Physical Interaction between Cyclin-Dependent Kinase 5 (CDK5) and Clock Factors Affects the Circadian Rhythmicity in Peripheral Oscillators
Abstract
:1. Introduction
2. Results
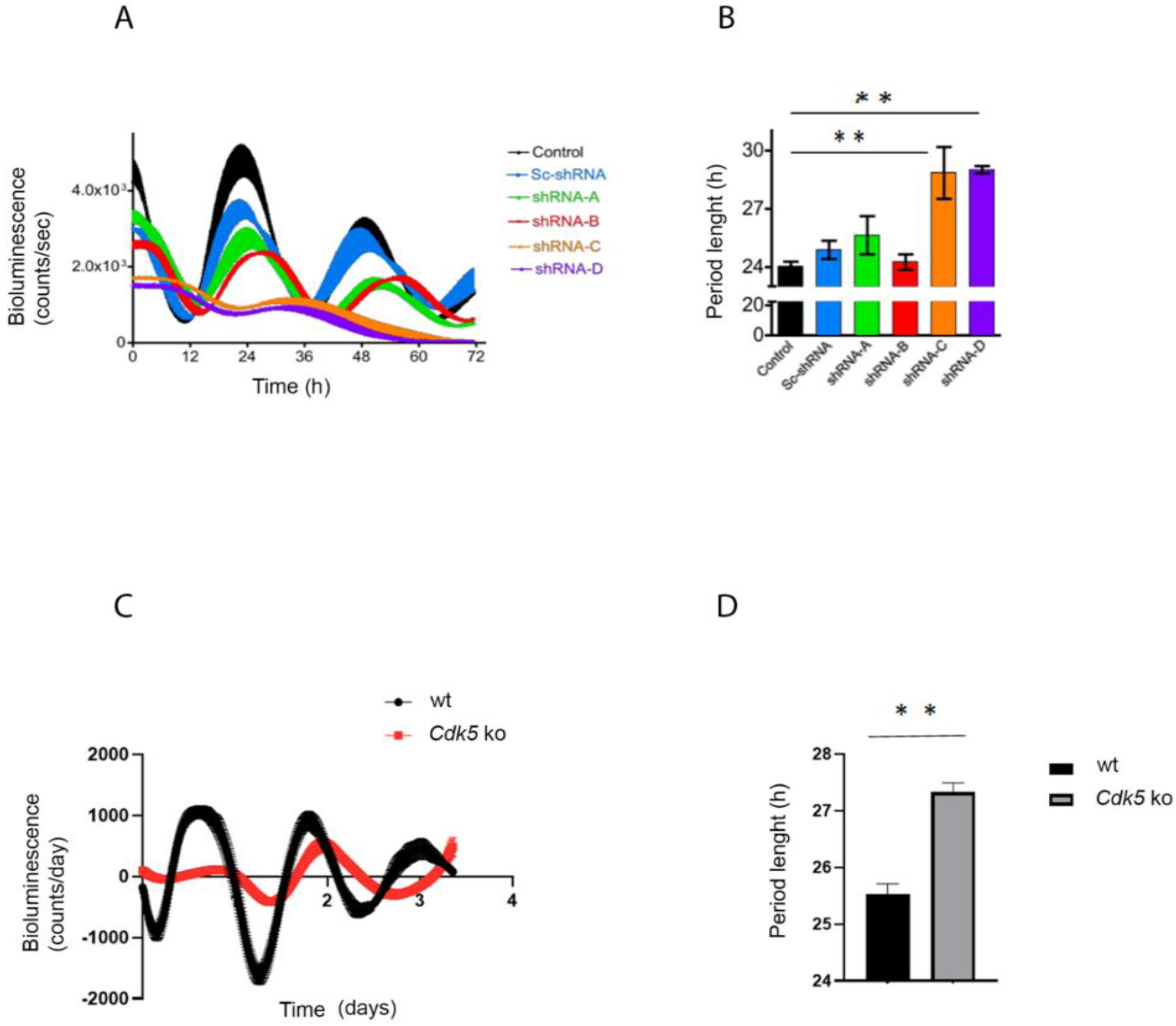
2.1. Lack of CDK5 Lengthens the Period Length of a Peripheral Clock
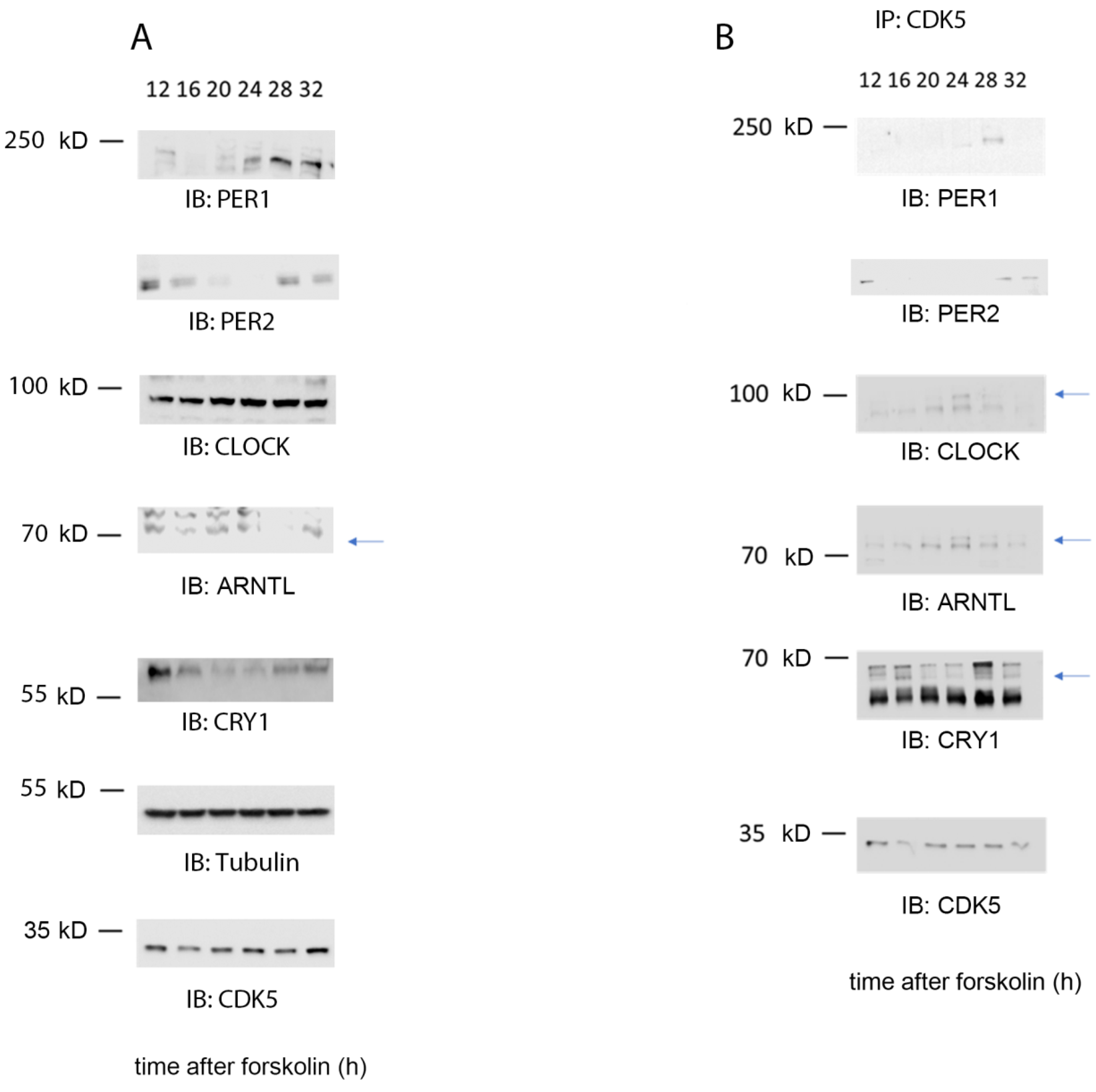
2.2. The Kinase Activity of CDK5 Is Circadian in Mouse Embryonic Fibroblasts
2.3. CDK5 Is Part of the Clock Machinery
2.4. CDK5 Influences the Nuclear Localization of Clock Factors
2.5. Delay of RNA Rhythmicity due to the Lack of CDK5 Is Mediated by a Shift of ARNTL and CLOCK Binding to Chromatin
3. Discussion
4. Materials and Methods
4.1. Cell Culture
4.2. Protein Extraction from Cells
4.3. Immunoprecipitation
4.4. In Vitro Kinase Assay Using Immunoprecipitated Cdk5 from Synchronized Cells
4.5. Nuclear/Cytoplasm Fractionation
4.6. Western Blot Analysis
4.7. RNA Extraction and Real-Time-PCR
- Sirt2 (normalization probe)
- FW: 5′-CAG GCC AGA CGG ACC CCT TC-3′
- RV: 5′-AGG CCA CGT CCC TGT AAG CC-3′
- TM: 5′-FAM-TGA TGG GCC TGG GAG GTG GCA TGG A-BHQ1-3′
- Nono (normalization probe)
- FW: 5′-TCT TTT CTC GGG ACG GTG GAG-3′
- RV: 5′-GTC TGC CTC GCA GTC CTC ACT-3′
- TM: 5′-FAM-CGT GCA GCG TCG CCC ATA CTC CGA GC-BHQ1-3′
- Tspo (normalization probe)
- FW: 5′-GGT CAG CTG GCT CTG AAC TG-3′
- RV: 5′-CAG TCG CCA CCC CAC TGA CA-3′
- TM: 5′-FAM-TGC CCG GCA GAT GGG CTG GGC-BHQ1-3′
- Tprkb (normalization probe)
- FW: 5′-GGC TGG CAT CAG ACC CAC AGA-3′
- RV: 5′-GGG CCC GTA GAG TCG GGA AA-3′
- TM: 5′-FAM-CCT GCG TCT GCC CTC TGA GGG CTG-BHQ1-3′
- Per1
- FW: 5′-GCC CCG CCT CCT TGC TAC A-3′
- RV: 5′-ACT GGG GCC ACC TCC AGT TC-3′
- TM: 5′-FAM-TCC TTC CCT GCC AGT CCC CAA ACC CC-BHQ1-3′
- Per2
- FW: 5′-TCC ACA GCT ACA CCA CCC CTT A-3′
- RV: 5′-TTT CTC CTC CAT GCA CTC CTG A-3′
- TM: 5′-FAM-CCG CTG CAC ACA CTC CAG GGC G-BHQ1-3′
- Per3
- FW: 5′-CGT CTG GCA TCA GCC AGT GC-3′
- RV: 5′-CTC AGG GCC CAC GGC TTA CA-3′
- TM: 5′-FAM-CCT CTG GCC ACG CTC CGC CCC T-BHQ1-3′
- Cry1
- FW: 5′-CTG GCG TGG AAG TCA TCG T-3′
- RV: 5′-CTG TCC GCC GAG TTC TAT G-3′
- TM: 5′-FAM-CGC ATT TCA CAT ACA CTG TAT GAC CTG GAC A-BHQ1-3′
- Cry2
- FW: 5′-TGT CCC TTC CTG TGT GGA AGA-3′
- RV: 5′-GCT CCC AGC TTG GCT TGA-3′
- TM: 5′-FAM-CAG TCA CCC TGT GGC AGA GCC TGG-BHQ1-3′
- Arntl
- FW: 5′-CCA AGA AAG TAT GGA CAC AGA CAA A-3′
- RV: 5′-GCA TTC TTG ATC CTT CCT TGG T-3′
- TM: 5′-FAM-TGA CCC TCA TGG AAG GTT AGA ATA TGC AGA A-BHQ1-3′
- Clock
- FW: 5′-TTG CTC CAC GGG AAT CCT T-3′
- RV: 5′-GGA GGG AAA GTG CTC TGT TGT AG-3′
- TM: 5′-FAM-ACA CAG CTC ATC CTC TCT GCT GCC TTT C-BHQ1-3′
- Npas2
- FW: 5′-CAG CCC TGA CTT CGG CCA TGA-3′
- RV: 5′-CAT CGC AGG ACC CAG GCA TCA-3′
- TM: 5′-FAM-CGG CAG CTC AGG CTG TTG CTG AGC C-BHQ1-3′
- Nr1d1
- FW: 5′-GAA GTG TCT CTC CGT TGG CAT GTC T-3′
- RV: 5′-CGC TCT GCA TCT CGG CAA GCA T-3′
- TM: 5′-FAM-CTG TGC GTT TTG GGC GCA TCC CCA AG-BHQ1-3′
- Nr1d2
- FW: 5′-GTG AGG GCC GCA CCC TGT-3′
- RV: 5′-CAG GGC TGG AGG CAG AGC T-3′
- TM: 5′-FAM-CCA GCG CCA TGG AGC TGA ACG CAG G-BHQ1-3′
4.8. Chromatin Immunoprecipitation
- Per1_E box
- FW: 5′-AGG CAC CAG AAA CCT CTT G-3′
- RV: 5′-GGC GTA GAT CTG ACA GGC TA-3′
- TM: 5′-FAM- TGC CAG AGT CTC CAA AGT ATG CCC AC-BHQ1-3′
4.9. Analysis of Circadian Rhythms (CircaCompare)
4.10. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Schibler, U.; Sassone-Corsi, P. A Web of Circadian Pacemakers. Cell 2002, 111, 919–922. [Google Scholar] [CrossRef] [Green Version]
- Brenna, A.; Albrecht, U. Phosphorylation and Circadian Molecular Timing. Front. Physiol. 2020, 11, 612510. [Google Scholar] [CrossRef]
- Reppert, S.M.; Weaver, D.R. Coordination of circadian timing in mammals. Nature 2002, 418, 935–941. [Google Scholar] [CrossRef]
- Dibner, C.; Schibler, U.; Albrecht, U. The Mammalian Circadian Timing System: Organization and Coordination of Central and Peripheral Clocks. Annu. Rev. Physiol. 2010, 72, 517–549. [Google Scholar] [CrossRef] [Green Version]
- Partch, C.L.; Green, C.B.; Takahashi, J.S. Molecular architecture of the mammalian circadian clock. Trends Cell Biol. 2014, 24, 90–99. [Google Scholar] [CrossRef] [Green Version]
- Kucera, N.; Schmalen, I.; Hennig, S.; Ollinger, R.; Strauss, H.M.; Grudziecki, A.; Wieczorek, C.; Kramer, A.; Wolf, E. Unwinding the differences of the mammalian PERIOD clock proteins from crystal structure to cellular function. Proc. Natl. Acad. Sci. USA 2012, 109, 3311–3316. [Google Scholar] [CrossRef] [Green Version]
- Chiou, Y.-Y.; Yang, Y.; Rashid, N.; Ye, R.; Selby, C.P.; Sancar, A. Mammalian Period represses and de-represses transcription by displacing CLOCK–BMAL1 from promoters in a Cryptochrome-dependent manner. Proc. Natl. Acad. Sci. USA 2016, 113, E6072–E6079. [Google Scholar] [CrossRef] [Green Version]
- Buhr, E.D.; Takahashi, J.S. Molecular Components of the Mammalian Circadian Clock. Handb. Exp. Pharmacol. 2013, 217, 3–27. [Google Scholar] [CrossRef] [Green Version]
- Panda, S.; Antoch, M.P.; Miller, B.H.; Su, A.I.; Schook, A.B.; Straume, M.; Schultz, P.G.; Kay, S.A.; Takahashi, J.S.; Hogenesch, J.B. Coordinated Transcription of Key Pathways in the Mouse by the Circadian Clock. Cell 2002, 109, 307–320. [Google Scholar] [CrossRef] [Green Version]
- Storch, K.-F.; Lipan, O.; Leykin, I.; Viswanathan, N.; Davis, F.C.; Wong, W.H.; Weitz, C.J. Extensive and divergent circadian gene expression in liver and heart. Nature 2002, 417, 78–83. [Google Scholar] [CrossRef]
- Bellet, M.M.; Sassone-Corsi, P. Mammalian circadian clock and metabolism–the epigenetic link. J. Cell Sci. 2010, 123, 3837–3848. [Google Scholar] [CrossRef] [Green Version]
- Hirano, A.; Fu, Y.H.; Ptacek, L.J. The intricate dance of post-translational modifications in the rhythm of life. Nat. Struct. Mol. Biol. 2016, 23, 1053–1060. [Google Scholar] [CrossRef]
- Robles, M.S.; Humphrey, S.J.; Mann, M. Phosphorylation Is a Central Mechanism for Circadian Control of Metabolism and Physiology. Cell Metab. 2017, 25, 118–127. [Google Scholar] [CrossRef] [Green Version]
- Iwasaki, H.; Nishiwaki, T.; Kitayama, Y.; Nakajima, M.; Kondo, T. KaiA-stimulated KaiC phosphorylation in circadian timing loops in cyanobacteria. Proc. Natl. Acad. Sci. USA 2002, 99, 15788–15793. [Google Scholar] [CrossRef] [Green Version]
- Tang, D.; Yeung, J.; Lee, K.-Y.; Matsushita, M.; Matsui, H.; Tomizawa, K.; Hatase, O.; Wang, J.H. An Isoform of the Neuronal Cyclin-dependent Kinase 5 (Cdk5) Activator. J. Biol. Chem. 1995, 270, 26897–26903. [Google Scholar] [CrossRef] [Green Version]
- Tsai, L.-H.; Delalle, I.; Caviness, V.S., Jr.; Chae, T.; Harlow, E. p35 is a neural-specific regulatory subunit of cyclin-dependent kinase 5. Nature 1994, 371, 419–423. [Google Scholar] [CrossRef]
- Brinkkoetter, P.T.; Olivier, P.; Wu, J.S.; Henderson, S.; Krofft, R.D.; Pippin, J.W.; Hockenbery, D.; Roberts, J.M.; Shankland, S.J. Cyclin I activates Cdk5 and regulates expression of Bcl-2 and Bcl-XL in postmitotic mouse cells. J. Clin. Investig. 2009, 119, 3089–3101. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kawauchi, T. Cdk5 regulates multiple cellular events in neural development, function and disease. Dev. Growth Differ. 2014, 56, 335–348. [Google Scholar] [CrossRef]
- Kwak, Y.; Jeong, J.; Lee, S.; Park, Y.-U.; Lee, S.-A.; Han, D.-H.; Kim, J.-H.; Ohshima, T.; Mikoshiba, K.; Suh, Y.-H.; et al. Cyclin-dependent Kinase 5 (Cdk5) Regulates the Function of CLOCK Protein by Direct Phosphorylation. J. Biol. Chem. 2013, 288, 36878–36889. [Google Scholar] [CrossRef] [Green Version]
- Brenna, A.; Olejniczak, I.; Chavan, R.; Ripperger, J.A.; Langmesser, S.; Cameroni, E.; Hu, Z.; De Virgilio, C.; Dengjel, J.; Albrecht, U. Cyclin-dependent kinase 5 (CDK5) regulates the circadian clock. Elife 2019, 8, e50925. [Google Scholar] [CrossRef]
- Contreras-Vallejos, E.; Utreras, E.; Gonzalez-Billault, C. Going out of the brain: Non-nervous system physiological and pathological functions of Cdk5. Cell Signal. 2012, 24, 44–52. [Google Scholar] [CrossRef] [PubMed]
- Pozo, K.; Bibb, J.A. The Emerging Role of Cdk5 in Cancer. Trends Cancer 2016, 2, 606–618. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sulli, G.; Lam, M.T.Y.; Panda, S. Interplay between Circadian Clock and Cancer: New Frontiers for Cancer Treatment. Trends Cancer 2019, 5, 475–494. [Google Scholar] [CrossRef] [PubMed]
- Hsu, F.-N.; Chen, M.-C.; Lin, K.-C.; Peng, Y.-T.; Li, P.-C.; Lin, E.; Chiang, M.-C.; Hsieh, J.-T.; Lin, H. Cyclin-dependent kinase 5 modulates STAT3 and androgen receptor activation through phosphorylation of Ser727 on STAT3 in prostate cancer cells. Am. J. Physiol. Metab. 2013, 305, E975–E986. [Google Scholar] [CrossRef] [PubMed]
- Yagita, K.; Okamura, H. Forskolin induces circadian gene expression of rPer1, rPer2 and dbp in mammalian rat-1 fibroblasts. FEBS Lett. 1999, 465, 79–82. [Google Scholar] [CrossRef] [Green Version]
- Tomov, N.; Surchev, L.; Wiedenmann, C.; Döbrössy, M.; Nikkhah, G. Roscovitine, an experimental CDK5 inhibitor, causes delayed suppression of microglial, but not astroglial recruitment around intracerebral dopaminergic grafts. Exp. Neurol. 2019, 318, 135–144. [Google Scholar] [CrossRef]
- Aryal, R.P.; Kwak, P.B.; Tamayo, A.G.; Gebert, M.; Chiu, P.-L.; Walz, T.; Weitz, C.J. Macromolecular Assemblies of the Mammalian Circadian Clock. Mol. Cell 2017, 67, 770–782. [Google Scholar] [CrossRef] [Green Version]
- Wang, Z.; Wu, Y.; Li, L.; Su, X.-D. Intermolecular recognition revealed by the complex structure of human CLOCK-BMAL1 basic helix-loop-helix domains with E-box DNA. Cell Res. 2012, 23, 213–224. [Google Scholar] [CrossRef] [Green Version]
- Ino, H.; Chiba, T. Intracellular localization of cyclin-dependent kinase 5 (CDK5) in mouse neuron: CDK5 is located in both nucleus and cytoplasm. Brain Res. 1996, 732, 179–185. [Google Scholar] [CrossRef]
- Qu, D.; Li, Q.; Lim, H.Y.; Cheung, N.S.; Li, R.; Wang, J.H.; Qi, R.Z. The protein SET binds the neuronal Cdk5 activator p35nck5a and modulates Cdk5/p35nck5a activity. J. Biol. Chem. 2002, 277, 7324–7332. [Google Scholar] [CrossRef] [Green Version]
- Parsons, R.; Parsons, R.; Garner, N.; Oster, H.; Rawashdeh, O. CircaCompare: A method to estimate and statistically support differences in mesor, amplitude, and phase, between circadian rhythms. Bioinformatics 2019, 36, 1208–1212. [Google Scholar] [CrossRef] [PubMed]
- Ripperger, J.A.; Schibler, U. Rhythmic CLOCK-BMAL1 binding to multiple E-box motifs drives circadian Dbp transcription and chromatin transitions. Nat. Genet. 2006, 38, 369–374. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zheng, B.; Larkin, D.W.; Albrecht, U.; Sun, Z.S.; Sage, M.; Eichele, G.; Lee, C.C.; Bradley, A. The mPer2 gene encodes a functional component of the mammalian circadian clock. Nature 1999, 400, 169–173. [Google Scholar] [CrossRef] [PubMed]
- Dhavan, R.; Tsai, L.H. A decade of CDK5. Nat. Rev. Mol. Cell Biol. 2001, 2, 749–759. [Google Scholar] [CrossRef]
- Dhariwala, F.A.; Rajadhyaksha, M.S. An Unusual Member of the Cdk Family: Cdk5. Cell. Mol. Neurobiol. 2008, 28, 351–369. [Google Scholar] [CrossRef]
- Wei, F.-Y.; Nagashima, K.; Ohshima, T.; Saheki, Y.; Lu, Y.-F.; Matsushita, M.; Yamada, Y.; Mikoshiba, K.; Seino, Y.; Matsui, H.; et al. Cdk5-dependent regulation of glucose-stimulated insulin secretion. Nat. Med. 2005, 11, 1104–1108. [Google Scholar] [CrossRef]
- Banks, A.S.; McAllister, F.E.; Camporez, J.P.; Zushin, P.J.; Jurczak, M.J.; Laznik-Bogoslavski, D.; Shulman, G.I.; Gygi, S.P.; Spiegelman, B.M. An ERK/Cdk5 axis controls the diabetogenic actions of PPARgamma. Nature 2015, 517, 391–395. [Google Scholar] [CrossRef] [Green Version]
- Lee, K.Y.; Rosales, J.L.; Lee, B.C.; Chung, S.H.; Fukui, Y.; Lee, N.S.; Lee, K.Y.; Jeong, Y.G. Cdk5/p35 expression in the mouse ovary. Mol. Cells 2004, 17, 17–22. [Google Scholar]
- Musa, F.R.; Tokuda, M.; Kuwata, Y.; Ogawa, T.; Tomizawa, K.; Konishi, R.; Takenaka, I.; Hatase, O. Expression of cyclin-dependent kinase 5 and associated cyclins in Leydig and Sertoli cells of the testis. J. Androl. 1999, 19, 657–666. [Google Scholar]
- Musa, F.R.; Takenaka, I.; Konishi, R.; Tokuda, M. Effects of luteinizing hormone, follicle-stimulating hormone, and epidermal growth factor on expression and kinase activity of cyclin-dependent kinase 5 in Leydig TM3 and Sertoli TM4 cell lines. J. Androl. 2000, 21, 392–402. [Google Scholar]
- Richards, J.; Gumz, M.L. Advances in understanding the peripheral circadian clocks. FASEB J. 2012, 26, 3602–3613. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Camacho, F.; Cilio, M.; Guo, Y.; Virshup, D.M.; Patel, K.; Khorkova, O.; Styren, S.; Morse, B.; Yao, Z.; Keesler, G.A. Human casein kinase Idelta phosphorylation of human circadian clock proteins period 1 and 2. FEBS Lett. 2001, 489, 159–165. [Google Scholar] [CrossRef] [Green Version]
- DeBruyne, J.P.; Noton, E.; Lambert, C.M.; Maywood, E.S.; Weaver, D.R.; Reppert, S.M. A Clock Shock: Mouse CLOCK Is Not Required for Circadian Oscillator Function. Neuron 2006, 50, 465–477. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- DeBruyne, J.P.; Weaver, D.R.; Reppert, S.M. CLOCK and NPAS2 have overlapping roles in the suprachiasmatic circadian clock. Nat. Neurosci. 2007, 10, 543–545. [Google Scholar] [CrossRef] [Green Version]
- Koike, N.; Yoo, S.-H.; Huang, H.-C.; Kumar, V.; Lee, C.; Kim, T.-K.; Takahashi, J.S. Transcriptional Architecture and Chromatin Landscape of the Core Circadian Clock in Mammals. Science 2012, 338, 349–354. [Google Scholar] [CrossRef] [Green Version]
- Gekakis, N.; Staknis, D.; Nguyen, H.B.; Davis, F.C.; Wilsbacher, L.D.; King, D.P.; Takahashi, J.S.; Weitz, C.J. Role of the CLOCK protein in the mammalian circadian mechanism. Science 1998, 280, 1564–1569. [Google Scholar] [CrossRef]
- Shearman, L.P.; Sriram, S.; Weaver, D.R.; Maywood, E.S.; Chaves, I.; Zheng, B.; Kume, K.; Lee, C.C.; van der Horst, G.T.; Hastings, M.H.; et al. Interacting Molecular Loops in the Mammalian Circadian Clock. Science 2000, 288, 1013–1019. [Google Scholar] [CrossRef]
- Kwon, I.; Lee, J.; Chang, S.H.; Jung, N.C.; Lee, B.J.; Son, G.H.; Kim, K.; Lee, K.H. BMAL1 Shuttling Controls Transactivation and Degradation of the CLOCK/BMAL1 Heterodimer. Mol. Cell. Biol. 2006, 26, 7318–7330. [Google Scholar] [CrossRef] [Green Version]
- Antoch, M.P.; Song, E.-J.; Chang, A.-M.; Vitaterna, M.H.; Zhao, Y.; Wilsbacher, L.D.; Sangoram, A.M.; King, D.P.; Pinto, L.H.; Takahashi, J.S. Functional Identification of the Mouse Circadian Clock Gene by Transgenic BAC Rescue. Cell 1997, 89, 655–667. [Google Scholar] [CrossRef] [Green Version]
- Sujino, M.; Asakawa, T.; Nagano, M.; Koinuma, S.; Masumoto, K.H.; Shigeyoshi, Y. CLOCKΔ19 mutation modifies the manner of synchrony among oscillation neurons in the suprachiasmatic nucleus. Sci. Rep. 2018, 8, 854. [Google Scholar] [CrossRef] [Green Version]
- Liang, Z.; Ye, T.; Zhou, X.; Lai, K.O.; Fu, A.K.; Ip, N.Y. Cdk5 Regulates Activity-Dependent Gene Expression and Dendrite Development. J. Neurosci. 2015, 35, 15127–15134. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schmutz, I.; Ripperger, J.A.; Baeriswyl-Aebischer, S.; Albrecht, U. The mammalian clock component PERIOD2 coordinates circadian output by interaction with nuclear receptors. Genes Dev. 2010, 24, 345–357. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Costa, S.S.F.; Wegmann, D.; Ripperger, J.A. Normalisation against Circadian and Age-Related Disturbances Enables Robust Detection of Gene Expression Changes in Liver of Aged Mice. PLoS ONE 2017, 12, e0169615. [Google Scholar] [CrossRef]


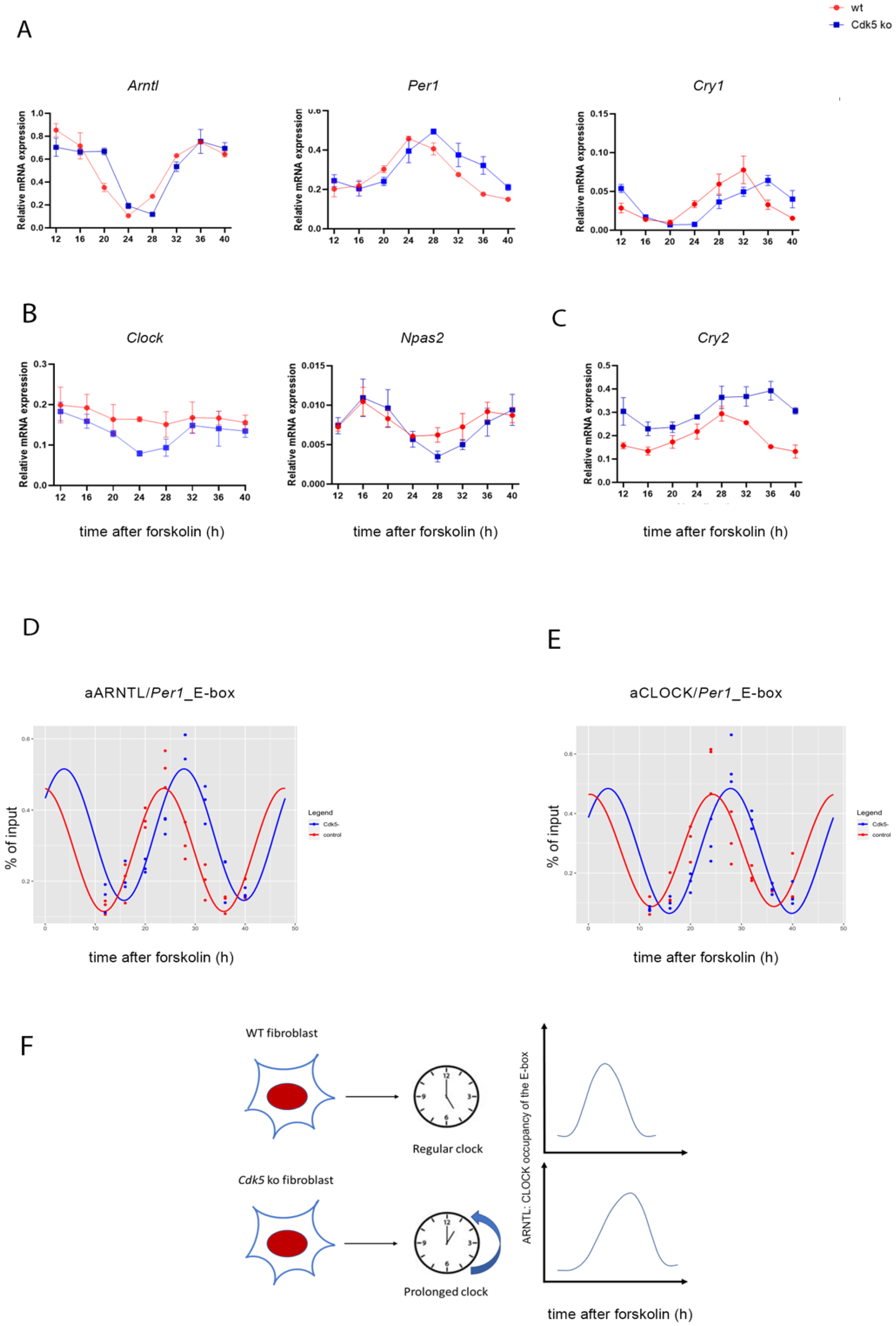
| mRNA Accumulation | Delta Mesor (AU) | p-Value | Delta Amplitude (AU) | p-Value | Delta Phase (h) | p-Value |
|---|---|---|---|---|---|---|
| Arntl | −0.004 | 0.860 | 0.043 | 0.199 | −2.02 | 0.000 |
| Clock | NA | NA | NA | NA | NA | NA |
| Cry1 | 0.005 | 0.075 | 0.003 | 0.465 | −3.71 | 0.000 |
| Cry2 | −0.108 | 0.000 | 0.005 | 0.765 | −3.53 | 0.000 |
| Npas2 | 0.001 | 0.107 | −0.002 | 0.006 | −1.41 | 0.194 |
| Nr1d1 | 0.167 | 0.156 | 0.016 | 0.918 | −2.55 | 0.000 |
| Nr1d2 | 0.000 | 0.763 | −0.001 | 0.687 | −3.71 | 0.000 |
| Per1 | −0.031 | 0.011 | 0.006 | 0.680 | −2.63 | 0.000 |
| Per2 | 0.064 | 0.382 | 0.147 | 0.155 | −2.76 | 0.000 |
| Per3 | 0.036 | 0.058 | 0.030 | 0.238 | −2.55 | 0.000 |
| ChIP | Delta Mesor (AU) | p-Value | Delta Amplitude (AU) | p-Value | Delta Phase (h) | p-Value |
|---|---|---|---|---|---|---|
| aARNTL | 0.002 | 0.944 | −0.021 | 0.491 | −3.50 | 0.000 |
| aCLOCK | −0.043 | 0.018 | −0.012 | 0.622 | −4.04 | 0.000 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ripperger, J.A.; Chavan, R.; Albrecht, U.; Brenna, A. Physical Interaction between Cyclin-Dependent Kinase 5 (CDK5) and Clock Factors Affects the Circadian Rhythmicity in Peripheral Oscillators. Clocks & Sleep 2022, 4, 185-201. https://doi.org/10.3390/clockssleep4010017
Ripperger JA, Chavan R, Albrecht U, Brenna A. Physical Interaction between Cyclin-Dependent Kinase 5 (CDK5) and Clock Factors Affects the Circadian Rhythmicity in Peripheral Oscillators. Clocks & Sleep. 2022; 4(1):185-201. https://doi.org/10.3390/clockssleep4010017
Chicago/Turabian StyleRipperger, Jürgen A., Rohit Chavan, Urs Albrecht, and Andrea Brenna. 2022. "Physical Interaction between Cyclin-Dependent Kinase 5 (CDK5) and Clock Factors Affects the Circadian Rhythmicity in Peripheral Oscillators" Clocks & Sleep 4, no. 1: 185-201. https://doi.org/10.3390/clockssleep4010017
APA StyleRipperger, J. A., Chavan, R., Albrecht, U., & Brenna, A. (2022). Physical Interaction between Cyclin-Dependent Kinase 5 (CDK5) and Clock Factors Affects the Circadian Rhythmicity in Peripheral Oscillators. Clocks & Sleep, 4(1), 185-201. https://doi.org/10.3390/clockssleep4010017






