Adipokines in Sleep Disturbance and Metabolic Dysfunction: Insights from Network Analysis
Abstract
:1. Introduction
2. Basic Concepts in Network Analysis
Network Metrics
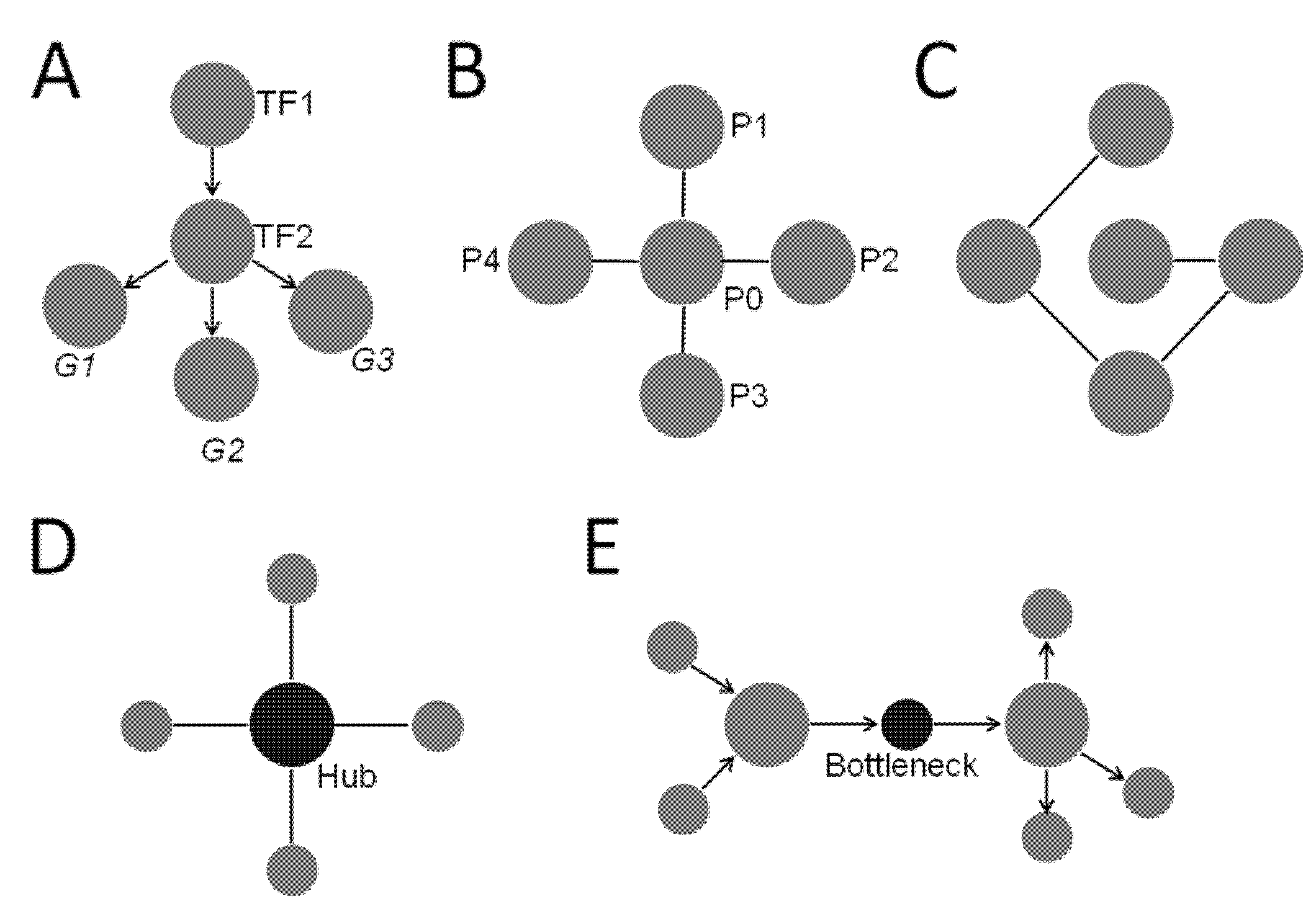
- Degree—Degree is a fundamental metric of a network that determines many other aspects of the network. Degree of a node is simply the number of edges each node is connected to. For network with directed edges, each node may have an in-degree, which is the number of edges going into the node, and/or an out-degree, which is the number of edges coming out of the node [10]. Degree can be used to define other properties of a network, such as centrality.
- Centrality—Centrality identifies the most “central” node of the network. However, being central may represent different properties in a network. For example, it could mean a node that is well-connected. That is, nodes with many edges connected to them are “central”. This is defined as degree centrality. The degree centrality of node i is calculated as where deg(i) is the node’s degree or edges [10]. Another important property of being “central” is a strategic position where the node lies in the cross paths of many shortest paths connecting other nodes. This is called betweenness centrality, defined as the extent to which a given node lies in the shortest paths connecting other nodes [10]. It is calculated as where is the total number of shortest paths from node x to node y and is the number of those paths that pass through node i [10]. A node with high betweenness lies on many shortest paths of other nodes which are not directly connected and has systems-level influence in the network (Figure 1; also see definition of bottlenecks below) [14,15].
- Hubs—Hubs are nodes with much higher degrees than those of the other nodes in the network, although not every network has hubs. In a random network (Figure 1C), every node is connected to a low number of nodes, and all have similar degrees. Therefore, there are no hubs in a random network. In a scale-free network, most nodes are connected to a small number of high degree nodes that are considered as hubs (Figure 1D). It turns out most biological networks have properties similar to a scale-free network, which has a “small word phenomenon” by which distances between nodes are shortened significantly by the presence of hubs. In biological systems, hub proteins/genes are enriched in essential/lethal genes, and are often evolutionarily conserved [10].
- Bottlenecks—Nodes with high betweenness centrality are known as bottlenecks (Figure 1E) [14]. They are similar to major highways in a transportation network with many shortest paths going through them (Figure 1E). Not surprisingly, bottleneck nodes control the information flow in a directed network, and disruption at these nodes causes widespread damage. In biological networks, bottleneck nodes are enriched in proteins/genes that are important for systems-level of phenotypes such as essentiality and virulence [15].
3. Methods
3.1. Construction of an Adipokine Network
- Suppose a physiologic perturbation or sleep disturbance has a positive/negative/uncertain association with an adipokine. In that case, a directed edge is created pointing from the perturbation/sleep disturbance to the adipokine with a label depicting a positive, negative, or uncertain relation. For instance, OSA is negatively associated with leptin level, so an edge from OSA to leptin is created with an edge label set as negative.
- Edges are then created starting from each of the adipokine to their molecular targets. For instance, the Janus kinase–signal transducer and activator of transcription 3 (JAK–STAT3) pathway is one of the molecular targets of leptin, so an edge from the leptin to the JAK–STAT3 is created. Note that the edge between an adipokine and its molecular target does not imply physical interactions between the two molecules, but rather information flow from one to the other.
- We then create the edge from a molecular target to a physiologic function of an adipokine if there is an association between them. For instance, the edge is created from AMPK to insulin signaling to indicate the information flow from AMPK to insulin signaling.
- If a physiologic function is associated with a disease process, then an edge starting from the physiologic function to the disease process is created. For instance, the edge from insulin signaling to metabolic syndrome is created to indicate the close association between insulin signaling and metabolic syndrome.
- We also create edges from an adipokine to its associated physiologic functions/diseases without identified molecular targets. For instance, leptin is associated with insulin signaling and metabolic syndrome. Thus, edges from leptin to insulin signaling and metabolic syndrome are created. This is to make sure the network has good “coverage”, even though some of the molecular targets/mechanisms of the linkage are unknown.
3.2. Degree Centrality Analysis
3.3. Betweenness Centrality Analysis
4. Results
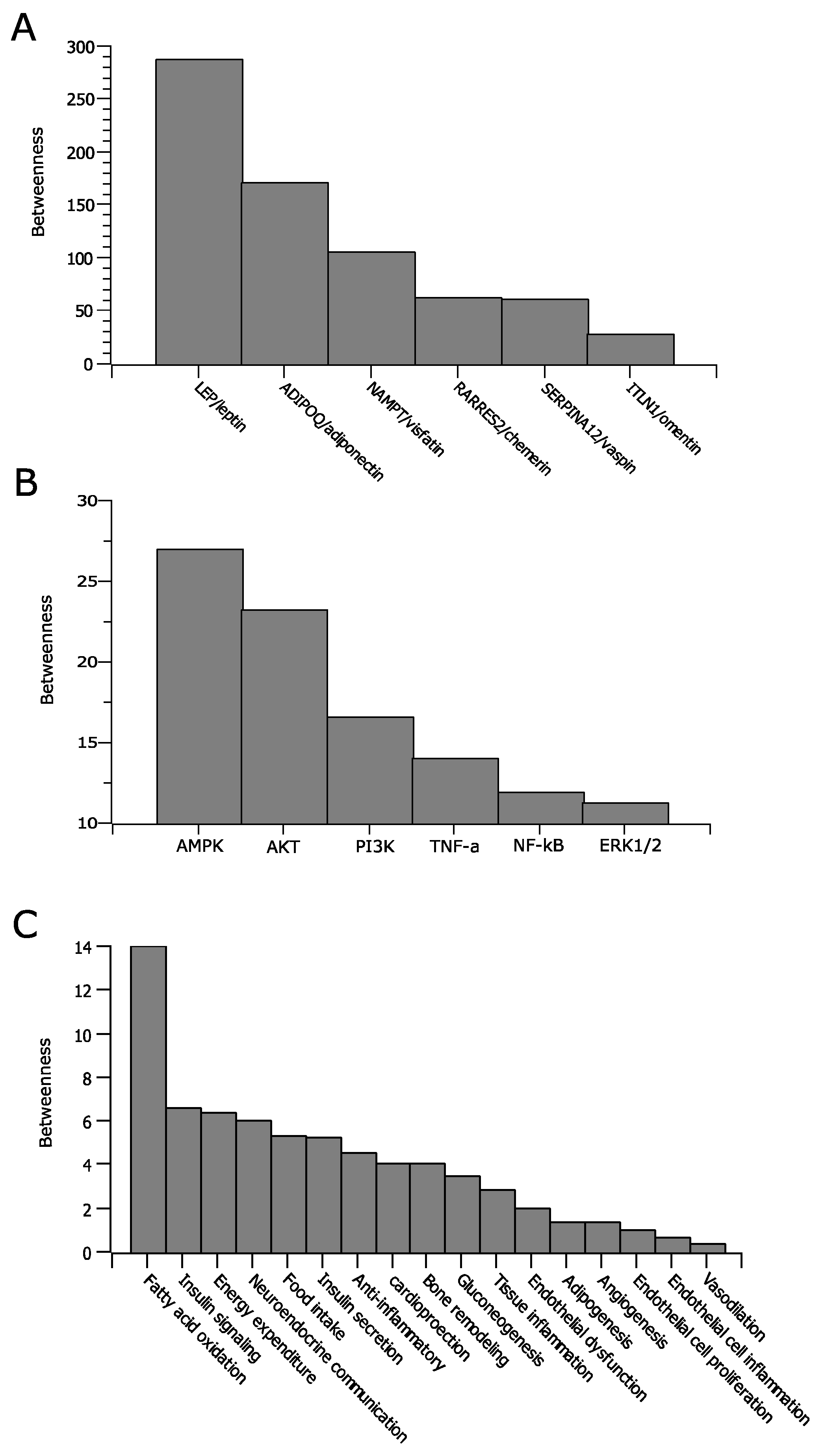
4.1. General Characteristics of the Adipokine Network
- Directed edges—The edges between two nodes are directed representing signal flow from one node to another.
- Weighted nodes—The nodes are weighted proportionally based on their degrees.
- Scale-free network—Most nodes in the network are connected to a small number of nodes which have higher degree than those of the rest of the network. The nodes with high degree centrality are considered hubs, which are important members of a network.
4.2. Analysis of the Local Environment of the Adipokine Network
4.3. Analysis of Global Relationships in the Adipokine Network
5. Discussion
6. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Conflicts of Interest
References
- Gaines, J.; Vgontzas, A.N.; Fernandez-Mendoza, J.; Bixler, E.O. Obstructive sleep apnea and the metabolic syndrome: The road to clinically-meaningful phenotyping, improved prognosis, and personalized treatment. Sleep Med. Rev. 2018, 42, 211–219. [Google Scholar] [CrossRef]
- Koren, D.; Taveras, E.M. Association of sleep disturbances with obesity, insulin resistance and the metabolic syndrome. Metabolism 2018, 84, 67–75. [Google Scholar] [CrossRef]
- Bonsignore, M.R.; Borel, A.L.; Machan, E.; Grunstein, R. Sleep apnoea and metabolic dysfunction. Eur. Respir. Rev. 2013, 22, 353–364. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jullian-Desayes, I.; Joyeux-Faure, M.; Tamisier, R.; Launois, S.; Borel, A.L.; Levy, P.; Pepin, J.L. Impact of obstructive sleep apnea treatment by continuous positive airway pressure on cardiometabolic biomarkers: A systematic review from sham CPAP randomized controlled trials. Sleep Med. Rev. 2015, 21, 23–38. [Google Scholar] [CrossRef] [PubMed]
- Vgontzas, A.N.; Papanicolaou, D.A.; Bixler, E.O.; Hopper, K.; Lotsikas, A.; Lin, H.M.; Kales, A.; Chrousos, G.P. Sleep apnea and daytime sleepiness and fatigue: Relation to visceral obesity, insulin resistance, and hypercytokinemia. J. Clin. Endocrinol. Metab. 2000, 85, 1151–1158. [Google Scholar] [CrossRef] [PubMed]
- Kritikou, I.; Basta, M.; Tappouni, R.; Pejovic, S.; Fernandez-Mendoza, J.; Nazir, R.; Shaffer, M.L.; Liao, D.; Bixler, E.O.; Chrousos, G.P.; et al. Sleep apnoea and visceral adiposity in middle-aged male and female subjects. Eur. Respir. J. 2013, 41, 601–609. [Google Scholar] [CrossRef] [PubMed]
- Lehr, S.; Hartwig, S.; Sell, H. Adipokines: A treasure trove for the discovery of biomarkers for metabolic disorders. Proteomics Clin. Appl. 2012, 6, 91–101. [Google Scholar] [CrossRef] [PubMed]
- Funcke, J.B.; Scherer, P.E. Beyond adiponectin and leptin: Adipose tissue-derived mediators of inter-organ communication. J. Lipid Res. 2019, 60, 1648–1684. [Google Scholar] [CrossRef]
- Wei, Z.; Chen, Y.; Upender, R.P. Sleep Disturbance and Metabolic Dysfunction: The Roles of Adipokines. Int. J. Mol. Sci. 2022, 23, 1706. [Google Scholar] [CrossRef]
- Koutrouli, M.; Karatzas, E.; Paez-Espino, D.; Pavlopoulos, G.A. A Guide to Conquer the Biological Network Era Using Graph Theory. Front. Bioeng. Biotechnol. 2020, 8, 34. [Google Scholar] [CrossRef]
- Baek, E.C.; Porter, M.A.; Parkinson, C. Social Network Analysis for Social Neuroscientists. Soc. Cogn. Affect. Neurosci. 2021, 16, 883–901. [Google Scholar] [CrossRef] [PubMed]
- McGillivray, P.; Clarke, D.; Meyerson, W.; Zhang, J.; Lee, D.; Gu, M.; Kumar, S.; Zhou, H.; Gerstein, M. Network Analysis as a Grand Unifier in Biomedical Data Science. Annu. Rev. Biomed. Data Sci. 2018, 1, 153–180. [Google Scholar] [CrossRef]
- Mechanick, J.I.; Zhao, S.; Garvey, W.T. The Adipokine-Cardiovascular-Lifestyle Network: Translation to Clinical Practice. J. Am. Coll. Cardiol. 2016, 68, 1785–1803. [Google Scholar] [CrossRef] [PubMed]
- Yu, H.; Kim, P.M.; Sprecher, E.; Trifonov, V.; Gerstein, M. The importance of bottlenecks in protein networks: Correlation with gene essentiality and expression dynamics. PLoS Comput. Biol. 2007, 3, e59. [Google Scholar] [CrossRef]
- McDermott, J.E.; Taylor, R.C.; Yoon, H.; Heffron, F. Bottlenecks and hubs in inferred networks are important for virulence in Salmonella typhimurium. J. Comput. Biol. 2009, 16, 169–180. [Google Scholar] [CrossRef] [Green Version]
- Wang, Z.V.; Scherer, P.E. Adiponectin, the past two decades. J. Mol. Cell Biol. 2016, 8, 93–100. [Google Scholar] [CrossRef]
- Park, H.K.; Ahima, R.S. Physiology of leptin: Energy homeostasis, neuroendocrine function and metabolism. Metabolism 2015, 64, 24–34. [Google Scholar] [CrossRef] [Green Version]
- Tan, B.K.; Adya, R.; Randeva, H.S. Omentin: A novel link between inflammation, diabesity, and cardiovascular disease. Trends Cardiovasc. Med. 2010, 20, 143–148. [Google Scholar] [CrossRef]
- Weiner, J.; Zieger, K.; Pippel, J.; Heiker, J.T. Molecular Mechanisms of Vaspin Action—From Adipose Tissue to Skin and Bone, from Blood Vessels to the Brain. Adv. Exp. Med. Biol. 2019, 1111, 159–188. [Google Scholar] [CrossRef] [Green Version]
- Saddi-Rosa, P.; Oliveira, C.S.; Giuffrida, F.M.; Reis, A.F. Visfatin, glucose metabolism and vascular disease: A review of evidence. Diabetol. Metab. Syndr. 2010, 2, 21. [Google Scholar] [CrossRef] [Green Version]
- Helfer, G.; Wu, Q.F. Chemerin: A multifaceted adipokine involved in metabolic disorders. J. Endocrinol. 2018, 238, R79–R94. [Google Scholar] [CrossRef] [PubMed]
- Bastian, M.; Heymann, S.; Jacomy, M. Gephi: An open source software for exploring and manipulating networks. In Proceedings of the Third International AAAI Conference on Weblogs and Social Media, San Jose, CA, USA, 17–20 May 2009. [Google Scholar]
- Cappuccio, F.P.; Miller, M.A. Sleep and Cardio-Metabolic Disease. Curr. Cardiol. Rep. 2017, 19, 110. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fu, J.; Wang, Y.; Li, G.; Han, L.; Li, Y.; Li, L.; Feng, D.; Wu, Y.; Xiao, X.; Li, M.; et al. Childhood sleep duration modifies the polygenic risk for obesity in youth through leptin pathway: The Beijing Child and Adolescent Metabolic Syndrome cohort study. Int. J. Obes. 2019, 43, 1556–1567. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wong, A.K.; Howie, J.; Petrie, J.R.; Lang, C.C. AMP-activated protein kinase pathway: A potential therapeutic target in cardiometabolic disease. Clin. Sci. 2009, 116, 607–620. [Google Scholar] [CrossRef] [Green Version]
- Martins, F.O.; Sacramento, J.F.; Olea, E.; Melo, B.F.; Prieto-Lloret, J.; Obeso, A.; Rocher, A.; Matafome, P.; Monteiro, E.C.; Conde, S.V. Chronic Intermittent Hypoxia Induces Early-Stage Metabolic Dysfunction Independently of Adipose Tissue Deregulation. Antioxidants 2021, 10, 1233. [Google Scholar] [CrossRef]
- Ruan, H.; Xun, P.; Cai, W.; He, K.; Tang, Q. Habitual Sleep Duration and Risk of Childhood Obesity: Systematic Review and Dose-response Meta-analysis of Prospective Cohort Studies. Sci. Rep. 2015, 5, 16160. [Google Scholar] [CrossRef] [Green Version]
- Castaneda, A.; Jauregui-Maldonado, E.; Ratnani, I.; Varon, J.; Surani, S. Correlation between metabolic syndrome and sleep apnea. World J. Diabetes 2018, 9, 66–71. [Google Scholar] [CrossRef]
- Xu, X.; Xu, J. Effects of different obesity-related adipokines on the occurrence of obstructive sleep apnea. Endocr. J. 2020, 67, 485–500. [Google Scholar] [CrossRef] [Green Version]
- Yao, Q.; Pho, H.; Kirkness, J.; Ladenheim, E.E.; Bi, S.; Moran, T.H.; Fuller, D.D.; Schwartz, A.R.; Polotsky, V.Y. Localizing Effects of Leptin on Upper Airway and Respiratory Control during Sleep. Sleep 2016, 39, 1097–1106. [Google Scholar] [CrossRef]
- Tankersley, C.; Kleeberger, S.; Russ, B.; Schwartz, A.; Smith, P. Modified control of breathing in genetically obese (ob/ob) mice. J. Appl. Physiol. (1985) 1996, 81, 716–723. [Google Scholar] [CrossRef]
- Laposky, A.D.; Shelton, J.; Bass, J.; Dugovic, C.; Perrino, N.; Turek, F.W. Altered sleep regulation in leptin-deficient mice. Am. J. Physiol. Regul. Integr. Comp. Physiol. 2006, 290, R894–R903. [Google Scholar] [CrossRef] [PubMed]
- Barabási, A.L.; Gulbahce, N.; Loscalzo, J. Network medicine: A network-based approach to human disease. Nat. Rev. Genet. 2011, 12, 56–68. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, X. Target network analysis of adiponectin, a multifaceted adipokine. J. Cell Biochem. 2013, 114, 1145–1152. [Google Scholar] [CrossRef] [PubMed]
- Huang, T.; Glass, K.; Zeleznik, O.A.; Kang, J.H.; Ivey, K.L.; Sonawane, A.R.; Birmann, B.M.; Hersh, C.P.; Hu, F.B.; Tworoger, S.S. A Network Analysis of Biomarkers for Type 2 Diabetes. Diabetes 2019, 68, 281–290. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, S.; Wang, L.; Zan, L. Investigation into the underlying molecular mechanisms of white adipose tissue through comparative transcriptome analysis of multiple tissues. Mol. Med. Rep. 2019, 19, 959–966. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Levy, E.D.; Landry, C.R.; Michnick, S.W. How perfect can protein interactomes be? Sci. Signal. 2009, 2, pe11. [Google Scholar] [CrossRef] [Green Version]
- Zhang, F.; Chen, Y.; Heiman, M.; Dimarchi, R. Leptin: Structure, function and biology. Vitam. Horm. 2005, 71, 345–372. [Google Scholar] [CrossRef]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wei, Z.; Chen, Y.; Upender, R.P. Adipokines in Sleep Disturbance and Metabolic Dysfunction: Insights from Network Analysis. Clocks & Sleep 2022, 4, 321-331. https://doi.org/10.3390/clockssleep4030027
Wei Z, Chen Y, Upender RP. Adipokines in Sleep Disturbance and Metabolic Dysfunction: Insights from Network Analysis. Clocks & Sleep. 2022; 4(3):321-331. https://doi.org/10.3390/clockssleep4030027
Chicago/Turabian StyleWei, Zhikui, You Chen, and Raghu P. Upender. 2022. "Adipokines in Sleep Disturbance and Metabolic Dysfunction: Insights from Network Analysis" Clocks & Sleep 4, no. 3: 321-331. https://doi.org/10.3390/clockssleep4030027
APA StyleWei, Z., Chen, Y., & Upender, R. P. (2022). Adipokines in Sleep Disturbance and Metabolic Dysfunction: Insights from Network Analysis. Clocks & Sleep, 4(3), 321-331. https://doi.org/10.3390/clockssleep4030027






