A Comparison of Different Natural Groundwaters from Repository Sites—Corrosivity, Chemistry and Microbial Community
Abstract
:1. Introduction
2. Materials and Methods
2.1. Steel Materials
2.2. Water Chemistry
- c(Cl−) The chloride concentration (mol/m3)
- c(SO42−) The sulphate concentration (mol/m3)
- c(HCO3−) The bicarbonate concentration (mol/m3).
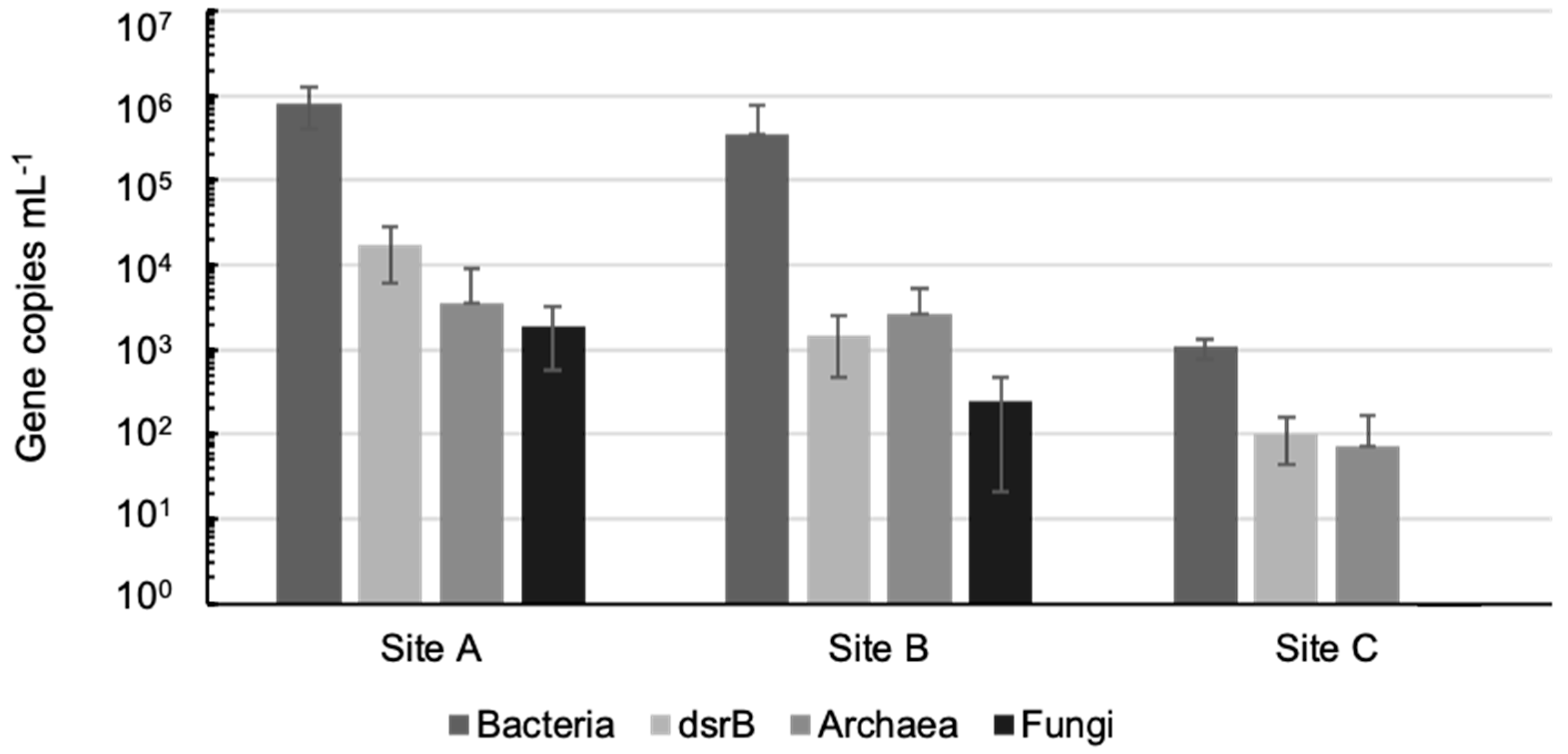
2.3. Estimation of Microbial Community Sizes in the Different Groundwaters by qPCR
2.4. Electrochemical Measurements
- CR The corrosion rate. Its units are given by the choice of K
- Icorr The corrosion current in amperes
- K A constant that defines the units for the corrosion rate
- EW The equivalent weight in grams/equivalent
- d Density in grams/cm³
- A Area of the sample in cm².
3. Results
3.1. Chemical Composition and Physical Properties of the Groundwaters
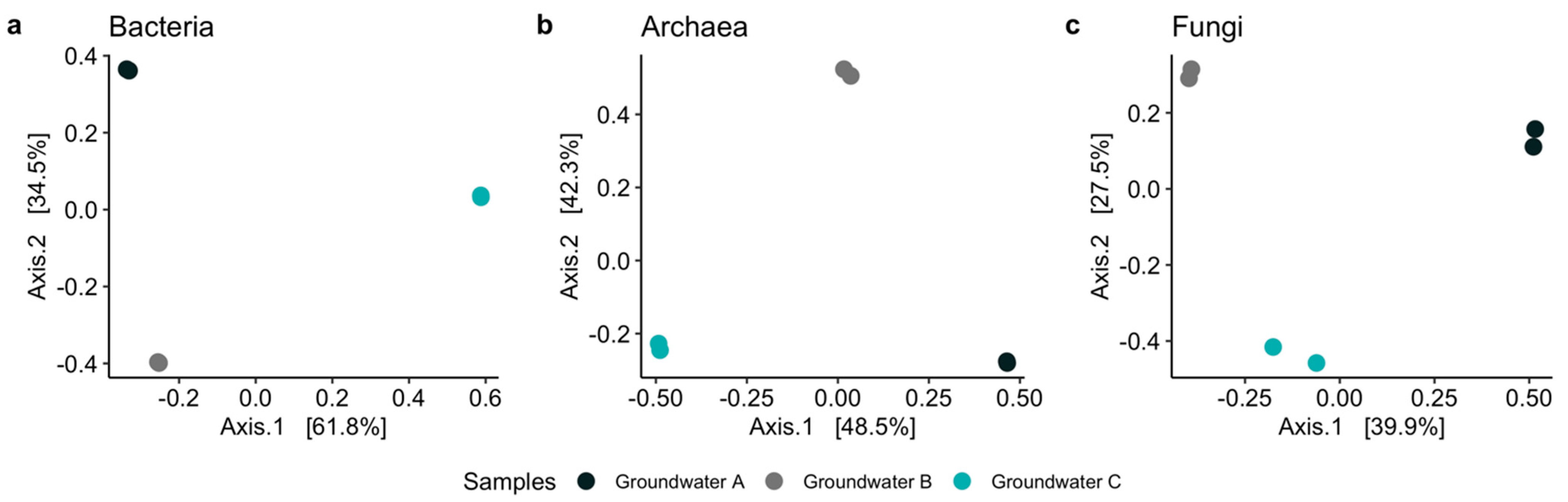
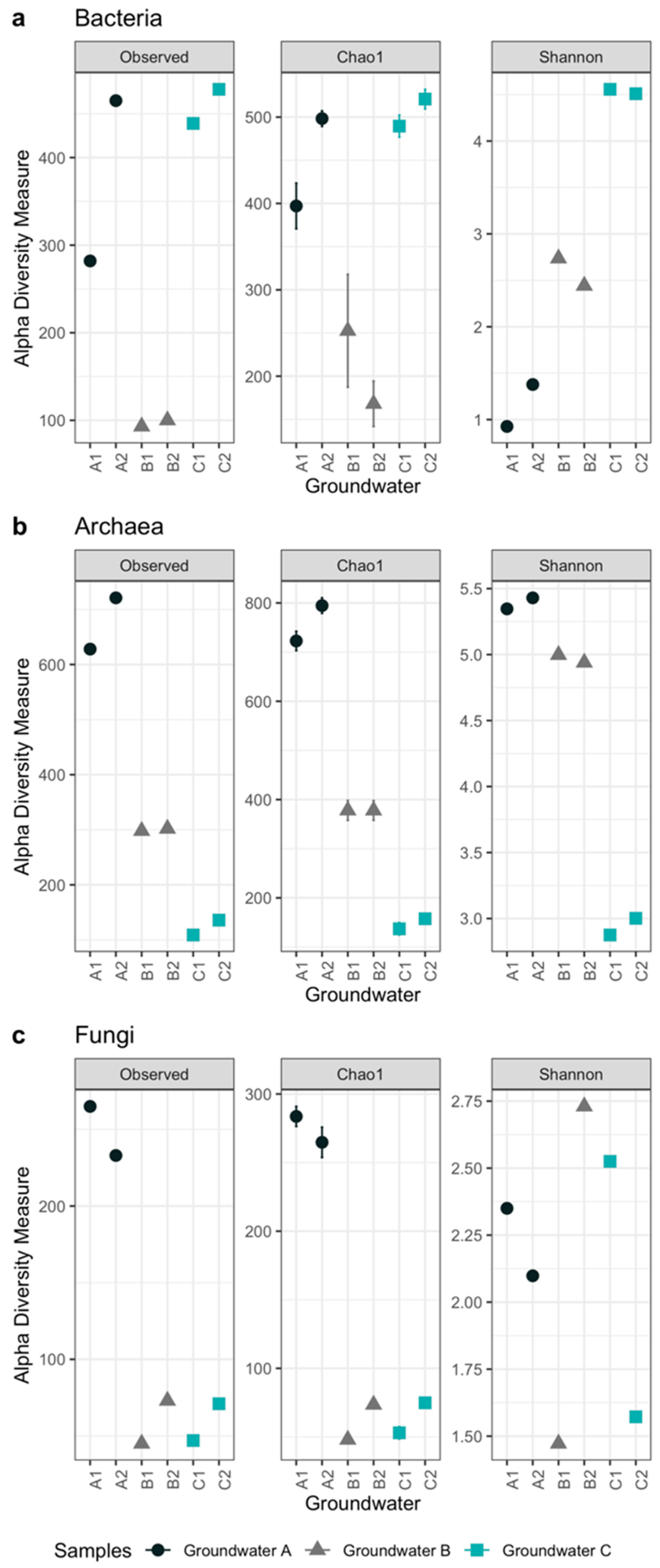
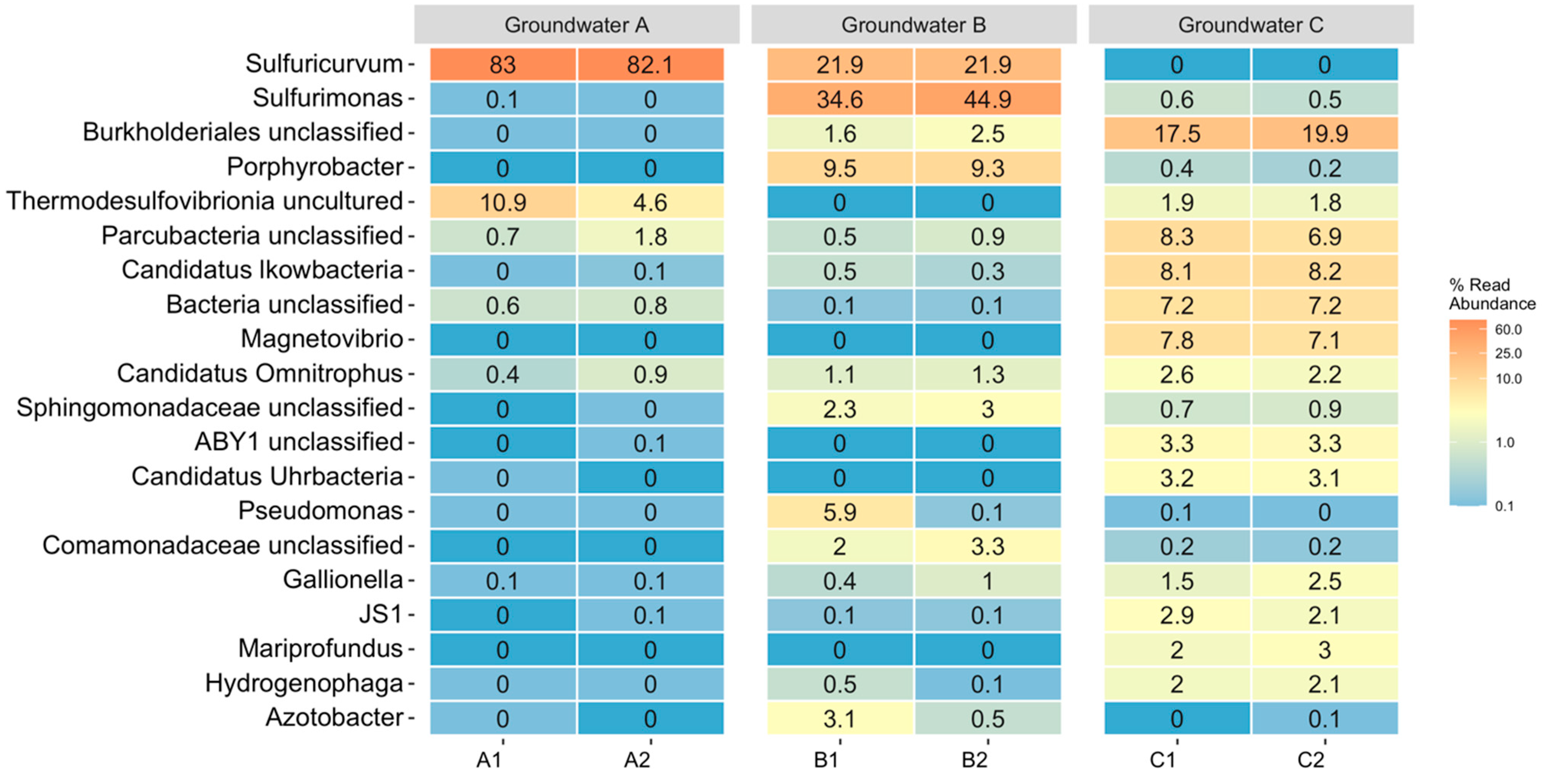
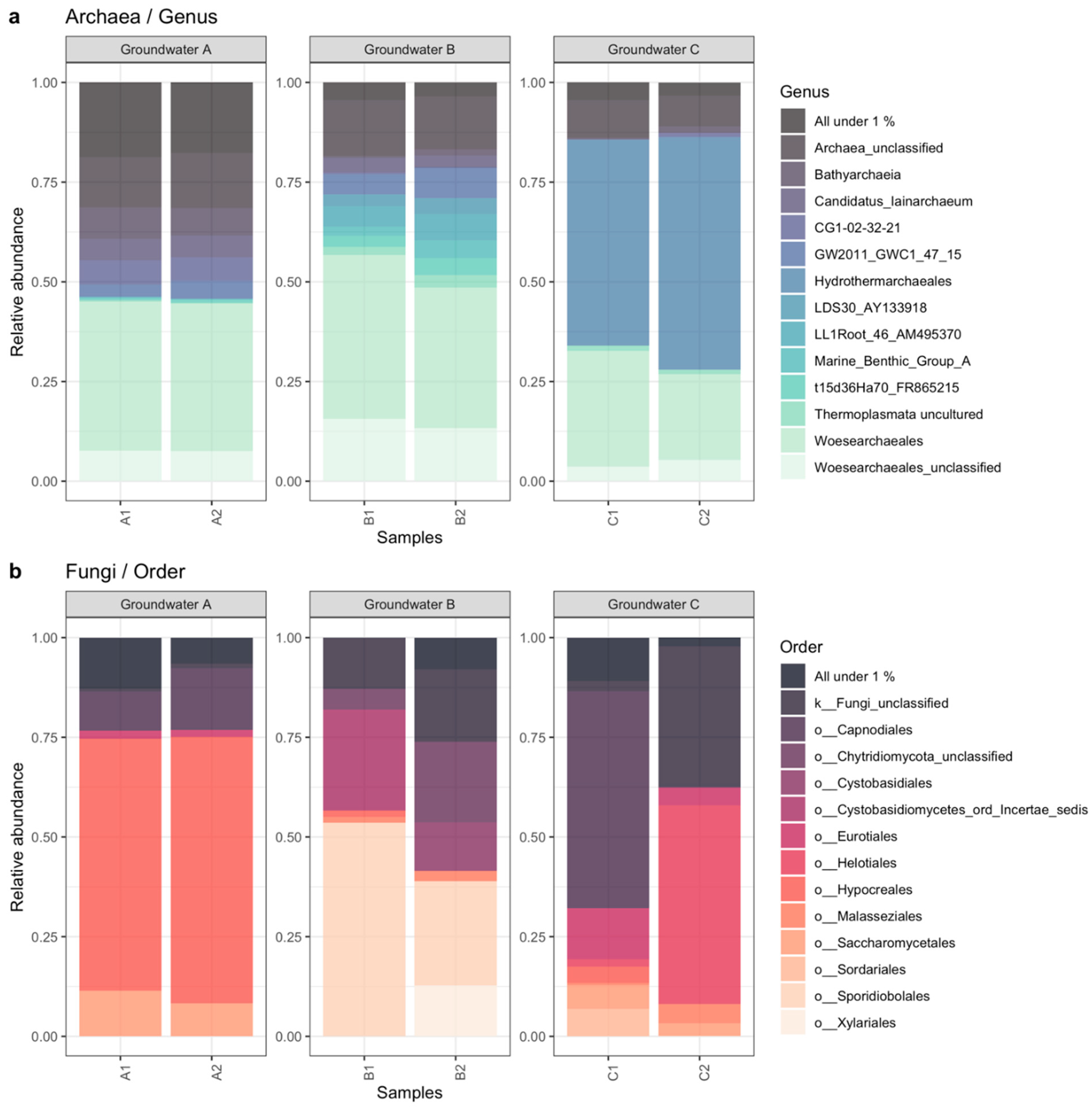
3.2. Microbiology of the Groundwaters
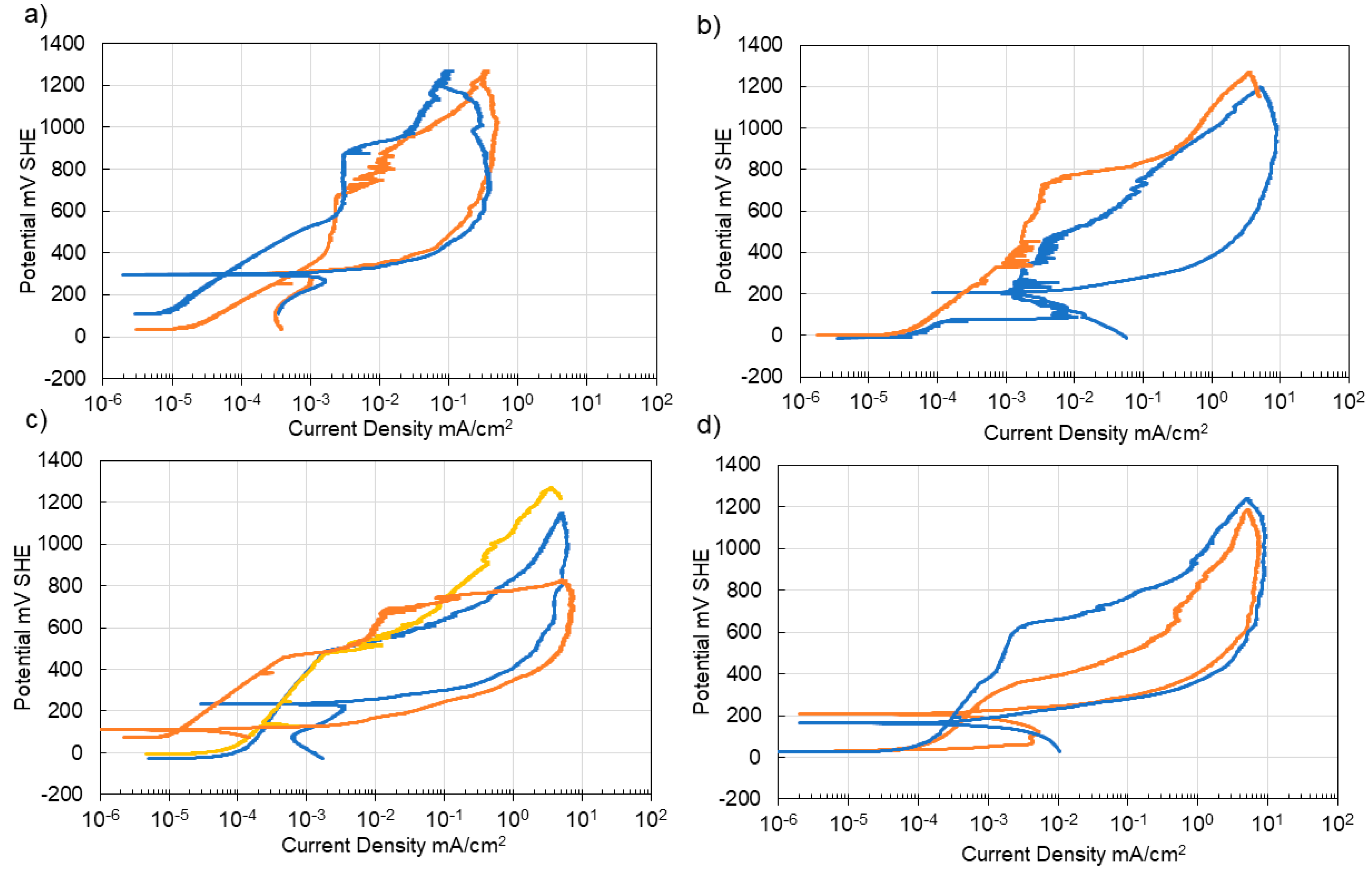
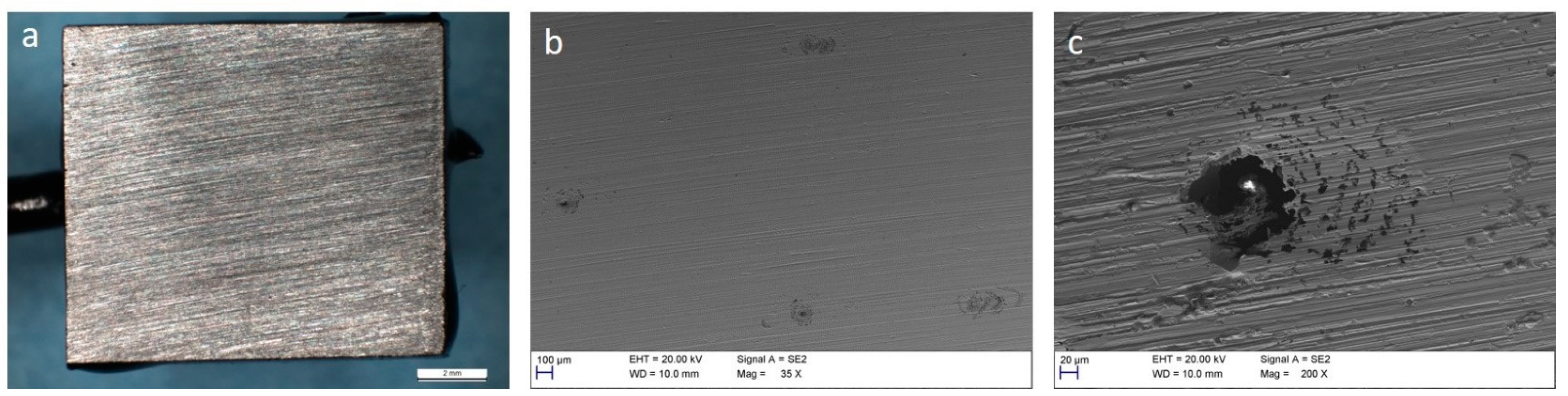
3.3. Corrosion of Carbon Steel
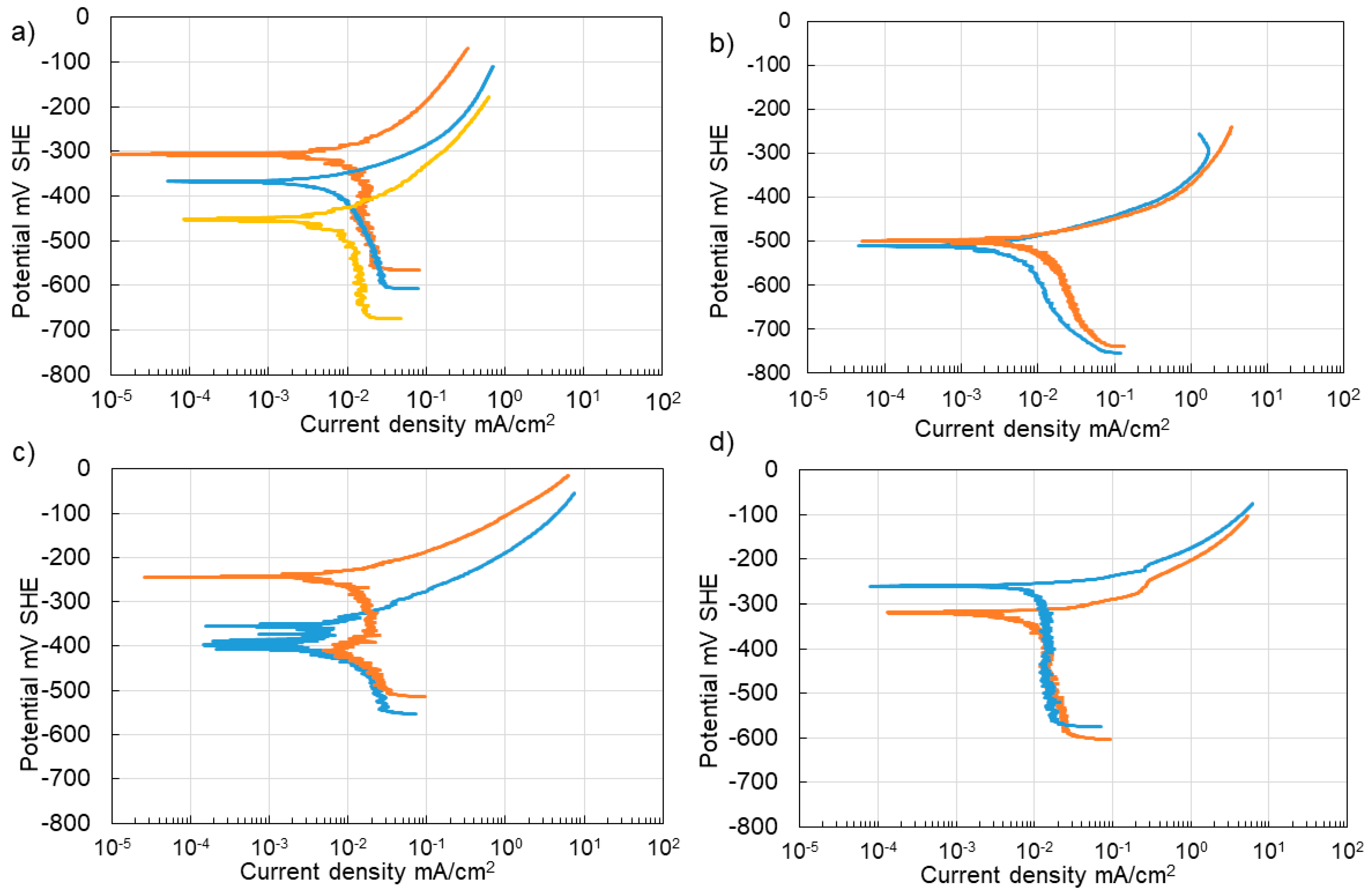
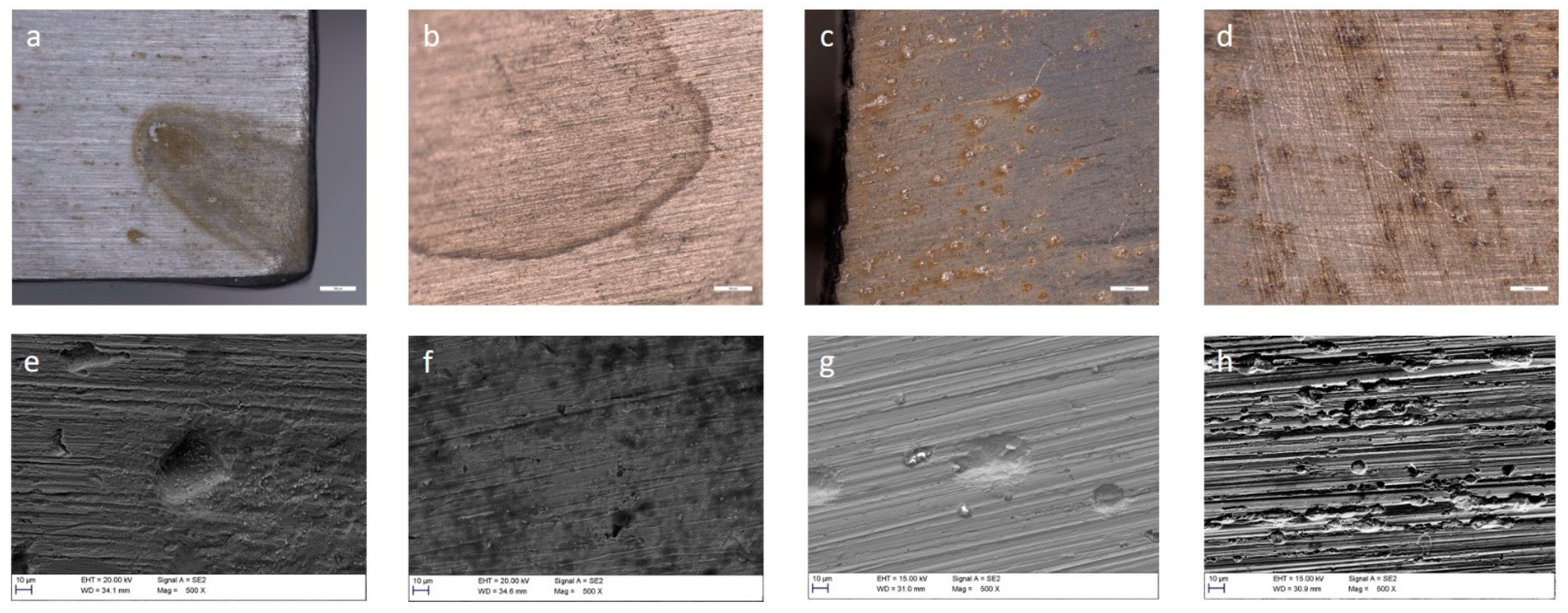
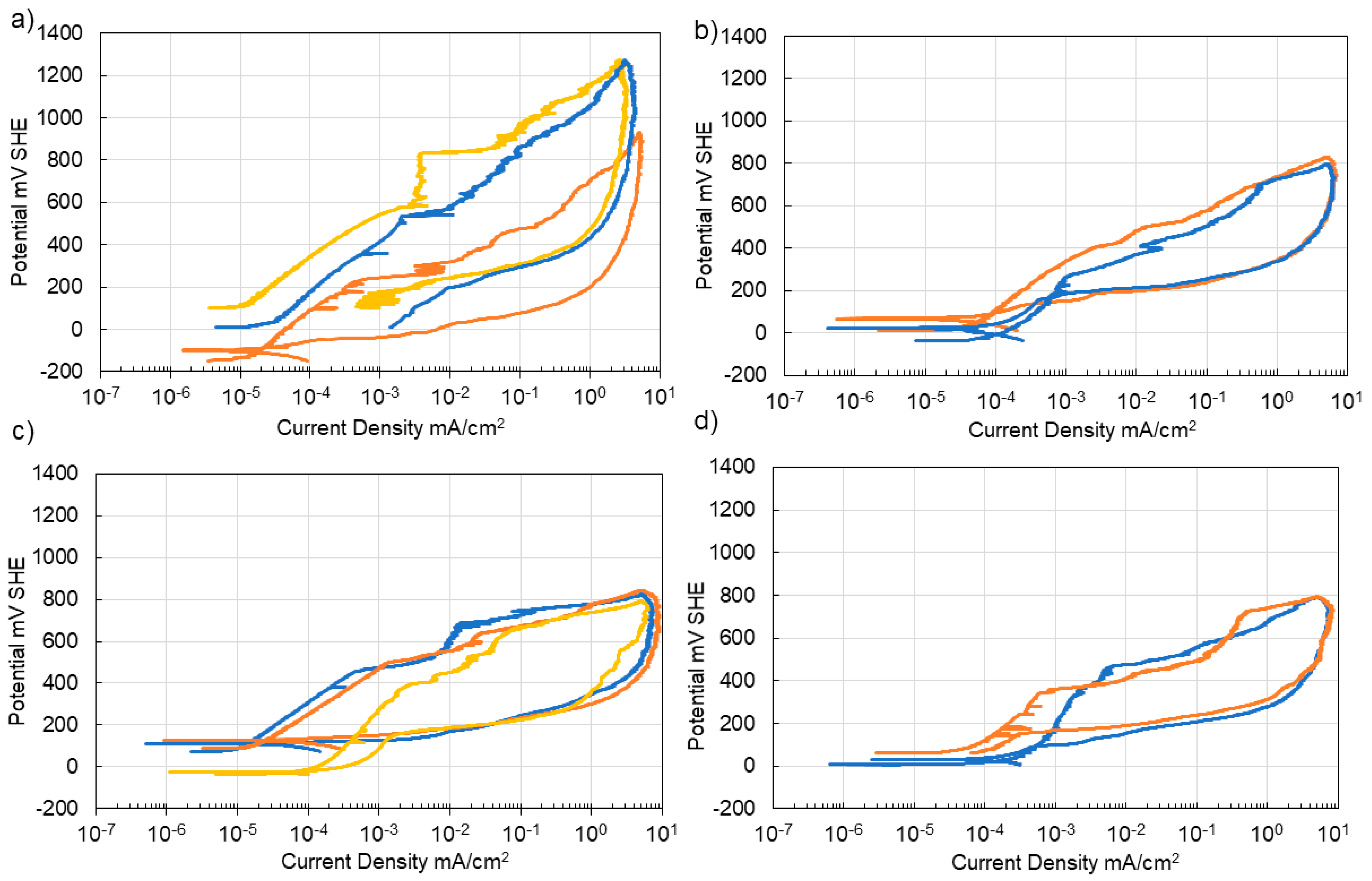
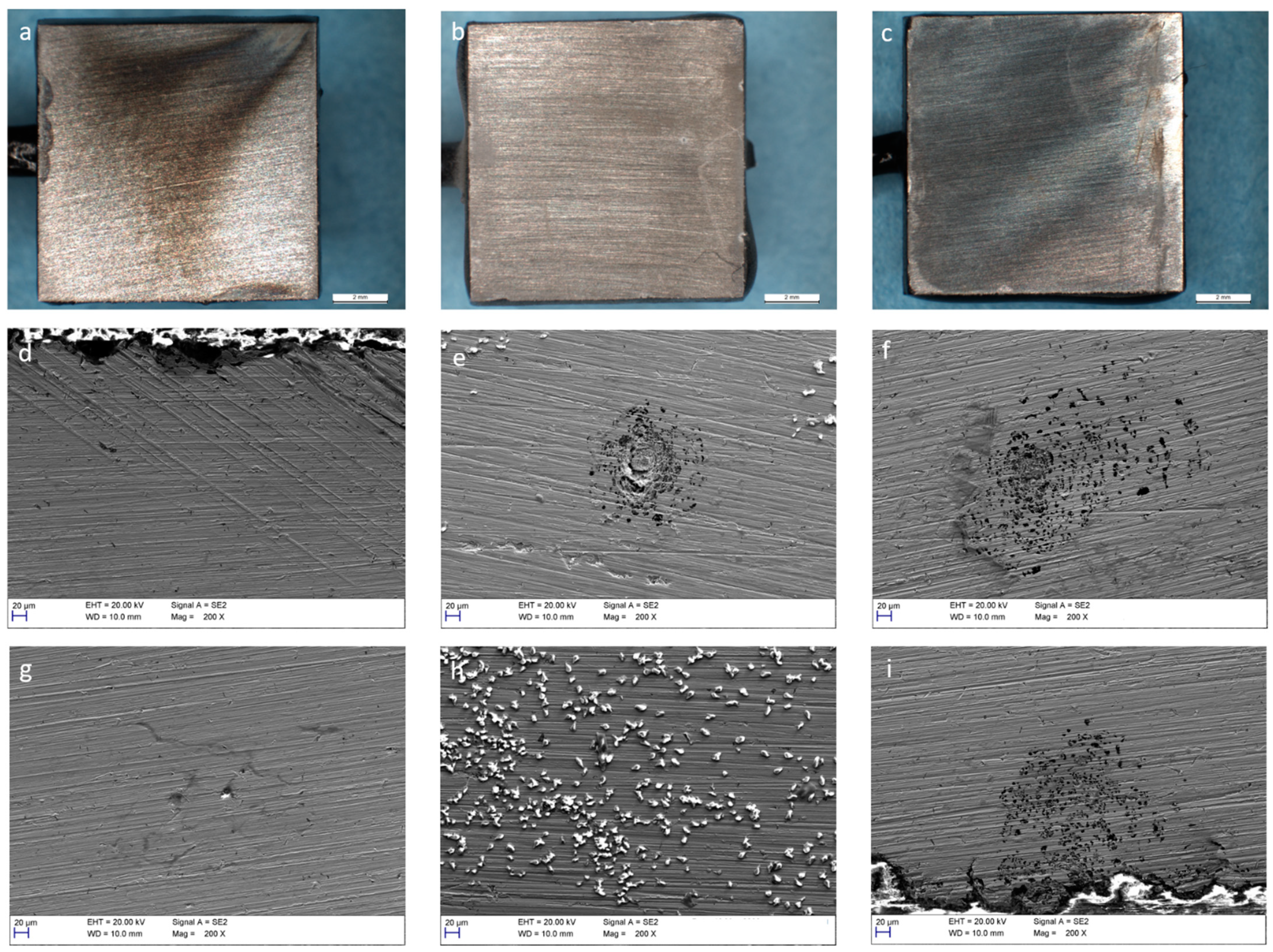
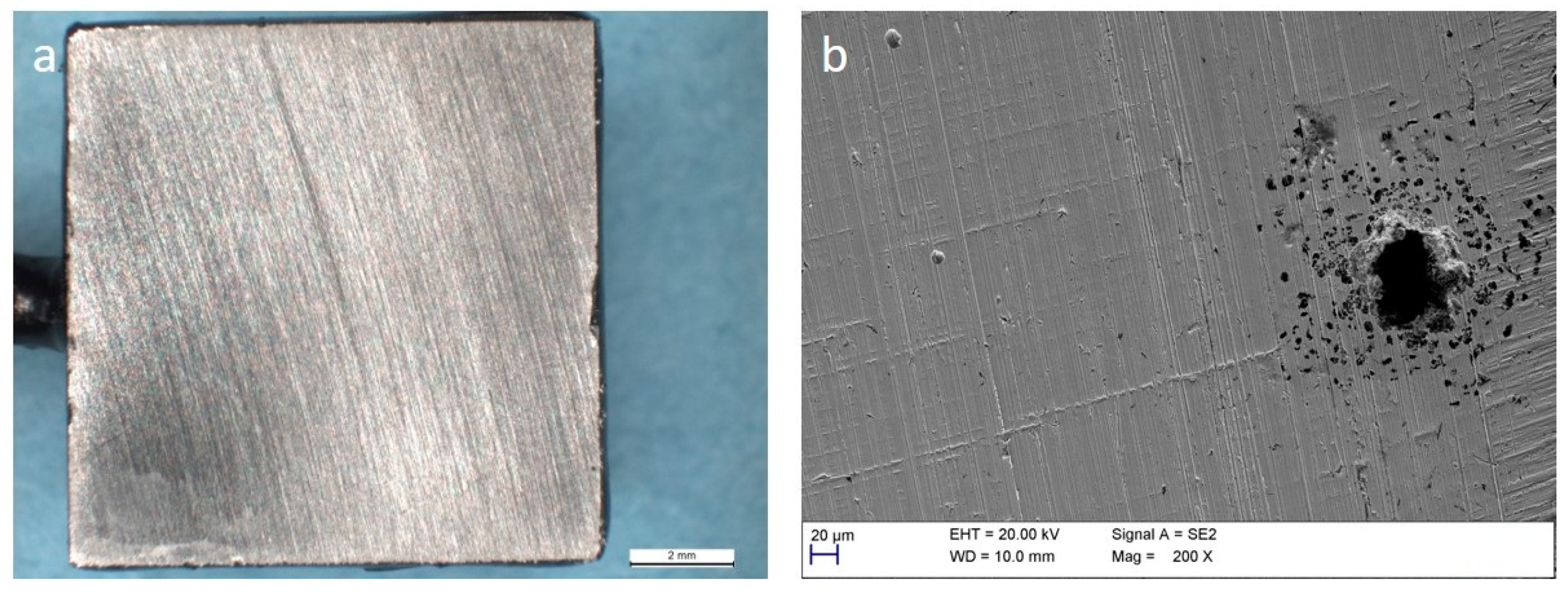
3.4. Corrosion of Stainless Steel
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Rajala, P.; Raulio, M.; Carpen, L. Sulphate Reducing Bacteria and Methanogenic Archaea Driving Corrosion of Steel in Deep Anoxic Ground Water. Corros. Sci. Technol. 2019, 18, 221–227. [Google Scholar]
- Rajala, P.; Huttunen-Saarivirta, E.; Bomberg, M.; Carpén, L. Corrosion and Biofouling Tendency of Carbon Steel in Anoxic Groundwater Containing Sulphate Reducing Bacteria and Methanogenic Archaea. Corros. Sci. 2019, 159, 108148. [Google Scholar] [CrossRef]
- Li, Y.; Xu, D.; Chen, C.; Li, X.; Jia, R.; Zhang, D.; Sand, W.; Wang, F.; Gu, T. Anaerobic Microbiologically Influenced Corrosion Mechanisms Interpreted Using Bioenergetics and Bioelectrochemistry: A Review. J. Mater. Sci. Technol. 2018, 34, 1713–1718. [Google Scholar] [CrossRef]
- Bale, S.J.; Goodman, K.; Rochelle, P.A.; Marchesi, J.R.; Fry, J.C.; Weightman, A.J.; Parkes, R.J. Desulfovibrio Profundus sp. nov., a Novel Barophilic Sulfate-Reducing Bacterium from Deep Sediment Layers in the Japan Sea. Int. J. Syst. Bacteriol. 1997, 47, 515–521. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rajala, P. Microbially-Induced Corrosion of Carbon Steel in a Geological Repository Environment; VTT Science: Espoo, Finland, 2017; Volume 155, ISBN 1-4458-83-159-879. [Google Scholar]
- Bomberg, M.; Lamminmäki, T.; Itävaara, M. Microbial Communities and Their Predicted Metabolic Characteristics in Deep Fracture Groundwaters of the Crystalline Bedrock at Olkiluoto, Finland. Biogeosciences 2016, 13, 6031–6047. [Google Scholar] [CrossRef] [Green Version]
- Hakkarainen, T. Effects of Sulphate Ions on Chloride Induced Pitting of Stainless Steel. In Proceedings of the 14th International Corrosion Congress, Cape Town, South Africa, 26 September–1 October 1999; p. 9. [Google Scholar]
- Rajala, P.; Bomberg, M.; Vepsäläinen, M.; Carpén, L. Microbial Fouling and Corrosion of Carbon Steel in Deep Anoxic Alkaline Groundwater. Biofouling 2017, 33, 195–209. [Google Scholar] [CrossRef]
- ISO. ISO 10523:2008—Water Quality—Determination of PH; ISO: Geneva, Switzerland, 2008. [Google Scholar]
- ISO. ISO 6878:2004—Water Quality—Determination of Phosphorus—Ammonium Molybdate Spectrometric Method; ISO: Geneva, Switzerland, 2004. [Google Scholar]
- ISO. ISO 11732:2005—Water Quality—Determination of Ammonium Nitrogen—Method by Flow Analysis (CFA and FIA) and Spectrometric Detection; ISO: Geneva, Switzerland, 2005. [Google Scholar]
- DIN. DIN EN 12506 Characterization of Waste—Analysis of Eluates—Determination of PH, As, Ba, Cd, CI-, Co, Cr, Cr(VI), Cu, Mo, Ni, NO2−, Pb, Total S, SO42−, V and Zn; DIN: Berlin, Germany, 2003. [Google Scholar]
- DIN. DIN EN 1484—Water Analysis—Guidelines for the Determination of Total Organic Carbon (TOC) and Dissolved Organic Carbon (DOC); DIN: Berlin, Germany, 2003. [Google Scholar]
- Revie, R.W.; Uhlig, H.H. Corrosion and Corrosion Control: An Introduction to Corrosion Science and Engineering, 4th ed.; Wiley-VCH Verlag GmbH: Weinheim, Germany, 2008; Volume 7, ISBN 978-0-471-73279-2. [Google Scholar]
- Herlemann, D.P.; Labrenz, M.; Jürgens, K.; Bertilsson, S.; Waniek, J.J.; Andersson, A.F. Transitions in Bacterial Communities along the 2000 Km Salinity Gradient of the Baltic Sea. ISME J. 2011, 5, 1571–1579. [Google Scholar] [CrossRef] [Green Version]
- Geets, J.; Borremans, B.; Diels, L.; Springael, D.; Vangronsveld, J.; van der Lelie, D.; Vanbroekhoven, K. DsrB Gene-Based DGGE for Community and Diversity Surveys of Sulfate-Reducing Bacteria. J. Microbiol. Methods 2006, 66, 194–205. [Google Scholar] [CrossRef]
- Wagner, M.; Roger, A.J.; Flax, J.L.; Brusseau, G.A.; Stahl, D.A. Phylogeny of Dissimilatory Sulfite Reductases Supports an Early Origin of Sulfate Respiration. J. Bacteriol. 1998, 180, 2975–2982. [Google Scholar] [CrossRef] [Green Version]
- Bano, N.; Ruffin, S.; Ransom, B.; Hollibaugh, J.T. Phylogenetic Composition of Arctic Ocean Archaeal Assemblages and Comparison with Antarctic Assemblages. Appl. Environ. Microbiol. 2004, 70, 781–789. [Google Scholar] [CrossRef] [Green Version]
- Barns, S.M.; Fundyga, R.E.; Jeffries, M.W.; Pace, N.R. Remarkable Archaeal Diversity Detected in a Yellowstone National Park Hot Spring Environment. Proc. Natl. Acad. Sci. USA 1994, 91, 1609–1613. [Google Scholar] [CrossRef] [Green Version]
- Takai, K.; Horikoshi, K. Rapid Detection and Quantification of Members of the Archaeal Community by Quantitative PCR Using Fluorogenic Probes. Appl. Environ. Microbiol. 2000, 66, 5066–5072. [Google Scholar] [CrossRef] [Green Version]
- Bomberg, M.; Miettinen, H. Data on the Optimization of an Archaea-Specific Probe-Based QPCR Assay. Data Brief 2020, 33, 106610. [Google Scholar] [CrossRef] [PubMed]
- Haugland, R.; Vespe, S.J. Method of Identifying and Quantifying Specific Fungi and Bacteria. U.S. Patent 6,387,652, 14 May 2002. [Google Scholar]
- Klindworth, A.; Pruesse, E.; Schweer, T.; Peplies, J.; Quast, C.; Horn, M.; Glöckner, F.O. Evaluation of General 16S Ribosomal RNA Gene PCR Primers for Classical and Next-Generation Sequencing-Based Diversity Studies. Nucleic Acids Res. 2013, 41, e1. [Google Scholar] [CrossRef] [PubMed]
- Gardes, M.; Bruns, T.D. ITS Primers with Enhanced Specificity for Basidiomycetes—Application to the Identification of Mycorrhizae and Rusts. Mol. Ecol. 1993, 2, 113–118. [Google Scholar] [CrossRef]
- White, T.J.; Bruns, T.; Lee, S.; Taylor, J. Amplification and Direct Sequencing of Fungal Ribosomal RNA Genes for Phylogenetics. In PCR Protocols. A Guide to Methods and Applicati; Innis, M.A., Gelfand, D.H., Sninsky, J.J., White, T.J., Eds.; Academic Press: San Diego, CA, USA, 1990; pp. 315–322. [Google Scholar]
- Schloss, P.D.; Westcott, S.L.; Ryabin, T.; Hall, J.R.; Hartmann, M.; Hollister, E.B.; Lesniewski, R.A.; Oakley, B.B.; Parks, D.H.; Robinson, C.J.; et al. Introducing Mothur: Open-Source, Platform-Independent, Community-Supported Software for Describing and Comparing Microbial Communities. Appl. Environ. Microbiol. 2009, 75, 7537–7541. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pruesse, E.; Quast, C.; Knittel, K.; Fuchs, B.M.; Ludwig, W.; Peplies, J.; Glöckner, F.O. SILVA: A Comprehensive Online Resource for Quality Checked and Aligned Ribosomal RNA Sequence Data Compatible with ARB. Nucleic Acids Res. 2007, 35, 7188–7196. [Google Scholar] [CrossRef] [Green Version]
- Quast, C.; Pruesse, E.; Yilmaz, P.; Gerken, J.; Schweer, T.; Yarza, P.; Peplies, J.; Glöckner, F.O. The SILVA Ribosomal RNA Gene Database Project: Improved Data Processing and Web-Based Tools. Nucleic Acids Res. 2012, 41, D590–D596. [Google Scholar] [CrossRef]
- RStudio Team. RStudio: Integrated Development for R 2015. 2015. Available online: http://www.rstudio.com/ (accessed on 18 October 2021).
- McMurdie, P.J.; Holmes, S. Phyloseq: An R Package for Reproducible Interactive Analysis and Graphics of Microbiome Census Data. PLoS ONE 2013, 8, e61217. [Google Scholar] [CrossRef] [Green Version]
- Oksanen, J.; Blanchet, F.G.; Friendly, M.; Kindt, R.; Legendre, P.; McGlinn, D.; Minchin, P.R.; O’Hara, R.B.; Simpson, G.; Solymos, P.; et al. Vegan: Community Ecology Package, R Package. 2016. Available online: https://cran.r-project.org/web/packages/vegan/index.html (accessed on 18 October 2021).
- Wickham, H. Ggplot2: Elegant Graphics for Data Analysis. 2016. Available online: http://ggplot2.org.Springer (accessed on 18 October 2021).
- Andersen, K.S.; Kirkegaard, R.H.; Karst, S.M.; Albertsen, M. Ampvis2: An R Package to Analyse and Visualise 16S RRNA Amplicon Data. bioRxiv 2018, 299537. [Google Scholar] [CrossRef] [Green Version]
- Hilbert, L.; Carpen, L.; Moller, P.; Fontenay, F.; T, M. Unexpected Corrosion of Stainless Steel in Low Chloride Waters—Microbial Aspects. In Proceedings of the Roceedings of the Eurocorr 2009, Nice, France, 6–10 September 2009. [Google Scholar]
- Purkamo, L.; Kietäväinen, R.; Miettinen, H.; Sohlberg, E.; Kukkonen, I.; Itävaara, M.; Bomberg, M. Diversity and Functionality of Archaeal, Bacterial and Fungal Communities in Deep Archaean Bedrock Groundwater. FEMS Microbiol. Ecol. 2018, 94, fiy116. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Magnabosco, C.; Lin, L.-H.; Dong, H.; Bomberg, M.; Ghiorse, W.; Stan-Lotter, H.; Pedersen, K.; Kieft, T.L.; Onstott, T.C. The Biomass and Biodiversity of the Continental Subsurface. Nat. Geosci. 2018, 11, 707–717. [Google Scholar] [CrossRef]
- Purkamo, L.; Bomberg, M.; Kietäväinen, R.; Salavirta, H.; Nyyssönen, M.; Nuppunen-Puputti, M.; Ahonen, L.; Kukkonen, I.; Itävaara, M. Microbial Co-Occurrence Patterns in Deep Precambrian Bedrock Fracture Fluids. Biogeosciences 2016, 13, 3091–3108. [Google Scholar] [CrossRef] [Green Version]
- Böttcher, M.E. Sulfur Cycle. Encycl. Earth Sci. Ser. 2011, 859–864. [Google Scholar] [CrossRef]
- Lau, M.C.Y.; Kieft, T.L.; Kuloyo, O.; Linage-Alvarez, B.; van Heerden, E.; Lindsay, M.R.; Magnabosco, C.; Wang, W.; Wiggins, J.B.; Guo, L.; et al. An Oligotrophic Deep-Subsurface Community Dependent on Syntrophy Is Dominated by Sulfur-Driven Autotrophic Denitrifiers. Proc. Natl. Acad. Sci. USA 2016, 19, 1230–1262. [Google Scholar] [CrossRef] [Green Version]
- Finster, K.; Liesack, W.; Thamdrup, B. Elemental Sulfur and Thiosulfate Disproportionation by Desulfocapsa Sulfoexigens sp. nov., a New Anaerobic Bacterium Isolated from Marine Surface Sediment. Appl. Environ. Microbiol. 1998, 64, 119–125. [Google Scholar] [CrossRef] [Green Version]
- Han, C.; Kotsyurbenko, O.; Chertkov, O.; Held, B.; Lapidus, A.; Nolan, M.; Lucas, S.; Hammon, N.; Deshpande, S.; Cheng, J.F.; et al. omplete Genome Sequence of the Sulfur Compounds Oxidizing Chemolithoautotroph Sulfuricurvum Kujiense Type Strain (YK-1(T)). Stand. Genom. Sci. 2012, 6, 94–103. [Google Scholar] [CrossRef] [Green Version]
- Kodama, Y.; Watanabe, K. Sulfuricurvum Kujiense gen. nov., sp. nov., a Facultatively Anaerobic, Chemolithoautotrophic, Sulfur-Oxidizing Bacterium Isolated from an Underground Crude-Oil Storage Cavity. Int. J. Syst. Evol. Microbiol. 2004, 54, 2297–2300. [Google Scholar] [CrossRef] [Green Version]
- Bazylinski, D.A.; Williams, T.J.; Lefèvre, C.T.; Trubitsyn, D.; Fang, J.; Beveridge, T.J.; Moskowitz, B.M.; Ward, B.; Schübbe, S.; Dubbels, B.L.; et al. Magnetovibrio blakemorei gen. nov., sp. nov., a Magnetotactic Bacterium (Alphaproteobacteria: Rhodospirillaceae) Isolated from a Salt Marsh. Int. J. Syst. Evol. Microbiol. 2013, 63, 1824–1833. [Google Scholar] [CrossRef] [Green Version]
- Nelson, W.C.; Stegen, J.C. The Reduced Genomes of Parcubacteria (OD1) Contain Signatures of a Symbiotic Lifestyle. Front. Microbiol. 2015, 6, 713. [Google Scholar] [CrossRef] [Green Version]
- Anantharaman, K.; Hausmann, B.; Jungbluth, S.P.; Kantor, R.S.; Lavy, A.; Warren, L.A.; Rappé, M.S.; Pester, M.; Loy, A.; Thomas, B.C.; et al. Expanded Diversity of Microbial Groups That Shape the Dissimilatory Sulfur Cycle. ISME J. 2018, 12, 1715–1728. [Google Scholar] [CrossRef] [Green Version]
- Liu, X.; Li, M.; Castelle, C.J.; Probst, A.J.; Zhou, Z.; Pan, J.; Liu, Y.; Banfield, J.F.; Gu, J.D. Insights into the Ecology, Evolution, and Metabolism of the Widespread Woesearchaeotal Lineages. Microbiome 2018, 6, 102. [Google Scholar] [CrossRef] [PubMed]
- Dombrowski, N.; Lee, J.H.; Williams, T.A.; Offre, P.; Spang, A. Genomic Diversity, Lifestyles and Evolutionary Origins of DPANN Archaea. FEMS Microbiol. Lett. 2019, 366, fnz008. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhou, Z.; Liu, Y.; Xu, W.; Pan, J.; Luo, Z.H.; Li, M. Genome- and Community-Level Interaction Insights into Carbon Utilization and Element Cycling Functions of Hydrothermarchaeota in Hydrothermal Sediment. mSystems 2020, 5, e00795-19. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Carr, S.A.; Jungbluth, S.P.; Eloe-Fadrosh, E.A.; Stepanauskas, R.; Woyke, T.; Rappé, M.S.; Orcutt, B.N. Carboxydotrophy Potential of Uncultivated Hydrothermarchaeota from the Subseafloor Crustal Biosphere. ISME J. 2019, 13, 1457–1468. [Google Scholar] [CrossRef]
- Sohlberg, E.; Bomberg, M.; Miettinen, H.; Nyyssönen, M.; Salavirta, H.; Vikman, M.; Itävaara, M. Revealing the Unexplored Fungal Communities in Deep Groundwater of Crystalline Bedrock Fracture Zones in Olkiluoto, Finland. Front. Microbiol. 2015, 6, 573. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Perkins, A.K.; Ganzert, L.; Rojas-Jimenez, K.; Fonvielle, J.; Hose, G.C.; Grossart, H.P. Highly Diverse Fungal Communities in Carbon-Rich Aquifers of Two Contrasting Lakes in Northeast Germany. Fungal Ecol. 2019, 41, 116–125. [Google Scholar] [CrossRef]
- Liu, Z.L.; Saha, B.C.; Slininger, P.J. Lignocellulosic Biomass Conversion to Ethanol by Saccharomyces. In Bioenergy; ASM Press: Washington, DC, USA, 2008; pp. 17–36. [Google Scholar] [CrossRef]
- Shao, Z.; Sun, F. Intracellular Sequestration of Manganese and Phosphorus in a Metal-Resistant Fungus Cladosporium Cladosporioides from Deep-Sea Sediment. Extremophiles 2007, 11, 435–443. [Google Scholar] [CrossRef]
- Lopez-Fernandez, M.; Romero-González, M.; Günther, A.; Solari, P.L.; Merroun, M.L. Effect of U(VI) Aqueous Speciation on the Binding of Uranium by the Cell Surface of Rhodotorula Mucilaginosa, a Natural Yeast Isolate from Bentonites. Chemosphere 2018, 199, 351–360. [Google Scholar] [CrossRef]
- Nuppunen-Puputti, M.; Kietäväinen, R.; Purkamo, L.; Rajala, P.; Itävaara, M.; Kukkonen, I.; Bomberg, M. Rock Surface Fungi in Deep Continental Biosphere—Exploration of Microbial Community Formation with Subsurface In Situ Biofilm Trap. Microorganisms 2020, 9, 64. [Google Scholar] [CrossRef]
- Kagami, M.; Miki, T.; Takimoto, G. Mycoloop: Chytrids in Aquatic Food Webs. Front. Microbiol. 2014, 5, 166. [Google Scholar] [CrossRef] [PubMed]
- Ortega-Arbulú, A.S.; Pichler, M.; Vuillemin, A.; Orsi, W.D. Effects of Organic Matter and Low Oxygen on the Mycobenthos in a Coastal Lagoon. Environ. Microbiol. 2019, 21, 374–388. [Google Scholar] [CrossRef] [PubMed]
- Little, B.J.; Ray, R.I.; Pope, R.K. Relationship between Corrosion and the Biological Sulfur Cycle: A Review. Corrosion 2000, 56, 433–443. [Google Scholar] [CrossRef]



| C | Si | Mn | P | S | Ni | Cr | Mo | V | |
|---|---|---|---|---|---|---|---|---|---|
| AISI 1005 1 | 0.025 | <0.005 | 0.19 | 0.007 | 0.007 | 0.02 | 0.03 | <0.01 | 0.002 |
| AISI 304 | 0.037 | 0.42 | 1.54 | 0.027 | 0.002 | 8.47 | 18.1 | 0.13 | 0.056 |
| AISI 316L | 0.024 | 0.39 | 0.94 | 0.032 | 0.002 | 10.1 | 17.1 | 2.01 | 0.06 |
| Element/Characteristic | Unit | Site A | Site B | Site C | Simulated Groundwater * |
|---|---|---|---|---|---|
| pH | 8.73 | 7.33 | 7.74 | 7 | |
| Alkalinity pH 4.5 | mmol/L | 3.15 | 5.41 | 1.82 | - |
| CO2 (total) | mg/L | 138 | 268 | 87 | - |
| HCO3− | mg/L | 192 | 330 | 111 | - |
| Phosphate | mg/L | <0.040 | 0.068 | <0.040 | - |
| Sulphate | mg/L | 116 | 482 | 566 | 500 |
| Sulphide | mg/L | 0.147 | <0.050 | <0.010 | - |
| Chloride | mg/L | 723 | 3210 | 4880 | 5500 |
| Ammonium | mg/L | 0.179 | 2.9 | 1.82 | - |
| Ca (soluble) | mg/L | 56.8 | 444 | 615 | - |
| Mg (soluble) | mg/L | 30.2 | 109 | 276 | - |
| Na (soluble) | mg/L | 489 | 1380 | 2210 | 3800 |
| S | mg/L | 44.4 | 140 | 204 | - |
| Si | mg/L | 2.28 | 7.23 | 4.56 | - |
| TOC | mg/L | 6.37 | 10.3 | 1.33 | - |
| Sulphate/Chloride Ratio | 0.16 | 0.15 | 0.12 | 0.09 | |
| Pitting Index | 7.25 | 18.60 | 82.14 | - | |
| Langelier Saturation Index | 1.04 | 1.13 | 0.80 | - |
| Alphadiversity Metrics | Groundwater Samples | |||||
|---|---|---|---|---|---|---|
| A1 | A2 | B1 | B2 | C1 | C2 | |
| Bacteria | ||||||
| Observed OTU number | 282 | 465 | 93 | 100 | 439 | 478 |
| Chao1 estimated OTU number | 397 | 498 | 235 | 168 | 490 | 521 |
| Se.chao1 | 26 | 9 | 65 | 26 | 13 | 11 |
| Shannon’s diversity index | 0.9 | 1.4 | 2.7 | 2.4 | 4.6 | 4.5 |
| Archaea | ||||||
| Observed OTU number | 628 | 721 | 298 | 302 | 109 | 136 |
| Chao1 estimated OTU number | 723 | 795 | 378 | 378 | 137 | 157 |
| Se.chao1 | 19.6 | 15.7 | 20.2 | 20.0 | 12.7 | 9.3 |
| Shannon’s diversity index | 5.3 | 5.4 | 5.0 | 4.9 | 2.9 | 3.0 |
| Fungi | ||||||
| Observed OTU number | 265 | 233 | 45 | 73 | 47 | 71 |
| Chao1 estimated OTU number | 284 | 265 | 48 | 74 | 53 | 75 |
| Se.chao1 | 7.3 | 11 | 2.9 | 0.9 | 4.5 | 3.2 |
| Shannon’s diversity index | 2.4 | 2.1 | 1.5 | 2.7 | 2.5 | 1.6 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Somervuori, M.; Isotahdon, E.; Nuppunen-Puputti, M.; Bomberg, M.; Carpén, L.; Rajala, P. A Comparison of Different Natural Groundwaters from Repository Sites—Corrosivity, Chemistry and Microbial Community. Corros. Mater. Degrad. 2021, 2, 603-624. https://doi.org/10.3390/cmd2040032
Somervuori M, Isotahdon E, Nuppunen-Puputti M, Bomberg M, Carpén L, Rajala P. A Comparison of Different Natural Groundwaters from Repository Sites—Corrosivity, Chemistry and Microbial Community. Corrosion and Materials Degradation. 2021; 2(4):603-624. https://doi.org/10.3390/cmd2040032
Chicago/Turabian StyleSomervuori, Mervi, Elisa Isotahdon, Maija Nuppunen-Puputti, Malin Bomberg, Leena Carpén, and Pauliina Rajala. 2021. "A Comparison of Different Natural Groundwaters from Repository Sites—Corrosivity, Chemistry and Microbial Community" Corrosion and Materials Degradation 2, no. 4: 603-624. https://doi.org/10.3390/cmd2040032
APA StyleSomervuori, M., Isotahdon, E., Nuppunen-Puputti, M., Bomberg, M., Carpén, L., & Rajala, P. (2021). A Comparison of Different Natural Groundwaters from Repository Sites—Corrosivity, Chemistry and Microbial Community. Corrosion and Materials Degradation, 2(4), 603-624. https://doi.org/10.3390/cmd2040032









