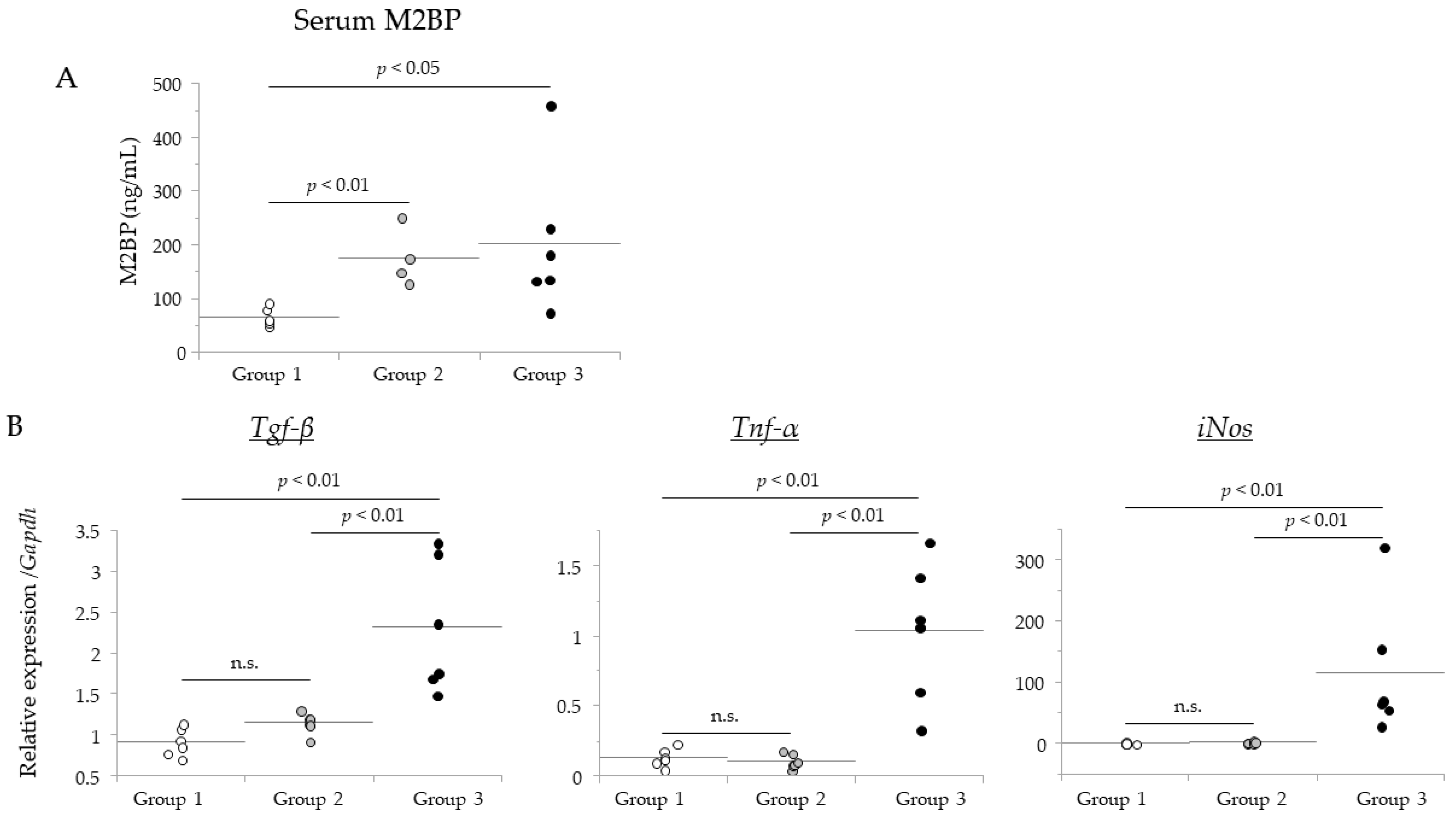
3.5. Comparison of SWE Results with Levels of a Liver Fibrosis Biomarker and Hepatic Genes
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Bataller, R.; Brenner, D.A. Liver fibrosis. J. Clin. Investig. 2005, 115, 209–218. [Google Scholar] [CrossRef] [PubMed]
- Sarin, S.K.; Kumar, M.; Eslam, M.; George, J.; Al Mahtab, M.; Akbar, S.M.F.; Jia, J.; Tian, Q.; Aggarwal, R.; Muljono, D.H.; et al. Liver diseases in the Asia-Pacific region: A Lancet Gastroenterology & Hepatology Commission. Lancet Gastroenterol. Hepatol. 2020, 5, 167–228. [Google Scholar] [PubMed] [Green Version]
- Angulo, P.; Kleiner, D.E.; Dam-Larsen, S.; Adams, L.A.; Bjornsson, E.S.; Charatcharoenwitthaya, P.; Mills, P.R.; Keach, J.C.; Lafferty, H.D.; Stahler, A.; et al. Liver Fibrosis, but No Other Histologic Features, Is Associated With Long-term Outcomes of Patients With Nonalcoholic Fatty Liver Disease. Gastroenterology 2015, 149, 389–397.e10. [Google Scholar] [CrossRef] [Green Version]
- Sugimoto, K.; Moriyasu, F.; Oshiro, H.; Takeuchi, H.; Yoshimasu, Y.; Kasai, Y.; Furuichi, Y.; Itoi, T. Viscoelasticity Measurement in Rat Livers Using Shear-Wave US Elastography. Ultrasound. Med. Biol. 2018, 44, 2018–2024. [Google Scholar] [CrossRef] [PubMed]
- Morin, J.; Swanson, T.A.; Rinaldi, A.; Boucher, M.; Ross, T.; Hirenallur-Shanthappa, D. Application of Ultrasound and Shear Wave Elastography Imaging in a Rat Model of NAFLD/NASH. J. Vis. Exp. 2021, 170, e62403. [Google Scholar] [CrossRef] [PubMed]
- Yeh, C.L.; Chen, B.R.; Tseng, L.Y.; Jao, P.; Su, T.H.; Li, P.C. Shear-wave elasticity imaging of a liver fibrosis mouse model using high-frequency ultrasound. IEEE Trans. Ultrason. Ferroelectr. Freq. Control. 2015, 62, 1295–1307. [Google Scholar] [CrossRef]
- Iwata, A.; Kamada, Y.; Ebisutani, Y.; Yamamoto, A.; Ueda, Y.; Arai, H.; Fujii, H.; Takamatsu, S.; Maruyama, N.; Maeda, M.; et al. Establishment of mouse Mac-2 binding protein enzyme-linked immunosorbent assay and its application for mouse chronic liver disease models. Hepatol. Res. 2017, 47, 902–909. [Google Scholar] [CrossRef]
- Kamada, Y.; Fujii, H.; Fujii, H.; Sawai, Y.; Doi, Y.; Uozumi, N.; Mizutani, K.; Akita, M.; Sato, M.; Kida, S.; et al. Serum Mac-2 binding protein levels as a novel diagnostic biomarker for prediction of disease severity and nonalcoholic steatohepatitis. Proteom. Clin. Appl. 2013, 7, 648–656. [Google Scholar] [CrossRef]
- Kamada, Y.; Ono, M.; Hyogo, H.; Fujii, H.; Sumida, Y.; Yamada, M.; Mori, K.; Tanaka, S.; Maekawa, T.; Ebisutani, Y.; et al. Use of Mac-2 binding protein as a biomarker for nonalcoholic fatty liver disease diagnosis. Hepatol. Commun. 2017, 1, 780–791. [Google Scholar] [CrossRef]
- Kamada, Y.; Morishita, K.; Koseki, M.; Nishida, M.; Asuka, T.; Naito, Y.; Yamada, M.; Takamatsu, S.; Sakata, Y.; Takehara, T.; et al. Serum Mac-2 Binding Protein Levels Associate with Metabolic Parameters and Predict Liver Fibrosis Progression in Subjects with Fatty Liver Disease: A 7-Year Longitudinal Study. Nutrients 2020, 12, 1770. [Google Scholar] [CrossRef]
- Kamada, Y.; Ono, M.; Hyogo, H.; Fujii, H.; Sumida, Y.; Mori, K.; Tanaka, S.; Yamada, M.; Akita, M.; Mizutani, K.; et al. A novel noninvasive diagnostic method for nonalcoholic steatohepatitis using two glycobiomarkers. Hepatology 2015, 62, 1433–1443. [Google Scholar] [CrossRef]
- Arihara, N.; Saito, S.; Sawaya, R.; Onishi, R.; Tsuji, K.; Ohki, A.; Ueda, J.; Morimoto-Ishiwaka, D. Evaluation of liver T(1rho) and T(2) values in acute liver inflammation models using 7T-MRI. Magn. Reson. Imaging 2022, 88, 20–24. [Google Scholar] [CrossRef] [PubMed]
- Zhao, F.; Yuan, J.; Deng, M.; Lu, P.X.; Ahuja, A.T.; Wang, Y.X. Further exploration of MRI techniques for liver T1rho quantification. Quant. Imaging Med. Surg. 2013, 3, 308–315. [Google Scholar] [PubMed]
- Kamada, Y.; Mori, K.; Matsumoto, H.; Kiso, S.; Yoshida, Y.; Shinzaki, S.; Hiramatsu, N.; Ishii, M.; Moriwaki, K.; Kawada, N.; et al. N-Acetylglucosaminyltransferase V regulates TGF-beta response in hepatic stellate cells and the progression of steatohepatitis. Glycobiology 2012, 22, 778–787. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Saito, S.; Takahashi, Y.; Ohki, A.; Shintani, Y.; Higuchi, T. Early detection of elevated lactate levels in a mitochondrial disease model using chemical exchange saturation transfer (CEST) and magnetic resonance spectroscopy (MRS) with 7T MR imaging. Radiol. Phys. Technol. 2019, 12, 232–233. [Google Scholar] [CrossRef]
- Luetkens, J.A.; Klein, S.; Träber, F.; Schmeel, F.C.; Sprinkart, A.M.; Kuetting, D.L.R.; Block, W.; Uschner, F.E.; Schierwagen, R.; Hittatiya, K.; et al. Quantification of Liver Fibrosis at T1 and T2 Mapping with Extracellular Volume Fraction MRI: Preclinical Results. Radiology 2018, 288, 748–754. [Google Scholar] [CrossRef]
- Zhao, F.; Wang, Y.X.; Yuan, J.; Deng, M.; Wong, H.L.; Chu, E.S.; Go, M.Y.; Teng, G.J.; Ahuja, A.T.; Yu, J. MR T1ρ as an imaging biomarker for monitoring liver injury progression and regression: An experimental study in rats with carbon tetrachloride intoxication. Eur. Radiol. 2012, 22, 1709–1716. [Google Scholar] [CrossRef]
- Xie, S.; Qi, H.; Li, Q.; Zhang, K.; Zhang, L.; Cheng, Y.; Shen, W. Liver injury monitoring, fibrosis staging and inflammation grading using T1rho magnetic resonance imaging: An experimental study in rats with carbon tetrachloride intoxication. BMC Gastroenterol. 2020, 20, 14. [Google Scholar] [CrossRef]
- Idilman, I.S.; Celik, A.; Savas, B.; Idilman, R.; Karcaaltincaba, M. The feasibility of T2 mapping in the assessment of hepatic steatosis, inflammation, and fibrosis in patients with non-alcoholic fatty liver disease: A preliminary study. Clin. Radiol. 2021, 76, e13–e709. [Google Scholar] [CrossRef]
- Kim, S.H.; Lee, S.J.; Yu, S.M. Study of lipid proton difference evaluation via 9.4T MRI analysis of fatty liver induced by exposure to methionine and choline-deficient (MCD) diet and high-fat diet (HFD) in an animal model. Chem. Phys. Lipids 2022, 242, 105164. [Google Scholar] [CrossRef]
- Petta, S.; Maida, M.; Macaluso, F.S.; Di Marco, V.; Cammà, C.; Cabibi, D.; Craxì, A. The severity of steatosis influences liver stiffness measurement in patients with nonalcoholic fatty liver disease. Hepatology 2015, 62, 1101–1110. [Google Scholar] [CrossRef] [PubMed]
- Karlas, T.; Petroff, D.; Sasso, M.; Fan, J.G.; Mi, Y.Q.; de Lédinghen, V.; Kumar, M.; Lupsor-Platon, M.; Han, K.H.; Cardoso, A.C.; et al. Impact of controlled attenuation parameter on detecting fibrosis using liver stiffness measurement. Aliment. Pharmacol. Ther. 2018, 47, 989–1000. [Google Scholar] [CrossRef] [PubMed]
- Wong, V.W.; Vergniol, J.; Wong, G.L.; Foucher, J.; Chan, H.L.; Le Bail, B.; Choi, P.C.; Kowo, M.; Chan, A.W.; Merrouche, W.; et al. Diagnosis of fibrosis and cirrhosis using liver stiffness measurement in nonalcoholic fatty liver disease. Hepatology 2010, 51, 454–462. [Google Scholar] [CrossRef]
- Tang, H.; Li, J.; Zinker, B.; Boehm, S.; Mauer, A.; Rex-Rabe, S.; Glaser, K.J.; Fronheiser, M.; Bradstreet, T.; Nakao, Y.; et al. Evaluation of a PEGylated Fibroblast Growth Factor 21 Variant Using Novel Preclinical Magnetic Resonance Imaging and Magnetic Resonance Elastography in a Mouse Mdel of Nonalcoholic Steatohepatitis. J. Magn. Reson. Imaging 2022, in press. [Google Scholar] [CrossRef]
- Bastard, C.; Bosisio, M.R.; Chabert, M.; Kalopissis, A.D.; Mahrouf-Yorgov, M.; Gilgenkrantz, H.; Mueller, S.; Sandrin, L. Transient micro-elastography: A novel non-invasive approach to measure liver stiffness in mice. World J. Gastroenterol. 2011, 17, 968–975. [Google Scholar] [CrossRef]
- Czernuszewicz, T.J.; Aji, A.M.; Moore, C.J.; Montgomery, S.A.; Velasco, B.; Torres, G.; Anand, K.S.; Johnson, K.A.; Deal, A.M.; Zukić, D.; et al. Development of a Robotic Shear Wave Elastography System for Noninvasive Staging of Liver Disease in Murine Models. Hepatol. Commun. 2022, 6, 1827–1839. [Google Scholar] [CrossRef] [PubMed]


| A. Comparison of factors of mice in groups 1 and 2. | ||||||||
| M2BP | Tgf-β | Tnf-α | iNos | |||||
| R | p | R | p | R | p | R | p | |
| SWE | 0.7 | <0.05 | 0.21 | n.s. | −0.50 | 0.1 | 0.02 | n.s. |
| T1rho | 0.9 | <0.001 | 0.27 | n.s. | −0.33 | n.s. | 0.60 | <0.05 |
| T2 | 0.53 | 0.08 | 0.11 | n.s. | −0.23 | n.s. | 0.64 | <0.05 |
| B. Comparison of factors of mice in groups 1 and 3. | ||||||||
| M2BP | Tgf-β | Tnf-α | iNos | |||||
| R | p | R | p | R | p | R | p | |
| SWE | −0.28 | n.s. | 0.17 | n.s. | 0.23 | n.s. | −0.21 | n.s. |
| T1rho | 0.6 | <0.05 | 0.77 | <0.005 | 0.83 | <0.001 | 0.66 | <0.05 |
| T2 | 0.6 | <0.05 | 0.77 | <0.005 | 0.73 | <0.01 | 0.80 | <0.005 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Futani, Y.; Hamano, M.; Matsumoto, R.; Hashimoto, S.; Nishimura, R.; Ueda, M.; Arihara, N.; Fujii, H.; Ono, M.; Miyoshi, E.; et al. Shear-Wave Elastography Using Commercially Available Ultrasound in a Mouse Model of Chronic Liver Disease. Gastrointest. Disord. 2022, 4, 153-164. https://doi.org/10.3390/gidisord4030015
Futani Y, Hamano M, Matsumoto R, Hashimoto S, Nishimura R, Ueda M, Arihara N, Fujii H, Ono M, Miyoshi E, et al. Shear-Wave Elastography Using Commercially Available Ultrasound in a Mouse Model of Chronic Liver Disease. Gastrointestinal Disorders. 2022; 4(3):153-164. https://doi.org/10.3390/gidisord4030015
Chicago/Turabian StyleFutani, Yoko, Megumi Hamano, Riku Matsumoto, Saya Hashimoto, Rikuto Nishimura, Mika Ueda, Narumi Arihara, Hideki Fujii, Masafumi Ono, Eiji Miyoshi, and et al. 2022. "Shear-Wave Elastography Using Commercially Available Ultrasound in a Mouse Model of Chronic Liver Disease" Gastrointestinal Disorders 4, no. 3: 153-164. https://doi.org/10.3390/gidisord4030015
APA StyleFutani, Y., Hamano, M., Matsumoto, R., Hashimoto, S., Nishimura, R., Ueda, M., Arihara, N., Fujii, H., Ono, M., Miyoshi, E., Saito, S., & Kamada, Y. (2022). Shear-Wave Elastography Using Commercially Available Ultrasound in a Mouse Model of Chronic Liver Disease. Gastrointestinal Disorders, 4(3), 153-164. https://doi.org/10.3390/gidisord4030015







