1. Background
Resampling and subsampling methods were widely utilized in many statistical and machine learning applications. In high-dimensional regression learning, for instance, bootstrap and subsampling play important roles in model selection uncertainty quantification and other model selection diagnostics (see, e.g., [
1] for references). Cross-validation (CV) (as a subsampling tool) and closely related methods were proposed to assess quality of variable selection performance in terms of F- and G-measures ([
2]) and to determine variable importances ([
3]).
In this paper, we focus on the examination of cross-validation as used for model selection. A core issue is how to properly quantify variabilities in subsamplings and associated evaluations, which is a really challenging issue that is yet to be solved, to the best of our knowledge. In application of variable and model selection, a popular practice is to use the “one standard error rule”. However, its validity hinges crucially on the goodness of the standard error formula used in the approach. Our aim in this work is to study the standard error estimation issue and its consequences on variable selection and regression estimation.
In this section, we provide a background on tuning parameter selection for high-dimensional regression.
1.1. Regularization Methods
Regularization methods are now widely used to tackle the problem of curse of dimensionality in high-dimensional regression analysis. By imposing a penalty on the complexity of the solution, it can solve an ill-posed problem or prevent overfitting. Examples include Lasso ([
4]) and Ridge regression ([
5,
6]), which add an
and
penalty on the coefficient estimates, respectively, to the usual least-squares fit objective function.
Specifically, given a response vector
, a matrix
of predictor variables and a vector of intercept
, the Lasso estimate is defined as
and the ridge estimate is defined as
The tuning parameter controls the strength of the penalty. As is well known, the nature of the penalty causes some coefficients to be shrunken to zero exactly while the penalty can shrink all of the coefficients towards zero, but it typically does not set any of them exactly to zero. Thus, an advantage of Lasso is that it makes the results simpler and more interpretable. In this work, ridge regression will be considered when regression estimation (i.e., the focus is on the accurate estimation of the regression function) or prediction is the goal.
1.2. Tuning Parameter Selection
The regularization parameter, or the tuning parameter, determines the extent of penalization. A traditional way to select a model is to use an information criterion such as Akaike information criterion (AIC) ([
7,
8]), Bayesian information criterion (BIC) and its extension ([
9,
10,
11]). A more commonly used way for tuning parameter selection is the cross-validation ([
12,
13,
14]). For recent work on cross-validation for consistent model selection for high-dimensional regression, see [
15].
The idea of
K-fold cross validation is to split the data into
K roughly equal-sized parts:
. For the
k-th part where
, we consider training on
and evaluation on
. For each value of the tuning parameter
, a set of candidate values, we compute the estimate
on the training set, and record the total error on the validation set:
For each tuning parameter value
, we compute the average error over all folds:
We then choose the value of tuning parameter that minimizes the above average error,
1.3. Goal for the One Standard Error Rule
The “one standard error rule” (1se rule) for model selection was first proposed for selecting the right sized tree ([
16]). It is also suggested to pick the most parsimonious model within one standard error of the minimum cross validation error ([
17]). Such a rule acknowledges the fact that the bias-variance trade-off curve is estimated with error, and hence takes a conservative approach: a smaller model is preferred when the prediction errors are more or less indistinguishable. To estimate the standard deviation of
at each
, one computes the sample standard deviation of validation errors in
:
Then, we estimate the standard deviation of
, which is declared the standard error of
, by
The organization of the rest of the paper is as follows.
Section 2 examines validity of the 1se rule from a theoretical perspective.
Section 3 describes the objectives and approaches of our numerical investigations.
Section 4 presents the experimental results of the 1se rule in three different aspects.
Section 5 applies the 1se rule on two real data sets and examine its performances via data guided simulations.
Section 6 concludes the paper.
Appendix A gives additional numerical results and
Appendix B provides the proof of the main theorem in the paper.
2. Theoretical Result on the One-Standard-Error Rule
2.1. When Is the Standard Error Formula Valid?
The 1se rule makes intuitive sense, but its validity is not clear at all. The issue is that the
errors completed at different folds are not independent, which invalidates the standard error formula for the sample mean of i.i.d. observations. Note that the validity of the standard error is equivalent to the validity of the sample standard deviation expressed in Equation (
6). Thus, it suffices to study if and to what extent the usual sample variance properly estimates the targeted theoretical variance.
In this section, we investigate the legitimacy of the standard error formula for a general learning procedure. We focus on the regression case, but similar results hold for classification as well.
Let be a learning procedure that produces the regression estimator based on the data . Throughout the section, we assume K is given and for simplicity, n is a multiple of K. Define , where denote the observations in that do not belong to and .
The
of the
errors of the learning procedure
is defined as
To study the
formula, given that
K is fixed, it is basically equivalent to studying the sample variance formula, i.e.,
The validity of these formulas (in an asymptotic sense) hinges on that the correlation between and for approaches zero as .
Definition 1. The sample variance is said to be asymptotically relatively unbiased (ARU) ifas . Clearly, if the property above holds, the formula is properly justified in terms of the relative estimation bias being negligible. Then, the 1se rule is sensible.
For
, define the
-norm
where
denotes the probability distribution of
X from which
are drawn. Let
denote the estimator
.
Theorem 1. If , then is asymptotically relatively unbiased.
Remark 1. The condition holds if the regression procedure is based on a parametric model under regularity conditions. For instance, if we consider a linear model with terms, then under sensible conditions (e.g., [15], p. 111), and consequently as long as , the variance estimator is asymptotically relatively unbiased. Remark 2. The above result suggests that when relatively simple models are considered, the 1se rule may be reasonably applicable. In the same spirit, when sparsity oriented variable selection methods are considered as candidate learning methods, if the most relevant selected models are quite sparse, the 1se rule may be proper.
2.2. An Illustrative Example
Our theorem shows that the standard error formula is asymptotically valid when the correlation between the errors at different folds is relatively small, which is related to the rate of convergence of the regression procedure. In this section, we illustrate how the dependence affects the validity of the standard error formula in a concrete example. For ease in highlighting the dependence among the errors at different folds, we consider a really simple setting, where the true regression function is a constant. The simplicity allows us to calculate the variance and covariance of the errors explicitly and then see the essence. The insight gained, however, is more broadly applicable.
The true data generating model is , where is unknown. Given data , for an integer r as a factor of n, the regression procedure considers only the first, r-th, -th, ..., -th observations and take the corresponding sample mean of Y to estimate .
Suppose
is an integer. Then, when applying the
K-fold
, the training sample size is
but only
observations are actually used to estimate
f. Let
,
denote the
errors on the
i-th and
j-th fold,
. Then, it can be shown that
and
for
, where ≍ denotes the same order.
Therefore, the standard error formula is ARU if and only if .
In this illustrating example, when
l is small (recall that only
l observations are used in each fold for training), the regression estimator obtained from excluding one fold respectively are highly dependent, which makes the sample variance formula problematic. In contrast, when
l is large, e.g.,
(i.e., all the observations are used), the dependence of the
errors at different folds becomes negligible compared to that of the randomness in the response. In the extreme case of
, we have
for
.
Note also that the choice of l determines the convergence rate of the regression estimator, i.e., is of order . Obviously, with large l, the estimator converges fast, which dilutes the dependence of the errors and helps the sample variance formula to be more reliable.
3. Numerical Research Objectives and Setup
One standard error rule is intended to improve the usual cross-validation method stated in
Section 1.2, where
is chosen to minimize the CV error. However, to date, no empirical or theoretical studies were conducted to justify the application of the one standard error rule in favor of parsimony, to the best of our knowledge. There are some interesting open questions that deserve investigations.
- (1)
Does the 1se itself provide a good estimation of the standard deviation of the cross validation error as intended?
- (2)
Does the model selected by the 1se rule (model with ) typically outperform the model selected by minimizing the CV error (model with ) in variable selection?
- (3)
What if estimating the regression function or prediction is the goal?
In this paper, we study the estimation accuracy of the one standard error rule and its application in regression estimation and variable selection under various regression scenarios, including both simulations and real data examples.
3.1. Simulation Settings
Our numerical investigations are done in a regression framework where our simulated response is generated by linear models. Both dependent and independent predictors are considered. The data generating process is shown as follows.
Denote the sample size as
n and the total number of predictors as
p. Firstly, the predictors are generated by normal distributions. In the independent case, the vector of the explanatory variables
has i.i.d.
components and in the dependent case, the vector follows
distribution, where
takes either
or
as follows
or
where
. Then, the response vector
is generated by
where
is the chosen coefficient vector (to be specified later) and the random error vector
has mean 0 and covariance matrix
for some
.
3.2. Regression Estimation
For estimation of the regression function, we intend to minimize the estimation risk. Given the data, we consider a regression method and its loss. For Lasso, we define
where
is chosen by the regular CV, i.e.,
. We let
be the parameter estimate obtained from the simplest model whose CV error is within the one standard error of the minimum CV error, i.e.,
. Given an estimate
, the loss for estimating the regression function is
where
is the true parameter vector and the expectation is taken with respect to the randomness of
that follows the same distribution as in the data generation. The loss can be approximated by
where
,
are i.i.d. with the same distribution as
in the data generation and
J is large (In the following numerical exercise, we set
). In the case of using real data to compare
and
, clearly, it is not possible to compute the estimation loss as described above. We will use cross-validation and data guided simulations (DGS) instead for comparison of
and
. See
Section 4 for details.
3.3. Variable Selection
Parameter value specification can substantially affect the accuracy of the variable selection by Lasso. In our simulation study, we investigate the performances of the model with and that with by comparing their probabilities of accurate variable selection. When considering real data, since we do not know the true data generation process, we employ data guided simulations to provide more insight. Let denote the set of variables in the true model and be the set of selected variables by a method. In the DGS over real data sets, we define the symmetric difference to be the variables that are either in but not in or in but not in . We use the size of the symmetric difference to measure the performance of the variable selection method. Performance of the model with and the model with in terms of variable selection will be compared this way for the data examples, where some plausible models based on the data are used to generate data.
Organization of the numerical results is as follows.
Section 4 presents the experimental results of the 1se rule in its estimation accuracy as well as its performances in regression estimation and variable selection, respectively.
Section 5 applies the 1se rule over two real data sets and investigates its performance relative to the regular CV via DGS. Some additional figures and tables are in
Appendix A. All of R codes used in this paper are available upon request.
4. Simulation Results
4.1. Is 1se on Target of Estimating the Standard Deviation?
This subsection presents a simulation study of the estimation accuracy of the 1se with respect to different parameter settings in three sampling settings, by varying value of the correlation () in the design matrix, error variance , sample size n, and the number of cross validation fold K.
4.1.1. Data Generating Processes (DGP)
We consider the following three sampling settings through which we first simulate the dependent ( with ) or independent design matrix , and then generate the response with either decaying or constant coefficient vector . Recall that p is the total number of predictors and let q denote the number of nonzero coefficients.
DGP 1: Y is generated by 5 predictors (out of 30, , ).
DGP 2: Y is generated by 20 predictors (out of 30, , ).
DGP 3: Y is generated by 20 predictors (out of 1000, , ).
For the coefficient = , it equals for decaying coefficient case, and it equals for constant coefficient case. In these three sampling settings, are i.i.d. . Therefore, the first and third configuration yield sparse models and the third configuration also represents the case of high-dimensional data.
4.1.2. Procedure
We apply the Lasso and Ridge procedures on the simulated data sets with tuning parameter selection by the two cross validation versions. The simulation results obtained with various parameter choices are summarized in
Table A1,
Table A2,
Table A3 and
Table A4 in the
Appendix A. The simulation choices are sample size
, the error variance
and
K-fold cross validation:
.
- (1)
Do the j-th simulation (). Simulate a data set of sample size n given the parameter values.
- (2)
K-fold cross validation. Randomly split the data set into K equal-sized parts: and record the cross validation error for each part where .
- (3)
Calculate the total cross validation error,
, by taking the mean of these
K numbers. Find
that minimizes the total CV error. Now, we take
throughout the remaining steps. Calculate the standard error of cross validation error
by one standard error rule:
- (4)
Repeat step 1 to step 3 for N times (). Calculate the standard deviation of the cross validation error , and call it , and the mean standard error of cross validation error (as used for 1se rule), .
- (5)
Performance assessment. Calculate the ratio of (the claimed) standard error over (the simulated true) standard deviation:
If the 1se works well, the ratio should be close to 1. However, the results in the
Table A1,
Table A2,
Table A3 and
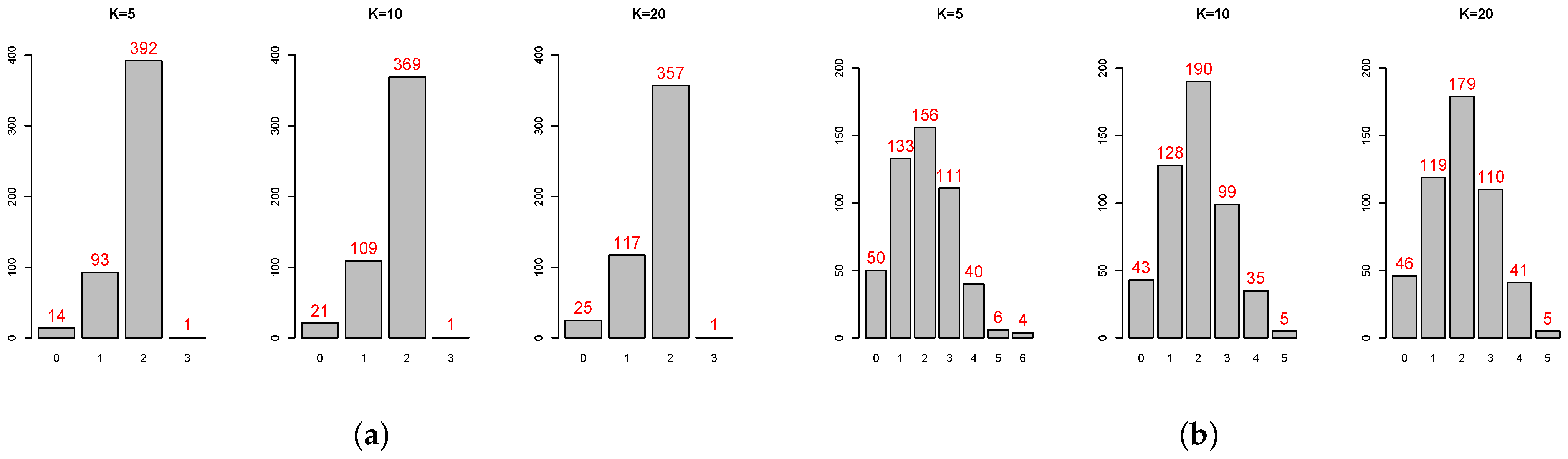
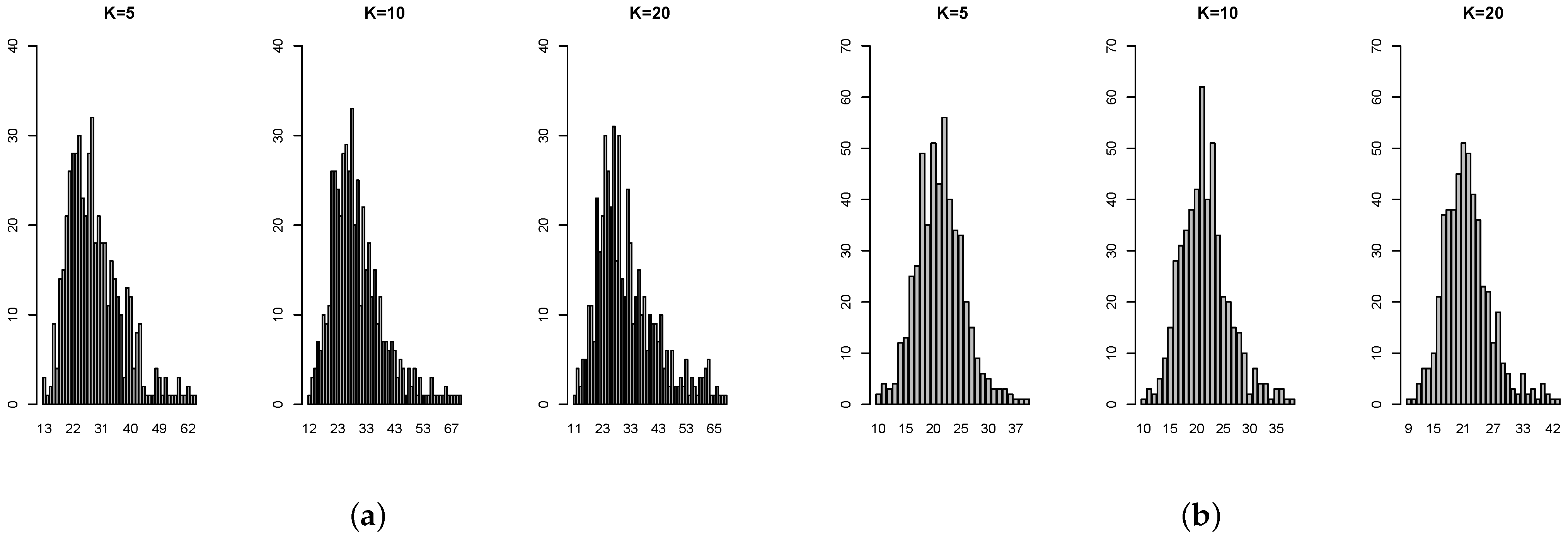
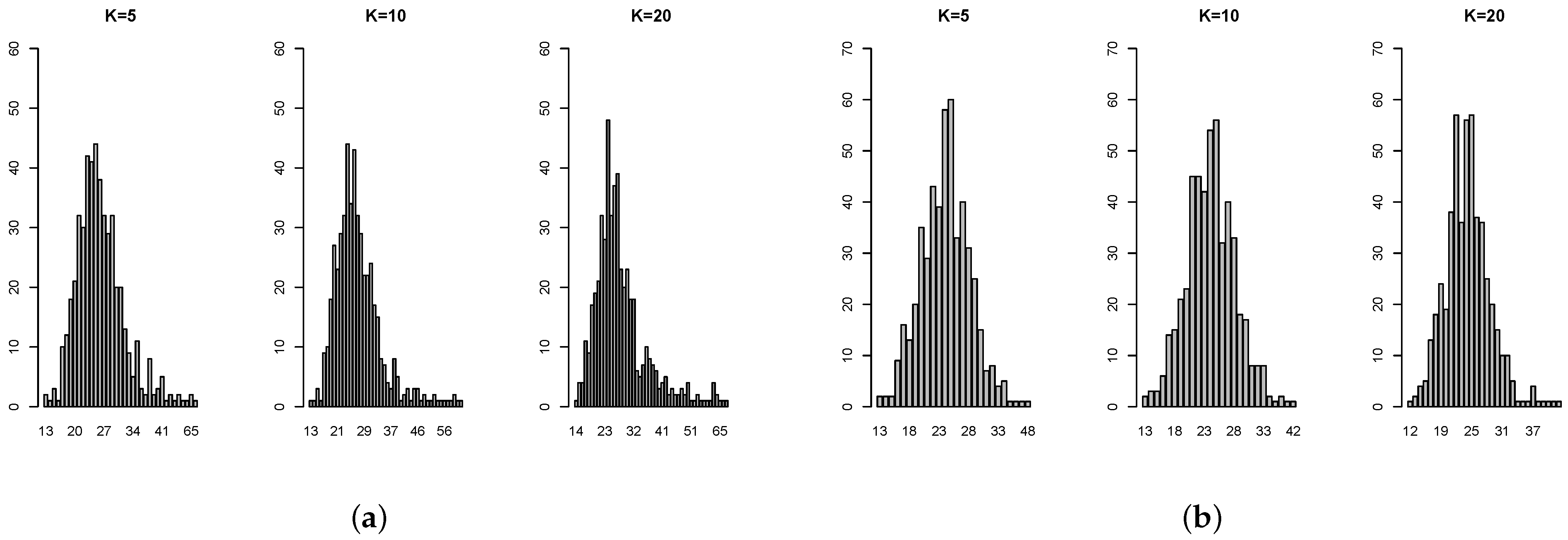
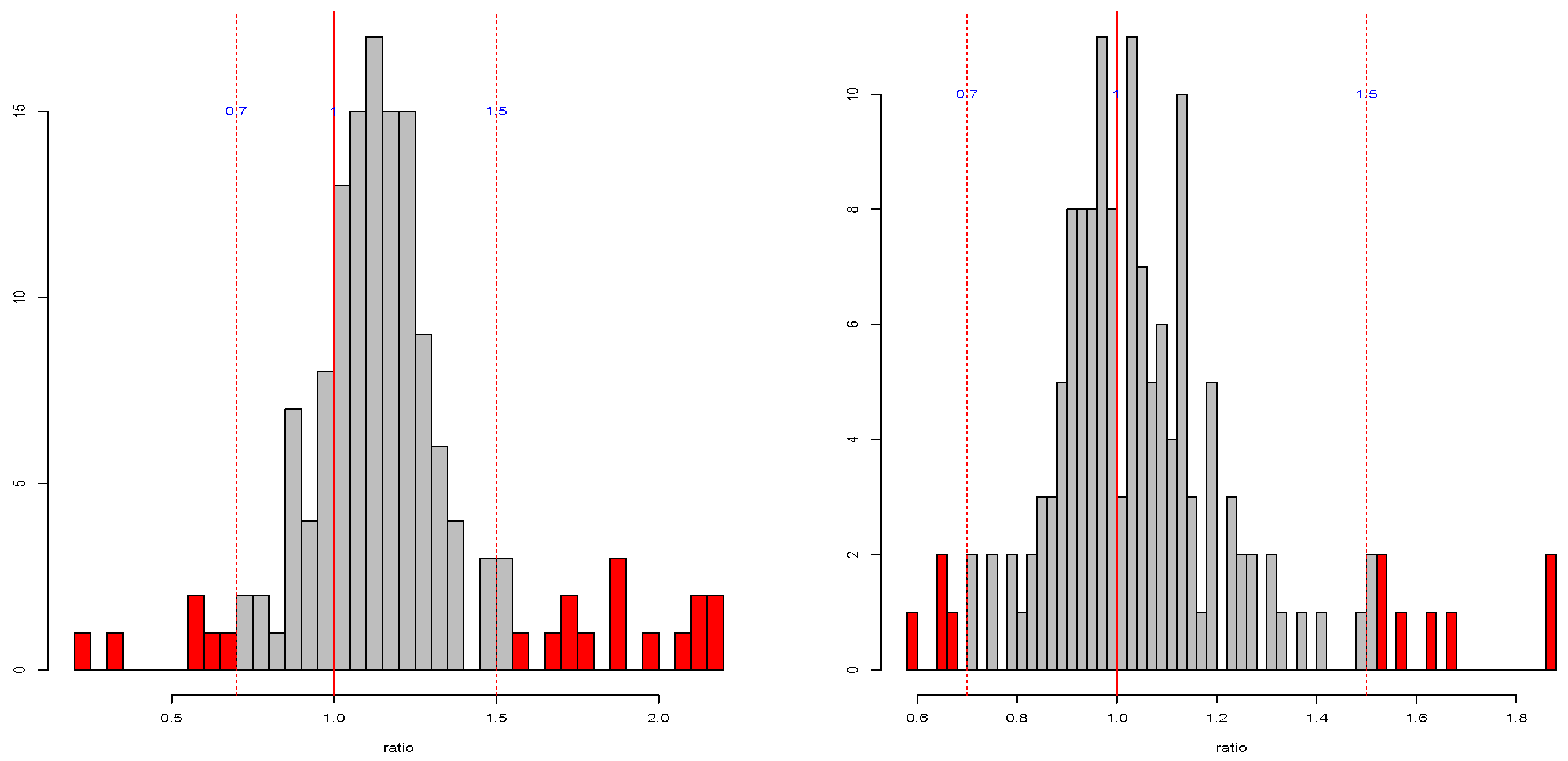
Table A4 show that, in some cases, the standard deviation is even more than 100% overestimated or more than 50% underestimated. The histograms of the ratios (
Figure A1 and
Figure A2) also show that the standard error estimates are not close to the targets. Not surprisingly, 2-fold is particularly untrustworthy. But for the common choice of 10-fold, even for independent predictors with constant coefficient, the standard error estimate by 1se can be 40% over or under the target. In addition, there is no obvious pattern on when the 1se is performing well or not. Overall, this simulation study provides evidence that the 1se fails to provide good estimation as intended (see [
18] for more discussions on the standard error issues).
4.2. Is 1se Rule Better for Regression Estimation?
Since the standard error estimation in 1se is not the end goal, the poor performance of 1se in the previous subsection does not necessary mean that the 1se rule is poor for regression estimation and variable selection. In this part, we study the performance of model in regression estimation and compare it with that of model . A large number of samples are drawn so that we know in which cases the 1se rule can consistently outperforms the regular CV in regression estimation.
By simulations, we compare and in term of accuracy in estimating the regression function. We consider several factors: nonzero coefficients , number of zero coefficients , sample size n, standard deviation of the noise and the dependency of the predictors.
4.2.1. Model Specification
We consider the data generation model:
, where
and
with
being the nonzero coefficients and
being the zero coefficients. The vector of explanatory variables is generated by the multivariate normal distribution,
where
.
4.2.2. Procedure
Simulate a fixed validation set of 500 observations of the predictors for estimation of the loss.
Each time randomly simulate a training set of n observations from the same data generating process.
Apply the 1se rule (10-fold cross validation) over the training set and record the selected model: model with and the model with . Calculate the estimation losses of these two models: , where is based on or respectively, and are independently generated from the same distribution used to generate X.
Repeat this process for M times () and calculate the fraction that model with has a smaller loss.
In our simulations, we make various comparisons: small or large nonzero coefficients; small or large error variance; small or large sample size; low- or high-dimensional data; independent or dependent design matrix. The parameter values used in our study are displayed in
Table 1 below. The results are reported in
Table A5 and
Table A6. Note that when
and
give identical selection over the
runs, the entry in the table is NaN.
The simulation results show that, in general, for the design matrix with either AR(1) correlation or constant correlation, the model with tends to provide better regression estimation (with smaller estimation errors), especially when the data is generated with relatively large coefficient. In addition, we consider two more settings below.
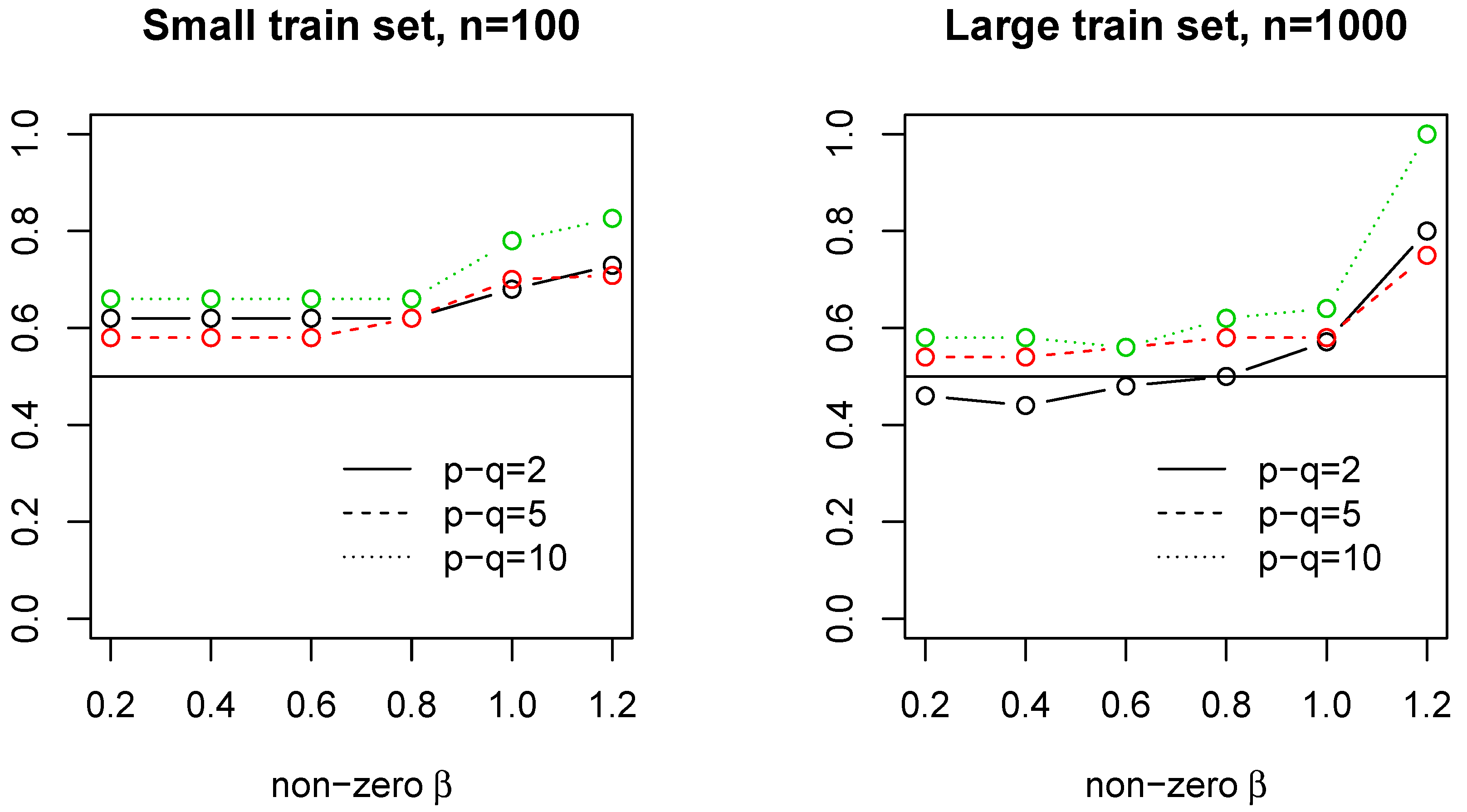
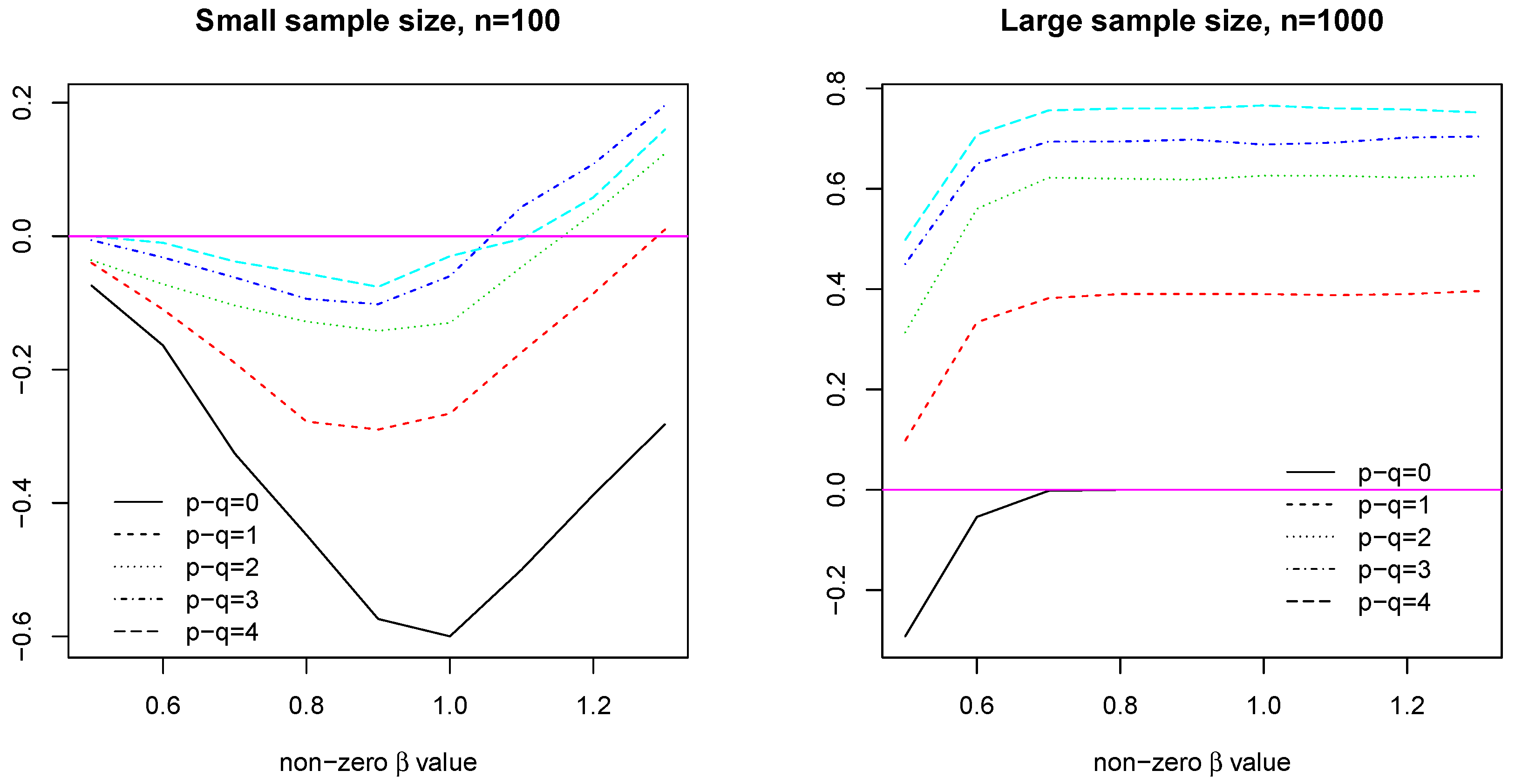
4.2.3. Case 1: Large
In this case, we consider high-dimensional data with AR(1) covariance design matrix,
, where
and
. We set
,
,
, constant nonzero coefficients with value
. We report the probability that model
estimates better. Clearly, in this case, the model
significantly outperforms the model
for a range of
values at both sample size (See
Figure 1).
4.2.4. Case 2: Small
In this case, we consider low-dimensional data with AR(1) covariance design matrix,
, where
and
. We set
,
,
, the common nonzero coefficient value
. We report the probability that model
estimates the regression function better. In this case, model
significantly outperforms model
as the common coefficient value increases over 1, getting close to or even over 0.8 (See
Figure 2).
4.3. Is 1se Rule Better for Variable Selection?
There are two main issues of the application of the 1se rule in variable selection. One is that the accuracy of variable selection by Lasso itself is very sensitive to the parameter values in the data generating process. In various situations (especially when the covariance are complicatedly correlated such as in gene expression type of data), Lasso may perform very poorly in accurate variable selection. The second issue is that and returned by the glmnet function are the same in many cases and then there is no difference in the result of variable selection. The cases where are eliminated in the following general study. To examine the real difference between and , we consider the difference of their probabilities of selecting the true model given that they have distinct select outcome.
4.3.1. Procedure
Randomly simulate a data set of sample size n given the parameter values.
Perform variable selection with and over the simulated data set, respectively. If the two returned models are the same, then discard the result and go back to step 1. Otherwise, check their variable selection results.
Repeat the above process for M times () and calculate the fraction of correct variable selection, denoted by and , respectively. We report the proportion difference . Positive means 1se rule is better.
We adopt the same framework and parameter settings (
Table 1) as that used for regression estimation. The results show that
tends to outperform
in most of the cases when the data are generated with AR(1) correlation design matrix (see
Table A7 and
Table A8). But with constant correlation design matrix, these two methods have no practically difference in variable selection.
Below we consider several special cases where model still needs to be considered, and although it can only do slightly better when the error variance is large, nonzero coefficients are small and no extra variables exist to interrupt the selection process.
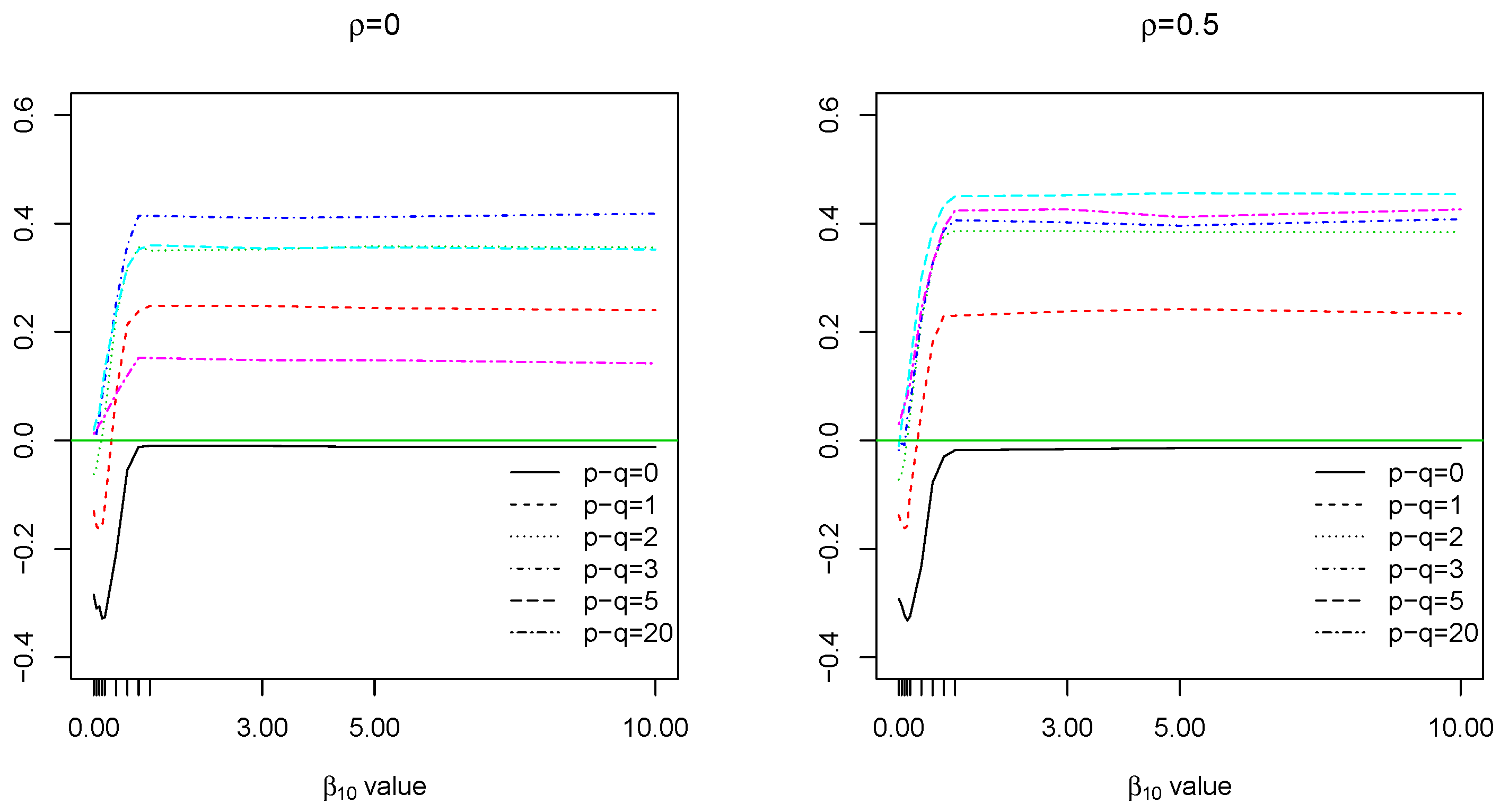
4.3.2. Case 1: Constant Coefficients
In this case, let
,
,
,
,
and the common nonzero
with
. The result is in
Figure 3. In the case of small sample size, say
, model
has higher probability of correct variable selection when the constant coefficient lies between (0.5,1). The largest difference appears when
, not surprisingly. As the common coefficient value becomes larger than 1, the probability increases from negative to positive, which indicates the reverse to be true: model
now works better. However, in the case of large sample size, say
, model
significantly outperforms model
unless when
.
4.3.3. Case 2: Decaying Coefficients
In this case, let
,
,
,
,
, and
,
with
.
Figure 4 indicates that, with decaying coefficients lie between (0,1) and large sample size
, model
has higher probability of correct variable selection when the number of zero-coefficient variables
is small. The largest difference appears when
, i.e., there is no extra variables provided to interrupt the variable selection process.
4.3.4. Case 3: Hybrid Case
Let
,
and
with
. In this data generating process, we set the first ten coefficients to be positive, i.e.,
, among which the first nine nonzero coefficients are constant 1, while the last one
increases from 0 to 10, i.e.,
. The number of zero coefficients
. From
Figure 5, when
is very small, i.e.,
and
,
performs better. For other situations,
is significantly better.
5. Data Examples
In this section, the performances of the 1se rule and the regular CV in regression estimation and variable selection are examined over real data sets: Boston Housing Price (an example of ) and Bardet–Biedl data (an example of ).
Boston Housing Price. The data were collected by [
19] for the purpose of discovering whether or not clean air influenced the value of houses in Boston. It consists of 506 observations and 14 non constant independent variables. Of these,
medv is the response variable while the other 13 variables are possible predictors.
Bardet–Biedl. The gene expression data are from the microarray experiments of mammalianeye tissue samples of [
20]. The data consists of 120 rats with 200 gene probes and the expression level of TRIM32 gene. It is interesting to know which gene probes are more able to explain the expression level of TRIM32 gene by regression analysis.
5.1. Regression Estimation
To test which method does better in regression estimation, we perform both cross validation and data guided simulation (DGS) over these two real data sets as follows.
5.1.1. Procedure for Cross Validation
We randomly select observations from the data sets as training set and the rest as validation set, for Boston Housing Price and for Bardet–Biedl data.
Apply K-fold cross validation , for Boston Housing Price and for Bardet–Biedl data over the training set and compute the mean square prediction error of model and model over the validation set.
Repeat the above process for 500 times and compute the proportion of better estimation for each method.
The results (
Table 2 and
Table 3) show that, for all the training size
considered, the proportion of model
doing better is about 52∼57% for Boston Housing Price; 56∼59% for Bardet–Biedl data. The consistent slight advantage of
agrees with our earlier simulation results in supporting
as the better method for regression estimation.
5.1.2. Procedure for DGS
Obtain model and model with K-fold cross validation, , over the data and also the coefficient estimates , , the standard deviation estimates , and the estimated responses , (fitted values).
Simulate the new responses in two scenarios:
Scenario 1: with ,
Scenario 2: with .
Here, apply Lasso with and (using the same K-fold CV) on the new data set (i.e., new response and the original design matrix) and get the new estimated responses and for each of the two scenarios.
Calculate the mean square estimation error and for scenario 1;
and for scenario 2.
Repeat the above resampling process for 500 times and compute the proportion of better estimation for each method.
The estimation results (
Table 4) show that, overall, the proportion of better estimation for the model
is between 46.8∼52.8% and for the model
, it is between 47.2∼53.2%. Therefore, based on the DGS, in term of regression estimation accuracy, there is not much difference before
and
: the observed properties are not significantly different from 50% at 0.05 level.
5.2. Variable Selection: DGS
To test which model does better in variable selection, we perform the DGS over the above real data sets. Since the exact variable selection might not be reached, especially when applied to the high-dimensional data, we use symmetric difference to measure their performance, which is the number of variables either in but not in or in but not in . Below is the algorithm for the DGS.
Procedure
Apply the 10-fold (default) cross validation over real data set and select the true set of variables : or , either by the model with or by the model with .
Do least square estimation by regressing the response on the true set of variables and obtain the estimated response and the residual standard error .
Simulate the new response by adding the error term randomly generated from to . Apply K-fold cross validation, , over the simulated data set (i.e., and the original design matrix) and select set of variables : or , either by model or by model . Repeat this process for 500 times.
Calculate the symmetric difference .
The distributions of the symmetric difference are shown in the appendix. (
Figure A3 and
Figure A4 for Boston Housing Price;
Figure A5 and
Figure A6 for Bardet–Biedl data) The mean of
is reported in
Table 5 and
Table 6. For Boston Housing Price, the size of the candidate variables is 13 while model
selects 11 variables and model
selects 8 variables on average, i.e., the mean of
is 11 and the mean of
is 8. For Bardet–Biedl data, the size of the candidate variables is 200, while model
selects 21 variables and model
selects 19 variables on average, i.e., the mean of
is 21 and the mean of
is 19.
On average, the 1se rule does better in variable selection since the mean of is more closed to . In the case of , if we want to have a perfect or near perfect variable selection result (i.e., ), then the 1se rule is a good choice, despite the fact that its selection result is more unstable: the range of is larger than that of . In the case of , both models fail to provide perfect selection result. But overall the 1se rule is more accurate and stable given that has smaller mean and range.
6. Conclusions
The one standard error rule is proposed to pick the most parsimonious model within one standard error of the minimum. Despite the fact that it was widely used as a conservative and alternative approach for cross validation, there is no evidence confirming that the 1se rule can consistently outperform the regular CV.
Our theoretical result shows that the standard error formula is asymptotically valid when the regression procedure converges relatively fast to the true regression function. Our illustrative example also shows that when the regression estimator converges slowly, the CV errors on the different evaluation folds may be highly dependent, which unfortunately makes the usual sample variance (or sample standard deviation) formula problematic. In such a case, the use of the 1se rule may easily lead to a choice of inferior candidates.
Our numerical results offer finite-sample understandings. First of all, the simulation study also casts doubt on the estimation accuracy of the 1se rule in terms of estimating the standard error of the minimum cross validation error. In some cases, the 1se is likely to have 100% overestimation or underestimation, depending on the number of cross validation fold and the dependency among the predictors. More specifically, for example, the 1se tends to have overestimation when used with 2-fold cross validation in case of dependent predictors; but it tends to have underestimation when used with 20-fold cross validation in case of independent predictors.
Secondly, the performances of the 1se rule and the regular CV in regression estimation and variable selection are compared via both simulated and real data sets. In general, on the one hand, the 1se rule often performs better in variable selection for sparse modeling; on the other hand, it often does worse in regression estimation.
While our theoretical and numerical results clearly challenge indiscriminate uses of the 1se rule in general model selection, its parsimony nature may still be appealing when a sparse model is highly desirable. For the real data sets, for regression estimation, the regular CV does better than the 1se rule but the difference is small. There is almost no difference in the estimation accuracy between these two methods when regression estimation is assessed by the DGS. Overall, considering the model simplicity and interpretability, the 1se rule would be a better choice here.
Future studies are encouraged to provide more theoretical understandings on the 1se rule. Also, it may be studied numerically in more complex settings (e.g., when used to compare several nonlinear/nonparametric classification methods) than those in this work. In a larger context, since it is well known that the penalty methods for model selection are often unstable (e.g., [
1,
21,
22]), alternative methods to strengthen reliability and reproductivity of variable selection may be applied (see reference in the above mentioned papers and [
23]).