Richard the Lionheart and the Ferocious Saladin Face to Face in Arsuf: A Proteomic Study
Abstract
1. Introduction
2. Materials and Methods
2.1. Samples
2.2. Chemicals and Materials
2.3. Synthesis and Characterization of the EVA Film
2.4. EVA Film Application
2.5. Mass Spectrometry Analysis for Protein Identifications
2.6. Extraction and Sample Preparation Tools for Archeology Field Analysis
2.7. Calorie Consumption and Diet Efficiency Estimations
3. Results
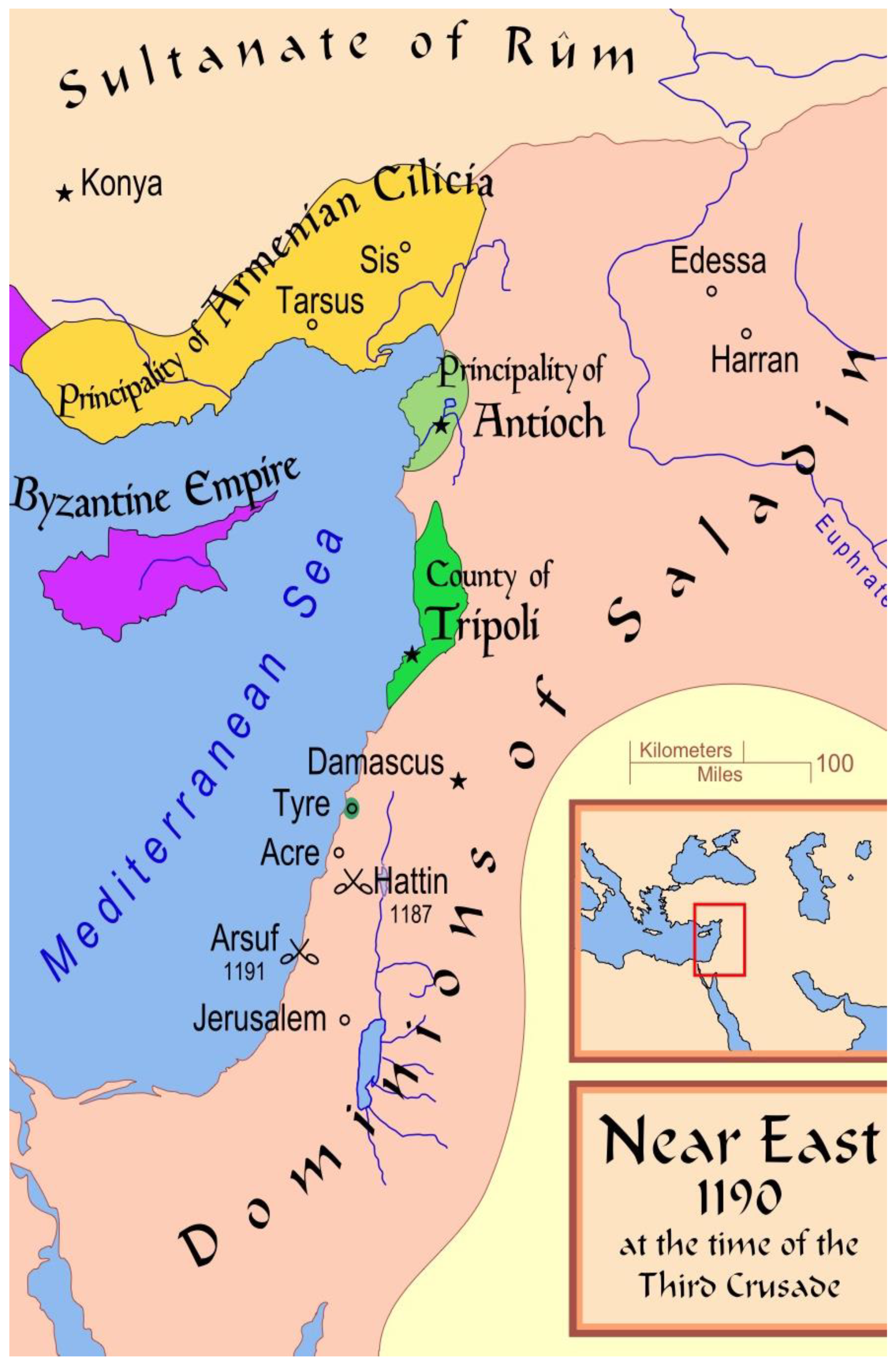
3.1. A Survey of the Territory
3.2. A Clue on the Crusader Diet
3.3. A Clue on the Saracen Diet
3.4. A Common Foodstuff to Both Armies
4. Discussion
4.1. Evaluation of our Extraction Procedure
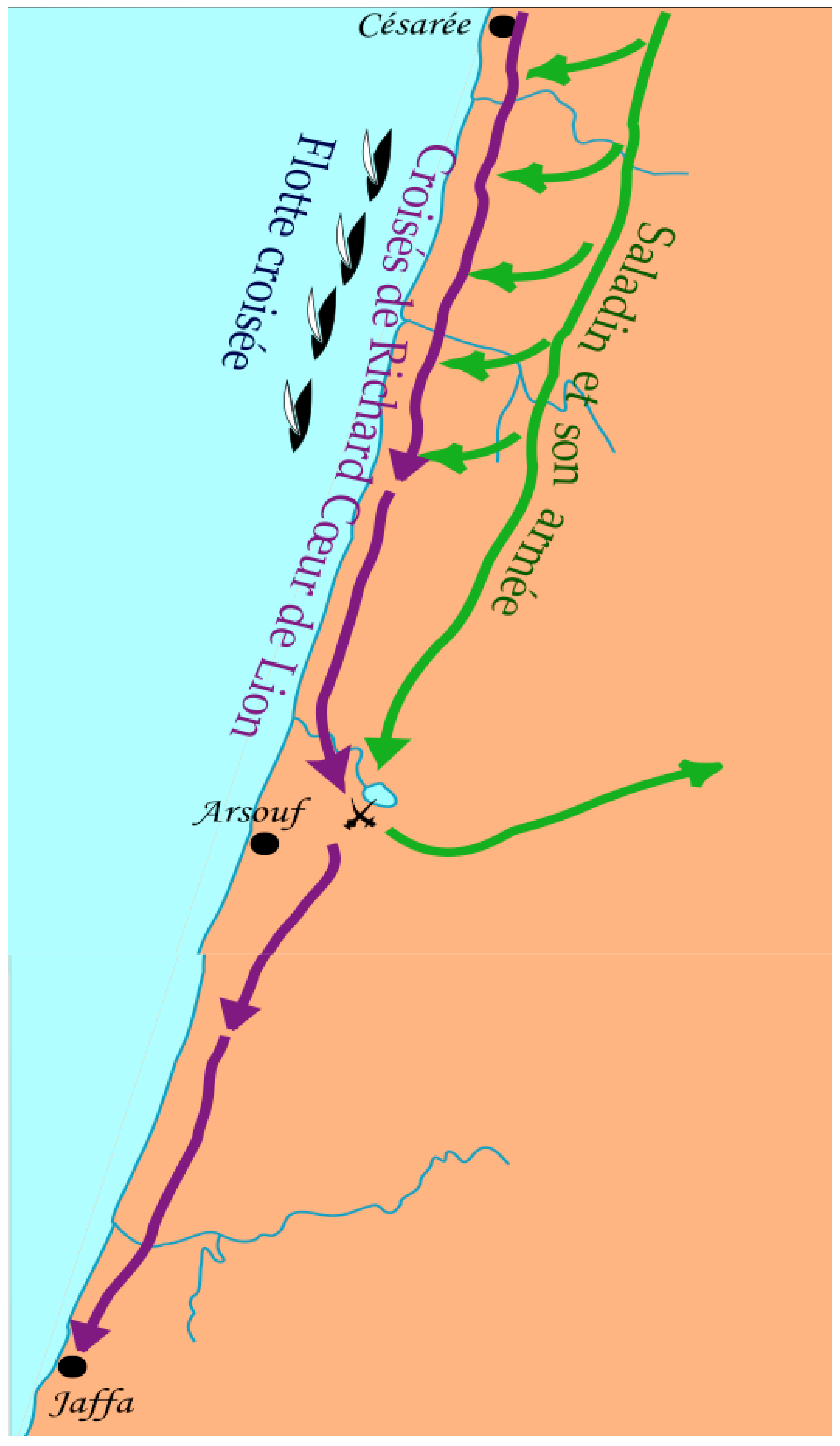
4.2. A Closer Survey of the Arsuf Battle
4.3. Evaluation of the Crusades over the Centuries
5. Conclusions
Post-Scriptum
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Barber, M. The Crusader States; Yale University Press: New Haven, CT, USA, 2012. [Google Scholar]
- Hindley, G. Saladin: Hero of Islam; Pen & Sword Military: Barnsley, UK, 2007. [Google Scholar]
- Hill, P. The Knights Templar at War. 1120–1312; Pen & Sword Military: Barnsley, UK, 2018. [Google Scholar]
- Runciman, S. A History of the Crusades 1: The First Crusade and the Foundation of the Kingdom of Jerusalem/Angla; Penguin Books: Westminster, UK, 2016. [Google Scholar]
- Riley-Smith, J. The Crusades: A History; Bloomsbury Academic: London, UK, 2014. [Google Scholar]
- Dossa, S.; Maalouf, A. The crusades through arab eyes. MERIP Middle East Rep. 1986, 42, 36–38. [Google Scholar] [CrossRef]
- Jones, T.; Ereira, A. Crusades; Penguin Group USA: New York, NY, USA, 1999. [Google Scholar]
- Bradford, E. The Great Betrayal; Open Road Media: New York, NY, USA, 2014. [Google Scholar]
- France, J. The Crusades and the Expansion of Catholic Christendom, 1000–1714; Routledge: Oxford, UK, 2006. [Google Scholar]
- Madden, T.F. The Crusades Controversy: Setting the Record Straight; North Palm Beach: Wellspring, UK, 2017. [Google Scholar]
- Jones, D. Crusaders: The Epic History of the Wars for the Holy Lands; Penguin Books: Westminster, UK, 2019. [Google Scholar]
- Stark, R. God’s Battalions: The Case for the Crusades; HarperOne: San Francisco, CA, USA, 2009. [Google Scholar]
- Carole, H. The Crusades Islam Perspectives; Endinburgh University Press: Edinburgh, UK, 1999. [Google Scholar]
- Richard, J. Histoire de Croisades; Fayard: Paris, France, 1996. [Google Scholar]
- Pines, M.; Sapir-Hen, L.; Tal, O. Crusader diet in times of war and peace: Arsur (Israel) as a case study. Oxf. J. Archaeol. 2017, 36, 307–328. [Google Scholar] [CrossRef]
- Pines, M.; Sapir-Hen, L.; Tal, O. Consumption and disposal practices in the southern levant in late antiquity: Animal bones from ’Aπολλωνία/Σώζουσα’s hinterland as a case study. Zeitschrift Deutschen Palästina Vereins 2017, 133, 186–204. [Google Scholar]
- Raphael, K.; Tepper, Y. The archaeological evidence from the mamluk siege of arsuf. Mamluk Stud. Rev. 2005, 9, 85–100. [Google Scholar]
- Roll, I. Medieval apollonia-arsuf: A Fortified coastal town in the levant of the early muslim and crusader periods. In Autour de la première Croisade. Actes du Colloque de la Society for the Study of the Crusades and the Latin East (Clermont-Ferrand, 22–25 juin 1995); Publications de la Sorbonne: Paris, France, 1996; pp. 597–606. [Google Scholar]
- Roll, I.; Ayalon, E. The market street at Apollonia-Arsuf. Bull. Am. Sch. Orient. Res. 1987, 267, 61–76. [Google Scholar] [CrossRef][Green Version]
- Roll, I.; Yohana, H.; Tepper, Y.; Harpak, T. Apollonia-Arsuf during the crusader period in light of new discoveries. Qadmoniot 2000, 33, 18–31. (In Hebrew) [Google Scholar]
- Sapir-Hen, L.; Pines, M.; Tal, O. Animal economy and social diversity in Byzantine Apollonia/Sozousa. Levant 2014, 46, 371–381. [Google Scholar] [CrossRef]
- Dallongeville, S.; Garnier, N.; Rolando, C.; Tokarski, C. Proteins in art, archaeology, and paleontology: From detection to identification. Chem. Rev. 2016, 116, 2–79. [Google Scholar] [CrossRef] [PubMed]
- Guasch-Jané, M.R.; Ibern-Gómez, M.; Andres-Lacueva, C.; Jáuregui, O.; Lamuela-Raventos, R.M. Liquid chromatography with mass spectrometry in tandem mode applied for the identification of wine markers in residues from ancient egyptian vessels. Anal. Chem. 2004, 76, 1672–1677. [Google Scholar] [CrossRef] [PubMed]
- Solazzo, C.; Fitzhugh, W.W.; Rolando, C.; Tokarski, C. Identification of protein remains in archaeological potsherds by pro-teomics. Anal. Chem. 2008, 80, 4590–4597. [Google Scholar] [CrossRef] [PubMed]
- Dallongeville, S.; Garnier, N.; Casasola, D.B.; Bonifay, M.; Rolando, C.; Tokarski, C. Dealing with the identification of protein species in ancient amphorae. Anal. Bioanal. Chem. 2010, 399, 3053–3063. [Google Scholar] [CrossRef] [PubMed]
- Hammann, S.; Cramp, L.J.E. Towards the detection of dietary cereal processing through absorbed lipid biomarkers in archaeological pottery. J. Archeol. Sci. 2018, 93, 74–81. [Google Scholar] [CrossRef]
- Hendy, J.; Colonese, A.C.; Franz, I.; Fernandes, R.; Fischer, R.; Orton, D.; Lucquin, A.; Spindler, L.; Anvari, J.; Stroud, E.; et al. Ancient proteins from ceramic vessels at Çatalhöyük West reveal the hidden cuisine of early farmers. Nature Commun. 2018, 9, 4064–4073. [Google Scholar] [CrossRef] [PubMed]
- Tanasi, D.; Cucina, A.; Cunsolo, V.; Saletti, R.; Di Francesco, A.; Greco, E.; Foti, S. Paleoproteomic profiling of organic residues on prehistoric pottery from Malta. Amino Acids 2021, 53, 295–312. [Google Scholar] [CrossRef] [PubMed]
- William, S. Itinerarium Peregrinorum et Gesta Regis Ricardi; Eyre & Spottiswoode: London, UK, 1864. [Google Scholar]
- Bulfinch, T. The Age of Chivalry; Reprinted by Dover Publications, 2012; Crosby, Nichols: Boston, MA, USA, 1859. [Google Scholar]
- Diderot, D.; D’Alembert, J.B. L’Encyclopédie, 1st ed.; Le Breton: Paris, France, 1751; Volume 4, pp. 502–505. [Google Scholar]
- Arouet, F.M. Essai sur les Mœurs et ! Esprit des Nations; Cramer: Paris, France, 1756. [Google Scholar]
- Atkins, R.C. Dr. Atkins New Diet Revolution: Revised and Improved; HarperCollins: New York, NY, USA, 2001. [Google Scholar]
- Charlier, P.; Poupon, J.; Jeannel, G.-F.; Favier, D.; Popescu, S.-M.; Weil, R.J.; Moulherat, C.; Huynh-Charlier, I.; Dorion-Peyronnet, C.; Lazar, A.-M.; et al. The embalmed heart of Richard the Lionheart (1199 A.D.): A biological and anthropological analysis. Sci. Rep. 2013, 3, 1296. [Google Scholar] [CrossRef]
- Ariosto, L. Orlando Furioso; Francesco Libri: Ferrara, Italy, 1516. [Google Scholar]
- Tasso, T. Gerusalemme Liberata; Cavalcalupo: Venezia, Italy, 1581. [Google Scholar]


| Accession Numbers | Protein Name | Mascot Score | Mr | Sequences | No. Peptides |
|---|---|---|---|---|---|
| P02670 | Kappa-CN | 225 | 21,441 | (1) MMKSFFLVVTILALTLPFLGAQEQNQEQPICCEK (2) MAIPPKKDQDK (3) LSRYPSYG LNYYQQRPVA LINNQFLPYP YYAKPVAVR | 3 |
| P02756 | Beta-lactoglobulin | 341 | 19,976 | (1) LEKFDKALKA LPMHIR (2) LLFCMENSAEPEQSLACQCLVRTPEVDK (3) LACGIQAIIVTQTMKGLDIQK | 3 |
| A0A5K6WAG1 | Kappa-casein | 764 | 21,455 | (1) VALINNQFLPYPYYAKPVAVR (2) VPAKSCQDQPTTLAR (3) QEQNQEQPICCEKDERFFDDKIAK (4) QPTTLARHPHPHLSFMAIP PK | 4 |
| P33048 | Beta-casein | 290 | 24,865 | (1) LGPVR (2) LQPEIMGVPKVKETMVPK HKEMPFPKYPVEPFTESQSL TLTDVEK (3) MFPPQSVLSLSQPKVLPVPQK (4) QPEIMGVPKVK | 4 |
| P85295 | Albumin | 192 | 10,055 | (1) DTHKS (2) DLGRHPEYAVS (3) VSVLLR (4) VRXXXKAPQVSTPTLVEISR (5) QTALVELLK | 5 |
| W5VVI0 | Cathelicidin-1 | 365 | 9799 | (1) DITCNNHQS (2) FKENGLLKRCEGT VTLDQVR (3) QCDFKENGLLK | 3 |
| P18626 | Alpha-S1-casein | 269 | 24,290 | (1) MEDAKQMKAGSS SSSEEIVPNSAEQK (2) LLRLKKYNVPQLEIVPKS AEEQLHSMK (3) MKAGSSSSSEEIVPNS AEQKYIQK | 3 |
| P33049 | Alpha-S2-casein | 199 | 26,389 | (1) MKFFIFTCLLAVALAK (2) MAIHPRKEK (3) LNPWDQVK (4) LTEEEKNRLNFLKKISQYYQK | 4 |
| Accession Numbers | Protein Name | Mascot Score | Mr | Sequences | No. Peptides |
|---|---|---|---|---|---|
| P16293 | Coagulation factor IX | 342 | 45,516 | (1) MDYENSTEVEPILDSL TESNQSSDDFIRIVGGENAK (2) QYLKVPLVDRATCLR STK (3) FSNMDYENSTEVEPILDSLTESNQSSDDFIR | 3 |
| P24964 | Cytochrome b | 201 | 42,782 | (1) LHANGASMFFICLFIHVGR (2) LILMPMLHTSK (3) MFFICLFIHVGR | 3 |
| O79876 | cytochrome c oxidase subunit 1 | 408 | 56,958 | (1) LYLLFGAWAGMVGTALSLLIR (2) QIYNVIVTAHAFVMIFFMV MPIMIGGFGNWLVPLMIGAP DMAFPR (3) LVPLMIGAPDMAFPR | 3 |
| P02189 | Myoglobin | 230 | 17,085 | (1) LSDGEWQLVLNVWGKVEA DVAGHGQEVLIR (2) LKKHGNTVLTALGGILKKK (3) LTALGGILKKKGHHEAELTPL AQSHATK | 3 |
| Q29548 | Beta-hexosaminidase subunit beta | 542 | 61,050 | (1) LQVHVESECDTFPSISSNESYVLHVK (2) QFMQEK (3) QSINFGVLSSK | 3 |
| Q29016 | Acrosin-binding protein | 213 | 60,827 | (1) MRQLAAGSLL SLLKVLLLPL APAPAQDANSASTPGSPLSPTEYER (2) LNNNVEELLQSSLSLGGQE QGQEHK (3) QSEFVSSNPFSFTPRVRE VESTPMMMENIQELIR (4) VASWLQTEFLSFQDGDFPT KICDTEYVQYPNYCAFKSQQ CMMR | 4 |
| A0A5G2QQE9 | Collagen type I alpha 1 chain | 277 | 140,883 | (1) MGPRGPPGPPGKNGDDG EAGKPGR (2) QAGVMGFPGPKGAAGEPGK (3) LPGAKGLTGSPGSPGPDGK | 3 |
| Q29218 | Keratin, type I cytoskeletal 20 | 678 | 12,528 | (1) QIQDLRNQIRDAQLQNARCV LQIDNAKLAAEDFRLK (2) MTMKNLNDRLASYLEK (3) QSNSKFELQIK | 3 |
| Accession Numbers | Protein Name | Mascot Score | Mr | Sequences | No. Peptides |
|---|---|---|---|---|---|
| Q6PWU2 | (-)-alpha-terpineol synthase | 590 | 68,883 | (1) MALSMLSSIP NLITHTRLPIIIK (2) MEQARDFAHRHLGKGLEQN IDQNLAIEVK (3) MVQATHQEDLRHMSSWWSSTRLGEK | 3 |
| A5C7X2 | Cytochrome P450 CYP76Y2 | 482 | 55,742 | (1) MPLILCFFLLQFLRPSSHATKLPPGPTGLPILGSLLEIGK (2) MFRRFDLLGVK (3) MQEQCGATLKK (4) MEGAGKFNISDYFPMFRRFDLLGVKRDTFSCYK | 4 |
| Q8S4W7 | DELLA protein GAI1 | 673 | 64,866 | (1) MKREYHHPHHPTCSTSPTGKGK (2) MWDADPQQDAGMDELLAVLGYNVK (3) VAVNSVFELHSLLARPGGER (4) VGWKLAQLAETIHVEFEYR | 4 |
| A8VPW4 | MYBC2-L1 protein | 230 | 25,255 | (1) VKNYWNSHLRRKLINMGIDPNNHRLSHNFPR (2) VKSVGDNDQTSDAGS CLDDNR (3) QDTNKGAWSKQEDQKLIDYIRK | 3 |
| Accession Numbers | Protein Name | Mascot Score | Mr | Sequences | No. Peptides |
|---|---|---|---|---|---|
| E3SU11 | Thaumatin-like protein | 458 | 24,727 | (1) VSLWAITFFAYTHAATFDIVNQCTYTVWAAASPGGGR (2) VFNTNEYCCTNGPGSCGPTPLSR (3) FALNQPNNLDFVDISNVDGFNIPLEFSPTTNVCR | 3 |
| A0A1D6ZNQ5 | Acyl-[acyl-carrier-protein] desaturase | 511 | 44,622 | (1) VAQRLGVYTAKDYADILEFLVGRWDVEKLTGLSGEGR (2) FQERATFISHGNTAR (3) VEKLFEIDPDGTVLALADMMR (4) VYTAKDYADILEFLVGRWDVEKLTGLSGEGR | 4 |
| E3NYV3 | Lipoxygenase | 297 | 101,717 | (1) VDKSYLPSNTPSGLKI YREKELQILRGDGTGERKTFER (2) VELPKQKSTGFLANIIPR (3) VEKNVLLFETPQLYER (4) VSHWLR (5) VIATNRQLSAMHPVYKLHPHLR | 5 |
| Accession Numbers | Protein Name | Mascot Score | Mr | Sequences | No. Peptides |
|---|---|---|---|---|---|
| P01501 | Melittin | 342 | 7585 | (1) VLTTGLPALISWIKR (2) MVVYISYIYAAPEPEPAPEPEAEADAEADPEAGIGAVLK (3) LTTGLPALISWIKR | 3 |
| O18330 | Major royal jelly protein 1 | 211 | 48,886 | (1) VSKSGVLFFGLVGDSLGCWNEHRTLER (2) VCQGTTGNILR (3) LPILHEWKFFDYDFGSDER (4) LSPMTNNLYYSPVASTSLYYVNTEQFR | 4 |
| Accession Numbers | Protein Name | Mascot Score | Mr | Sequences | No. Peptides |
|---|---|---|---|---|---|
| P17314 | Alpha-amylase/trypsin inhibitor CM3 | 123 | 18,221 | (1) MACKSSCSLLLLAAVLLSVLAAASASGSCVPGVAFR (2) LPEWMTSASIYSPGKPYLAKLYCCQELAEISQQCR (3) LYCCQELAEISQQCRCEALR | 3 |
| Q10464 | Puroindoline-B | 221 | 16,792 | (1) LFLLALLALVASTTFAQYSEVGGWYNEVGGGGGSQQCPQERPK (2) QERPK (3) VIQGRLG GFLGIWRGEVFKQLQR (4) LFLLALLALVASTTFAQYSEVGGWYNE VGGGGGSQQCPQER | 4 |
| P82900 | Non-specific lipid-transfer protein 2G | 291 | 9832 | (1) QGCFCQYAKD (2) LRAQQGCFCQYAKPTYGQYIR (3) QYAKDPTYGQYIR | 3 |
| P01544 | Alpha-1-purothionin | 652 | 13,526 | (1) LALESNSDEPDTIEYCN (2) LGCRLCRARGAQKLCAGVCRCK (3) QLQVEGKSCCRSTLGRNCYNLCR | 3 |
| P10388 | Glutenin high molecular mass subunit | 299 | 90,293 | (1) QVSYYPGQASPQR (2) QLRDISPECHPVVVSPVAGQYEQQIVPK (3) LQQPAQGQQGQQLAQGQQGQQPAQVQQGQR | 3 |
| A0A3B6C437 | Beta-amylase | 331 | 67,919 | (1) LEEDFRAAATDAGHPEWELPDDAGEYNDAPDDTR (2) LAAKVSGIHWWYR (3) LDGRDGYRPIAR (4) LARHDGAVLNFTCAEMR | 4 |
| P01543 | Purothionin A-1 | 441 | 14,625 | (1) LCANVCR (2) LSCPKDPKLVLESNSDEPDTMEYCNLGCR (3) LVLEQVQVEGKSCCK (4) LVLESNSD EPDTMEYCNLGCR | 4 |
| P17314 | Alpha-amylase/trypsin inhibitor | 226 | 18,221 | (1) LPEWMTSASIYSPGKPYLAK (2) LIDLPGCPREMQWDFVR (3) LPVPSQPVDPRSGNVGESGLIDLPGCPREMQWDFVR | 3 |
| P83207 | Chymotrypsin inhibitor | 270 | 12,944 | (1) QPPLAPRCPTEVKR (2) LQGCHYYVTSQTCGFVPLLPIEVMK (3) QGCHYYVTSQTCGFVPLLPIEVM KDRCCR | 3 |
| P93692 | Serpin-Z2B | 378 | 42,981 | (1) QYIS SSDGLKVLKLPYKQGGDK (2) LIKDILPAGSIDNTTR (3) QLAATLGEGEVEGLHALAEQVVQFVLADASNIGGPR | 3 |
| P01084 | Alpha-amylase inhibitor | 497 | 13,185 | (1) VPALPGCRPLKLQCNG SQVPEAVLR (2) LYSMLDSMYK (3) QAFQVPALPGCRPLLKLQCNGSQVPEAVLR | 3 |
| Accession Numbers | Protein Name | Mascot Score | Mr | Sequences | No. Peptides |
|---|---|---|---|---|---|
| A0A446NJU5 | Ribokinase | 234 | 36,611 | (1) LEARLRLPLAGPTPATAFLSGSNPK (2) LDLIRQAGVLLLQR (3) QNSIIIVGGANMEGWAPEVDQEDLDLIR | 3 |
| A0A446KRB8 | UMP-CMP kinase | 211 | 26,949 | (1) LIDGFPRNE ENRAAFENVTK (2) LIKTTIFVFPTIRIVLQGR (3) LMFGLCILIK | 3 |
| A0A446WFI0 | NAD(P)H-hydrate epimerase | 231 | 53,127 | (1) LIICGPGNNGGDGLVAAR (2) LTAPKLCAKKFTGPHHFLGGRFVPPPISSK (3) LTTVNKAGKPSSRMVLLKGVDK | 3 |
| A0A446K973 | Flap endonuclease 1 | 402 | 42,744 | (1) LQGMFSRTIRLLEAGI KPVYVFDGKPPEMKK (2) LENINKDKYQIPEDWPYQEARRMFKEPDVTLDIPELK (3) QHGSIEGILENINKDKYQ IPEDWPYQEAR | 3 |
| A0A446X1P5 | ATP-dependent 6-phosphofructokinase | 478 | 50,586 | (1) LKVAVAGIPKTIDNDIPVIDK (2) LAHSVVHGAMAGYTGFTVGQVNGR (3) LSSTNQPSFLSKQDVDDAVEDER | 3 |
| Accession Numbers | Protein Name | Mascot Score | Mr | Sequences | No. Peptides |
|---|---|---|---|---|---|
| P82993 | Beta-amylase | 308 | 59,639 | (1) VNIPIPQWVRDVGTRDPDIFYTDGHGTR (2) MEVNVK (3) MLPLDAVSVNNRFEKGDELRAQLRK (4) QAMSAPEELVQQVLSAGWR | 4 |
| P45850 | Oxalate oxidase 1 | 211 | 21,203 | (1) VSFNSQNPGIVFVPLTLFGSDPPIPTPVLTKALR (2) VAEWPGTNTLGVSMNRVDFAPGGTNPPHIHPR (3) QNPGIVFVPLTLFGSDPPIPTPVLTKALR | 3 |
| P27337 | Peroxidase 1 | 201 | 32,976 | (1) VSCADILTVAARDSVVALGGPSWTVPLGRR (2) VQGCDASVLLSGMEQNAIPNAGSLR (3) QGQIRLSCSR | 3 |
| P55307 | Catalase isozyme 1 | 309 | 56,586 | (1) VRFSTVVHERGSPETLRDPRGFAVKFYTR (2) VHAFKPSPKTNMQENWRVVDFFSHHPESLHMFTFLF DDVGIPLNYR (3) VNAPKCAHHNNHHDGLMNFIHR (4) VSQLTCADFLR | 4 |
| F2CTM5 | Ribokinase | 266 | 37,073 | (1) VEVNRLPLVGETVAASAGHSLAGGKGANQAACGGR (2) VALVEGKPKKECMRFAAAAASLCVRVKGAIPSMPDR (3) VPVLDAGGMDAPVPRELLELVDIFSPNETELAR | 3 |
| Accession Numbers | Protein Name | Mascot Score | Mr | Sequences | No. Peptides |
|---|---|---|---|---|---|
| P21814 | Uterine milk protein | 265 | 49,223 | (1) QPNLTQKEDFF LNDKTKVQVDMMRK (2) VHLGRELVKQKQLR (3) QIRKLQKMDIQMIDFSDTEKAKKAISHHVAEKTHTKIR | 3 |
| R9WQI9 | Prolactin | 390 | 25,778 | (1) VLMSLILGLLRSWNDPLYHLVTEVR (2) FGQVIPGAKETEPYPVW SGLPSLQTKDEDAR (3) VMVSHYIHNLSSEMFNEFDKR | 3 |
| C7DLN1 | Fatty acid synthase | 241 | 9,232 | (1) VEDAFRYMAQGKHIGKVVIQVREEER (2) VTFHGILLDALFEENSTMWQEVSSLLKAGIREGVVQPLKR (3) QVEDAFRYMAQGKHIGK VVIQVR | 3 |
| P04654 | Alpha-s2-casein | 322 | 26,332 | (1) VFTKKTKLTEEEKNR (2) MKFFIFTCLLAVALAKHKMEHVSSSEEPINISQEIYKQEK (3) QKFPQYLQYLYQGPIVLNPWDQVKR | 3 |
| Accession Numbers | Protein Name | Mascot Score | Mr | Sequences | No. Peptides |
|---|---|---|---|---|---|
| Q29524 | Lipoprotein lipase | 207 | 53,588 | (1) VIAERGLGDVDQLVKCSHERSVHLFIDSLLNEENPSKAYR (2) VRAKRSSKMYLKTR (3) VIFVKCHDKSLNR | 3 |
| P67930 | Somatotropin | 386 | 24,631 | (1) VTPRAGQILKQTYDKFDTNMR (2) VFTNSLVFGTSDR (3) QILKQTYDKFDTNMR | 3 |
| Q8SQG7 | Hyaluronidase-2 | 265 | 54,229 | (1) VHLEMLKGHVEHYIRTQEPAGLAVIDWEDWR (2) VYLEETLASSTHGRNFVSFRVQEALR (3) VADVHHANHALPVYVFTR (4) MDLISTIGES AALGAAGVIL WGDAGFTTSNETCRRLK (5) VASHHPDWPPERIVKEAQYEFEFAAR | 5 |
| P56591 | Cytochrome P450 1A1 | 497 | 59,230 | (1) VANVICAICFGRR (2) VFVNQWQINHDQKLWEDPSEFRPER (3) QLPYLEAFILETFRHSSFVPFTIPHSTTR | 3 |
| P43477 | Dipeptidase 1 | 567 | 45,096 | (1) VAVCTADQFRDNAVR (2) VSKYPDLVAELLRRQWTEEEVRGALAENLLR (3) QAPGEEPIPLGQLEASCR | 3 |
| Q6YNX6 | Calmodulin | 345 | 16,838 | (1) VDADGNGTIDFPEFLTMMARKMKDT DSEEEIREAF (2) VMTNLGEKLTDEEVDEMIR (3) VDEMIR | 3 |
| P02190 | Myoglobin | 522 | (1) VLNAWGKVEA DVAGHGQEVL IR (2) VKYLEFISDAIIHVLHAKHPSDFGADAQGAMSKALELFR (3) VFDKDGNGISAAELR | 3 | |
| W5P481 | Collagen | 455 | 140,644 | (1) VWKPVPCQICVCDNGNVLC DDVICDELKD CPNAKVPTPPR (2) QDGRPGPPGPPGARGQAGVMGFPGPK (3) V PGDLGAPGPSGARGER | 3 |
| P02446 | Keratins | 387 | (1) VRTGPATTICSSDKFCR (2) VYPDTYVPTCFLLNSSHPTPGLSGINLTTFIQPGCENVCEPR (3) LSGINLTTFIQPGCENVCEPR | 3 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zilberstein, G.; Zilberstein, S.; Righetti, P.G. Richard the Lionheart and the Ferocious Saladin Face to Face in Arsuf: A Proteomic Study. Heritage 2021, 4, 3382-3401. https://doi.org/10.3390/heritage4040188
Zilberstein G, Zilberstein S, Righetti PG. Richard the Lionheart and the Ferocious Saladin Face to Face in Arsuf: A Proteomic Study. Heritage. 2021; 4(4):3382-3401. https://doi.org/10.3390/heritage4040188
Chicago/Turabian StyleZilberstein, Gleb, Svetlana Zilberstein, and Pier Giorgio Righetti. 2021. "Richard the Lionheart and the Ferocious Saladin Face to Face in Arsuf: A Proteomic Study" Heritage 4, no. 4: 3382-3401. https://doi.org/10.3390/heritage4040188
APA StyleZilberstein, G., Zilberstein, S., & Righetti, P. G. (2021). Richard the Lionheart and the Ferocious Saladin Face to Face in Arsuf: A Proteomic Study. Heritage, 4(4), 3382-3401. https://doi.org/10.3390/heritage4040188






