1. Introduction
Modern prediction algorithms such as random forests and deep learning use training sets, often very large ones, to produce rules for predicting new responses from a set of available predictors. A second question—right after “How should the prediction rule be constructed?”—is “How accurate are the rule’s predictions?” Resampling methods have played a central role in the answer. This paper is intended to provide an overview of what are actually several different answers, while trying to keep technical complications to a minimum.
This is a Special Issue of
STATS devoted to resampling, and before beginning on prediction rules it seems worthwhile to say something about the general effect of resampling methods on statistics and statisticians.
Table 1 shows the
law school data [
1], a small data set but one not completely atypical of its time. The table reports average scores of the 1973 entering class at 15 law schools on two criteria: undergraduate grade point average (GPA) and result on the “LSAT”, a national achievement test. The observed Pearson correlation coefficient between GPA and LSAT score is
How accurate is
?
Suppose that Dr. Jones, a 1940s statistician, was given the data in
Table 1 and asked to attach a standard error to
; let’s say a nonparametric standard error since a plot of (LSAT,GPA) looks definitely non-normal. At his disposal is the
nonparametric delta method, which gives a first-order Taylor series approximation formula for
. For the Pearson correlation coefficient this turns out to be
where
and
is the mean of
, the bars indicating averages.
Table 1.
Average scores for admitees to 15 American law schools, 1973. GPA is undergraduate grade point average, LSAT “law boards” score. Pearson correlation coefficient between GPA and LSAT is 0.776.
Table 1.
Average scores for admitees to 15 American law schools, 1973. GPA is undergraduate grade point average, LSAT “law boards” score. Pearson correlation coefficient between GPA and LSAT is 0.776.
| GPA | LSAT |
|---|
| 3.39 | 576 |
| 3.30 | 635 |
| 2.81 | 558 |
| 3.03 | 578 |
| 3.44 | 666 |
| 3.07 | 580 |
| 3.00 | 555 |
| 3.43 | 661 |
| 3.36 | 651 |
| 3.13 | 605 |
| 3.12 | 653 |
| 2.74 | 575 |
| 2.76 | 545 |
| 2.88 | 572 |
| 2.96 | 594 |
Jones either looks up or derives (
2), evaluates the six terms on his mechanical calculator, and reports
after which he goes home with the feeling of a day well spent.
Jones’ daughter, a 1960s statistician, has a much easier go of it. Now she does not have to look up or derive Formula (
2). A more general resampling algorithm, the Tukey–Quenouille
jackknife is available, and can be almost instantly evaluated on her university’s mainframe computer. It gives her the answer
Dr. Jones is envious of his daughter:
- 1.
She does not need to spend her time deriving arduous formulas such as (
2).
- 2.
She is not restricted to traditional estimates such as that have closed-form Taylor series expansions.
- 3.
Her university’s mainframe computer is a million times faster than his old Marchant calculator (though it is across the campus rather than on her desk).
If now, 60 years later, the Jones family is still in the statistics business they’ll have even more reason to be grateful for resampling methods. Faster, cheaper, and more convenient computation combined with more aggressive methodology have pushed the purview of resampling applications beyond the assignment of standard errors.
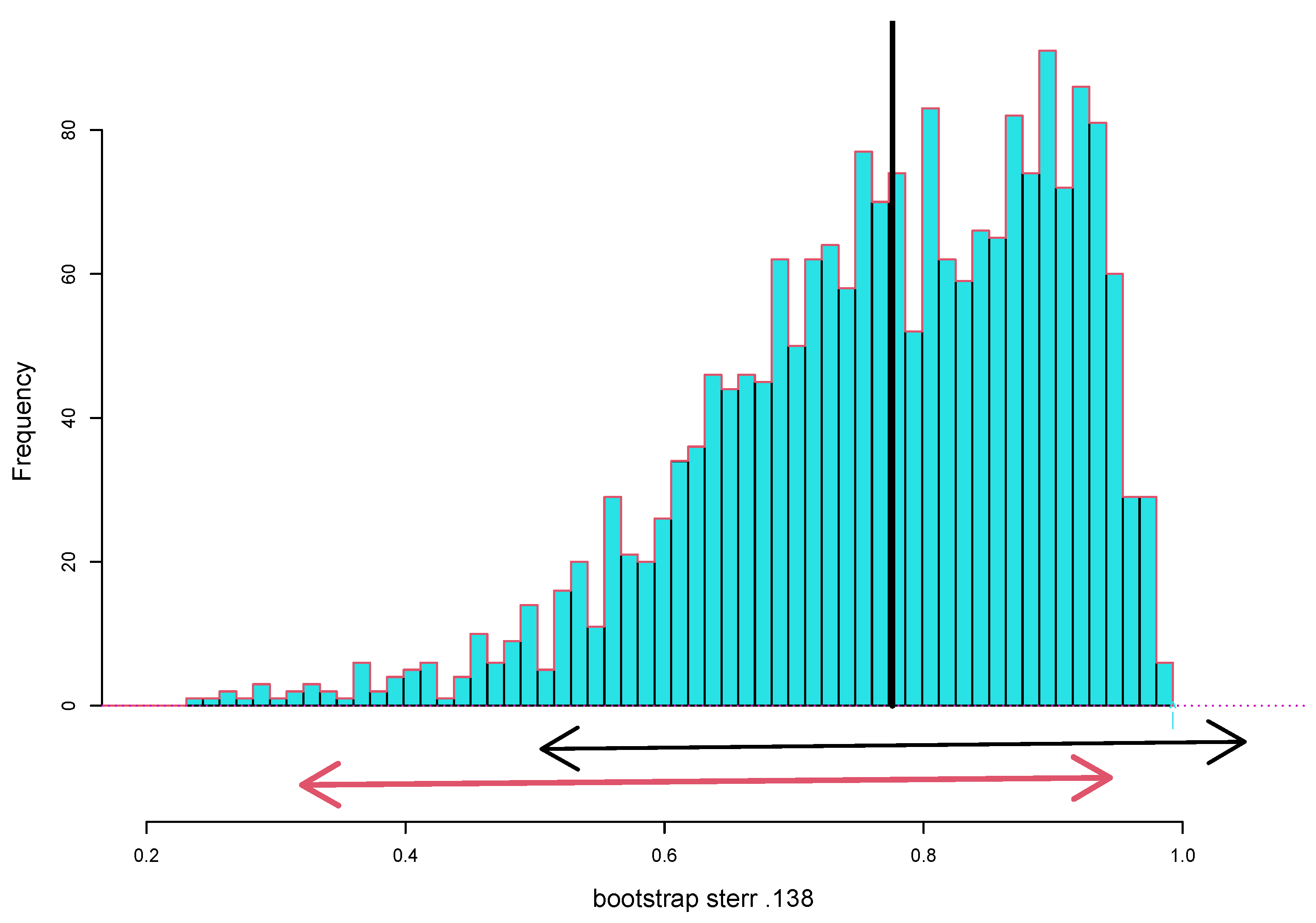
Figure 1 shows 2000 nonparametric bootstrap replications
from the law school data (Each
is the correlation from a bootstrapped data matrix obtained by resampling the 15 rows of the
matrix in
Table 1 15 times with replacement. See Chapter 11 of [
2]). Their empirical standard deviation is the nonparametric bootstrap estimate of standard error for
,
Two thousand is 10 times too many replications needed for a standard error, but it is not too many for a bootstrap confidence interval. The arrowed segments in
Figure 1 compare the standard approximate 95% confidence limit
, with the nonparametric bootstrap interval. (This is the
bca interval, constructed using program
bcajack from the CRAN package
bcaboot [
3]. Chapter 11 of [
2] shows why bca’s “second-order corrections”, here very large, improve on the standard method.)
The standard method does better if we first make Fisher’s
z transformation
compute the standard interval on the
scale, and transform the endpoints back to the
scale. This gives 0.95 interval
not so different from the bootstrap interval (
7), and at least not having its upper limit above 1.00!
This is the kind of trick Dr. Jones would have known. Resampling, here in the form of the bca algorithm, automates devices such as (
8) without requiring Fisher-level insight for each new application.
If there was a statistics’ “word of the year” it would be two words: deep learning. This is one of a suite of prediction algorithms that input data sets, often quite massive ones, and output prediction rules. Others in the suite include random forests, support vector machines, and gradient boosting.
Having produced a prediction rule, it is natural to wonder how accurately it will predict future cases, our subject in what follows:
Section 2 gives a careful definition of the prediction problem, and describes a class of loss functions (the “
Q class”) that apply to discrete as well as continuous response variables.
Section 3 concerns nonparametric estimates of prediction loss (cross-validation and the bootstrap “632 rule”) as well as Breiman’s
out-of-bag error estimates for random forests.
Covariance penalties, including Mallows’
and the Akaike Information Criterion, are parametric methods discussed in
Section 4 along with the related concept of
degrees of freedom.
Section 5 briefly discusses
conformal inference, the most recent addition to the resampling catalog of prediction error assessments.
2. The Prediction Problem
Statements of the prediction problem are often framed as follows:
A data set
of
n pairs is observed,
where the
are
p-dimensional predictor vectors and the
are one-dimensional responses.
The
pairs are assumed to be independent and identically distributed draws from an unknown
-dimensional distribution
F,
Using some algorithm
, the statistician constructs a prediction rule
that provides a prediction
,
for any vector
x in the space of possible predictors.
A new pair
is independently drawn from
F,
but with only
x observable.
The statistician predicts the unseen
y by
and wishes to assess prediction error. Later the prediction error will turn out to directly relate to the estimation error of
for the true conditional expectation of
y given
x,
Prediction error is assessed as the expectation of loss under distribution
F,
for a given loss function such as squared error:
.
Here,
indicates expectation over the random choice of all
pairs
and
in (
11) and (
13). The
u in
reflects the unconditional definition of error in (
15). The resampling algorithms we will describe calculate an estimate of
from the observed data. (One might hope for a more conditional error estimate, say one applying to the observed set of predictors
, a point discussed in what follows.)
Naturally, the primary concern within the prediction community has been with the choice of the algorithm that produces the rule . Elaborate computer-intensive algorithms such as random forests and deep learning have achieved star status, even in the popular press. Here, however, the “prediction problem” will focus on the estimation of prediction error. To a large extent the prediction problem has been a contest of competing resampling methods, as discussed in the next three sections.
Figure 2 illustrates a simple example:
pairs
have been observed, in this case with
x real. A fourth-degree polynomial
has been fit by ordinary least squares applied to
with the heavy curve tracing out
.
In the usual OLS notation, we’ve observed
from the notional model
where
is the
matrix with
ith row
,
the unknown 5-dimensional vector of regression coefficients, and
a vector of 20 uncorrelated errors having mean 0 and variance
,
The fitted curve
is given by
for
and
(the OLS estimate), this being algorithm
.
The
apparent error, what will be called
in what follows, is
which equals 1.99 for
. The usual unbiased estimate for the noise parameter
, not needed here, modifies the denominator in (
19) to take account of fitting a 5-vector
to
,
Dividing the sum of squared errors by
rather than
n can be thought of as a classical prediction error adjustment; err usually underestimates future prediction error since the
have been chosen to fit the observations
.
Because this is a simulation, we know the true function
(
14), the light dotted curve in
Figure 2:
the points
were generated with normal errors, variance 2,
Given model (
22), we can calculate the true prediction error for estimate
(
18). If
is a new observation, independent of the original data
which gave
, then the true prediction error
is
the notation
indicating expectation over
. Let
denote the average true error over the
observed values
,
equaling 2.40 in this case.
figures prominently in the succeeding sections. Here, it exceeds the apparent error
(
19) by 21%. (
is not the same as
(
15).)
Prediction algorithms are often, perhaps most often, applied to situations where the responses are dichotomous,
or 0; that is, they are
Bernoulli random variables, binomials of sample size 1 each,
Here
is the probability that
given prediction vector
x,
The probability model
F in (
11) can be thought of in two steps: as first selecting
x according to some
p-dimensional distribution
G and then “flipping a biased coin” to generate
.
Squared error is not appropriate for dichotomous data. Two loss (or “error”) functions
are in common use for measuring the discrepancy between
and
, the true and estimated probability that
in (
25). The first is
counting error,
For
or 1,
equals 0 or 1 if
y and
are on the same or different sides of
.
The second error function is
binomial deviance (or twice the Kullback–Leibler divergence),
Binomial deviance plays a preferred role in maximum likelihood estimation. Suppose that
is a
p-parameter family for the true vector of means in model (
25),
Then, the maximum likelihood estimate (MLE)
is the minimizer of the average binomial deviance (
28) between
and
,
see Chapter 8 of [
2]. Most of the numerical examples in the following sections are based on binomial deviance (
30). (If
equals 0 or 1 then (
30) is infinite. To avoid infinities, our numerical examples truncate
to
.)
Squared error, counting error, and binomial deviance are all members of the
Q-class, a general construction illustrated in
Figure 3 ([
4] Section 3). The construction begins with some concave function
; for the dichotomous cases considered here,
with
. The error
between a true value
and an estimate
is defined by the illustrated tangency calculation:
(equivalent to the “Bregman divergence” [
5]).
The entropy function
makes
equal binomial deviance. Two other common choices are
for counting error and
for squared error.
Working within the
Q-class (
31), it is easy to express the
true error of prediction
at predictor value
x where the true mean is
. Letting
be an independent realization from the distribution of
y given
x, the true error at
x is, by definition,
only
being random in the expectation.
Lemma 1. The true error at x (33) iswith in the dichotomous case. Proof. From definition (
31) of the
Q-class,
giving (
34) with
. In the dichotomous case
for
equal 0 or 1, so
. ☐
To simplify notation, let
and
, with
and
. The
average true error is defined to be
It is “true” in the desirable sense of applying to the given prediction rule
. If we average
over the random choice of
in (
11), we get the less desirable unconditional error
(
15).
is minimized by
,
Subtraction from (
36) gives
This is an exact analogue of the familiar squared error relationship. Suppose
and
are independently
vectors, and that
produces an estimate
. Then
which is (
38) if
.
At a given value of
x, say
, a prediction
can also be thought of as an estimate of
. Lemma 1 shows that the optimum choice of a prediction rule
is also the optimum choice of an estimation rule: for any rule
,
(
as in (
11)), so that the rule that minimizes the expected prediction error also minimizes the expected estimation error
.
Predicting y and estimating its mean μ are equivalent tasks within the Q-class. 3. Cross-Validation and Its Bootstrap Competitors
Resampling methods base their inferences on recomputations of the statistic of interest derived from systematic modifications of the original sample. This is not a very precise definition, but it cannot be if we want to cover the range of methods used in estimating prediction error. There are intriguing differences among the methods concerning just what the modifications are and how the inferences are made, as discussed in this and the next two sections.
Cross-validation has a good claim to being the first resampling method. The original idea was to randomly split the sample into two halves, the training and test sets, and . A prediction model is developed using only , and then validated by its performance on . Even if we cheated in the training phase, say by throwing out “bad” points, etc., the validation phase guarantees an honest estimate of prediction error.
One drawback is that inferences based on
data points are likely to be less accurate than those based on all
n, a concern if we are trying to accurately assess prediction error. “One-at-a-time“ cross-validation almost eliminates this defect: let
be data set (
10) with point
removed, and define
the prediction for case
i based on
using the rule
, constructed using only the data in
. The cross-validation estimate of prediction error is then
(This assumes that we know how to apply the construction rule
to subsets of size
). Because
is not involved in
, overfitting is no longer a concern. Under the independent-draws model (
11),
is a nearly unbiased estimate of
(
15) (“nearly” because it applies to samples of size
rather than
n).
The little example in
Figure 2 has
points
. Applying (
42) with
gave
, compared with apparent error
(
19) and true error
(
24). This not-very-impressive result has much to do with the small sample size resulting in large differences between the original estimates
and their cross-validated counterparts
. The single data point at
accounted for 40% of
.
Figure 4 concerns a larger data set that will serve as a basis for simulations comparing cross-validation with its competitors: 200 transplant recipients were followed to investigate the subsequent occurrence of anemia; 138 did develop anemia (coded as
) while 62 did not (
). The goal of the study was to predict
y from
x, a vector of
baseline variables including the intercept term (The predictor variables were body mass index, sex, race, patient and donor age, four measures of matching between patient and donor, three baseline medicine indicators, and four baseline general health measures).
A standard logistic regression analysis gave estimated values of the anemia probability
for
that we will denote as
Figure 4 shows a histogram of the 200
values. Here we will use
as the “ground truth” for a simulation study (rather than analyzing the transplant study itself). The
will play the role of the true mean
in Lemma 1 (
34), enabling us to calculate true errors for the various prediction error estimates.
A
matrix
of dichotomous responses
was generated as independent Bernoulli variables (that is, binomials of sample size 1),
for
and
. The
jth column of
,
is a simulated binomial response vector (
25) having true mean vector
.
provides 100 such response vectors.
Figure 4.
Logistic regression estimated anemia probabilities for the 200 transplant patients.
Figure 4.
Logistic regression estimated anemia probabilities for the 200 transplant patients.
For each one, a logistic regression was run,
in the language R, with
the
matrix of predictors from the transplant study. Cross-validation (“10-at-a-time” cross-validation rather than one-at-a-time: the 200 (x,y) pairs were randomly split into 20 groups of 10 each; each group was removed from the prediction set in turn and its 10 estimates obtained by logistic regression based on the other 190) gave an estimate of prediction error for the
jth simulation,
while (
36) gave true error
where
was the estimated mean from (
46).
In terms of mean ± standard deviation the 100 simulations gave
averaged 8% more than
(a couple percent of which came from having sample size 190 rather than 200). The standard deviation of
is not much bigger than the standard deviation of
, which might suggest that
was tracking
as it varied across the simulations.
Sorry to say, that was not at all the case. The pairs
,
, are plotted in
Figure 5. It shows
actually
decreasing as the true error
increases. The unfortunate implication is that
is not estimating the true error, but only its expectation
(
15). This is not a particular failing of cross-validation. It is habitually observed for all prediction error estimates—see for instance Figure 9 of [
4]—though the phenomenon seems unexplained in the literature.
Cross-validation tends to pay for its low bias with high variability. Efron [
1] proposed bootstrap estimates of prediction error intended to decrease variability without adding much bias. Among the several proposals the most promising was the
632 rule. (An improved version
632+ was introduced in Efron and Tibshirani [
6], designed for reduced bias in overfit situations where
(
19) equals zero. The calculations here use only
632.) The 632 rule is described as follows:
Nonparametric bootstrap samples
are formed by drawing
n pairs
with replacement from the original data set
(
10).
Applying the original algorithm
to
gives prediction rule
and predictions
as in (
12).
B bootstrap data sets
are independently drawn, giving predictions
for
and
.
Two numbers are recorded for each choice of
i and
j, the error of
as a prediction of
,
and
the number of times
occurs in
.
The
zero bootstrap is calculated as the average value of
for those cases having
,
Finally, the 632 estimate of prediction error is defined to be
being the apparent error rate (
19).
was calculated for the same 100 simulated response vectors
(
45) used for
(each using
replications), the 100 simulations giving
an improvement on
at (
49). This is in line with the 24 sampling experiments reported in [
6]. (There, rule
was used, with loss function
counting error (
27) rather than binomial deviance.
is discontinuous for counting error, which works to the advantage of 632 rules.)
Table 2 concerns the rationale for the 632 rule. The
80,000 values
for the first of the 100 simulated
vectors were averaged according to how many times
appeared in
,
for
. Not surprisingly,
decreases with increasing
k.
(
55), which would seem to be the bootstrap analogue of
, is seen to exceed the true error
, while
is below
. The intermediate linear combination
is motivated in Section 6 of [
1], though in fact the argument is more heuristic than compelling. The 632 rules
do usually reduce variability of the error estimates compared to cross-validation, but bias can be a problem.
The 632 rule recomputes a prediction algorithm by nonparametric bootstrap resampling of the original data.
Random forests [
7], a widely popular prediction algorithm, carries this further: the algorithm itself as well as estimates of its accuracy depend on bootstrap resampling calculations.
Regression trees are the essential component of random forests.
Figure 6 shows one such tree (constructed using
rpart, the R version of
CART [
8]; Chapters 9 and 15 of [
9] describe CART and random forests) as applied to
, the first of the 100 response vectors for the transplant data simulation (
44);
consists of 57 0 s and 143 1 s, average value
. The tree-making algorithm performs successive splits of the data, hoping to partition it into bins that are mostly 0 s or 1 s. The bin at the far right—comprising
cases having low body mass index, low age, and female gender—has just one 0 and 28 1 s, for an average of 0.97. For a new transplant case
, with only
x observable, we could follow the splits down the tree and use the terminal bin average as a quantitative prediction of
y.
Random forests improves the predictive accuracy of any one tree by
bagging (“bootstrap aggregation”), sometimes also called
bootstrap smoothing:
B bootstrap data sets
are drawn at random (
51), each one generating a tree such as that in
Figure 6. (Some additional variability is added to the tree-building process: only a random subset of the
p predictors is deemed eligible at each splitting point.) A new
x is followed down each of the
B trees, with the random forest prediction being the average of
x’s
B terminal values. Letting
, the prediction at
for the tree based on
, the random forest prediction at
is
Figure 6.
Regression tree for transplant data, simulation 1.
Figure 6.
Regression tree for transplant data, simulation 1.
The predictive accuracy for
uses a device such as that for
(
55): Let
be the average value of
for bootstrap samples
not containing
,
called the “out-of-bag” (oob) estimate of
. The oob error estimate is then
The overall oob estimate of prediction error is
(Notice that the leave-out calculations here are for the estimates
, while those for
(
56) are for the errors
.) Calculated for the 100 simulated response vectors
(
45), this gave
a better match to the true error
than either
(
49) or
(
57). In fact the actual match was even better than (
63) suggests, as shown in
Table 3 of
Section 4. This is all the more surprising given that, unlike
and
,
is fully nonparametric: it makes no use of the logistic regression model
(
46), which was involved in generating the simulated response vectors
(
44) (It has to be added that
is not an estimate for the prediction error of the logistic regression model
(
46), but rather for the random forest estimates
).
4. Covariance Penalties and Degrees of Freedom
A quite different approach to the prediction problem was initiated by Mallows’
formula [
10]. An observed
n-dimensional vector
is assumed to follow the homoskedastic model
the notation indicating uncorrelated errors of mean 0 and variance
as in (
17);
is known. A linear rule
is used to estimate
with
a fixed and known matrix. How accurate is
as a predictor of future observations?
The apparent error
is likely to underestimate the true error of
given a hypothetical new observation vector
independent of
,
the
notation indicating that
is fixed in (
67). Mallows’
formula says that
is an unbiased estimator of
,
that is,
is not unbiased for
but
is unbiased for
(
fixed in the expectation) under model (
64)–(
65).
One might wonder what has happened to the covariates
in
(
10)? The answer is that they are still there but no longer considered random–rather as fixed ancillary quantities such as the sample size
n. In the OLS model
for
Figure 3 (
16)–(
18) the covariates
determine
,
and
Formula (
65). We could, but do not, write
as
.
Mallows’
Formula (
68) can be extended to the
Q class of error measures,
Figure 4. An unknown probability model
—notice that
is not the same as
f in (
12)— is assumed to have produced
and its true mean vector
,
an estimate
has been calculated using some algorithm
,
the apparent error
(
19) and true error
(
33) are defined as before,
with the
fixed and
independently of
. Lemma 1 for the true error (
36) still applies,
The
Q-class version of Mallows’ Formula (
68) is derived as
Optimism Theorem 1 in Section 3 of [
4]:
Theorem 1. DefineThenwhere indicates covariance under model (72), is an unbiased estimate of in the same sense as (69), The covariance terms in (
76) measure how much each
affects its own estimate. They sum to a
covariance penalty that must be added to the apparent error to account for the fitting process. If
is binomial deviance then
the logistic parameter; the theorem still applies as stated whether or not
is logistic regression.
as stated in (
76) is not directly usable since the covariance terms
are not observable statistics. This is where resampling comes in.
Suppose that the observed data
provides an estimate
of
. For instance in a normal regression model
we could take
to be
for some estimate
. We replace (
72) with the
parametric bootstrap model
and generate
B independent replications
, from which are calculated
B pairs,
for
as in (
75). The covariances in (
76) can then be estimated as
and
, yielding a useable version of (
76),
“cp” standing for “covariance penalty”.
Table 3 compares the performances of cross-validation, covariance penalties, the 632 rule, and the random forest out-of-bag estimates on the 100 transplant data simulations. The results are given in terms of
total prediction error, rather than average error as in (
47). The bottom line shows their root-mean-square differences from true error
,
is highest, with
,
, and
respectively 71%, 83%, and 65% as large.
Table 3.
Transplant data simulation experiment. Top: First 5 of 100 estimates of total prediction error using cross-validation, covariance penalties (cp), the 632 rule, the out-of-bag random forest results, and the apparent error; degrees of freedom (df) estimate from cp. Bottom: Mean of the 100 simulations, their standard deviations, correlations with Errtrue, and root mean square differences from Errtrue. Cp and 632 rules used bootstrap replications per simulation.
Table 3.
Transplant data simulation experiment. Top: First 5 of 100 estimates of total prediction error using cross-validation, covariance penalties (cp), the 632 rule, the out-of-bag random forest results, and the apparent error; degrees of freedom (df) estimate from cp. Bottom: Mean of the 100 simulations, their standard deviations, correlations with Errtrue, and root mean square differences from Errtrue. Cp and 632 rules used bootstrap replications per simulation.
| | Errtrue | ErrCv | ErrCp | Err632 | Erroob | err | df |
| | 194 | 216.5 | 232.9 | 260.6 | 240.3 | 180.4 | 26.2 |
| | 199 | 245.8 | 220.9 | 241.0 | 230.9 | 173.8 | 23.5 |
| | 192 | 221.2 | 225.9 | 247.6 | 232.7 | 180.6 | 22.7 |
| | 234 | 197.4 | 200.9 | 215.2 | 219.7 | 162.2 | 19.4 |
| | 302 | 175.7 | 178.3 | 187.1 | 211.2 | 144.6 | 16.9 |
| | Errtrue | ErrCv | ErrCp | Err632 | Erroob | err | df |
| mean | 214.0 | 231.1 | 216.1 | 229.4 | 223.8 | 169.9 | 23.1 |
| stdev | 23.0 | 28.7 | 15.8 | 19.0 | 14.6 | 13.5 | 4.0 |
| cor.true | | −0.58 | | | | | |
| rms | | 48.8 | 34.8 | 40.7 | 31.6 | 54.2 | |
For the
jth simulation vector
(
45) the parametric bootstrap replications (
79) were generated as follows: the logistic regression estimate
was calculated from (
46),
; the bootstrap replications
had independent Bernoulli components
independent replications
were generated for each
, giving
according to (
81)–(
82).
Because the resamples were generated by a model of the same form as that which originally gave the
’s (
43)–(
44),
is unbiased for
. In practice our resampling model
in (
79) might not match
in (
72), causing
to be downwardly biased (and raising its rms).
’s nonparametric resamples make it nearly unbiased for the unconditional error rate
(
15) irrespective of the true model, accounting for the overwhelming popularity of cross-validation in applications.
4.1. A Few Comments
In computing
it is not necessary for
in (
79) to be constructed from the original estimate
. We might base
on a bigger model than that which led to the choice of
for
; in the little example of
Figure 3, for instance, we could take
to be
where
is the OLS sixth degree polynomial fit, while still taking
to be fourth degree as in (
18). This reduces possible model-fitting bias in
, while increasing its variability.
A major conceptual difference between
and
concerns the role of the covariates
, considered as random in model (
10) but fixed in (
72). Classical regression problems have usually been analyzed in a fixed-
framework for three reasons:
- 1.
mathematical tractability;
- 2.
not having to specify ’s distribution;
- 3.
inferential relevance.
The reasons come together in the classic covariance formula for the linear model
,
A wider dispersion of the
’s in
Figure 3 would make
more accurate, and conversely.
It can be argued that because
is estimating the conditional error
given it is more relevant to what is being estimated. See [
11,
12] for a lot more on this question.
On the other hand, “fixed-
” methods such as Mallows’
can be faulted for missing some of the variability contributing to the unconditional prediction error
(
15). Let
be the conditional error given
for predicting
y given
x,
according to Lemma 1 (
34). Then
where
is the marginal density of
x and
E indicates expectation over the choice of the training data
.
In the fixed-
framework of (
36),
replaces the integrand in (
87) with its
average
. We expect
since
typically increases for values of
x farther away from
. Rosset and Tibshirani [
12] give an explicit formula for the difference in the case of normal-theory ordinary least squares when they show that the factor 2 for the penalty term in Mallows’
formula should be increased to
. Cross-validation effectively estimates
while
estimates the fixed-
version of
.
With (
88) in mind,
and
are often contrasted as
This is dangerous terminology if it’s taken to mean that
applies to prediction errors
at specific points
x outside of
. In
Figure 3 for instance it seems likely that
exceeds
, but this is a fixed-
question and beyond the reach of the random-
assumptions underlying (
88). See the discussion of Figure 8 in
Section 5.
The sad story told in
Figure 5 shows
negatively correlated with the true
. The same is the case for
and
, as can be seen from the negative correlations in the
cor.true row of
Table 3.
is also negatively correlated with
, but less so. In terms of
rms, the bottom row shows that the fully nonparametric
estimates beat even the parametric
ones.
Figure 7 compares the 200
estimates from the logistic regression estimates (
46) with those from random forests, for
y the first of the 100 transplant simulations. Random forests is seen to better separate the
from the
cases.
relates to error prediction for random forest estimates,
not for logistic regression, but this does not explain how
could provide excellent estimates of the true error
–which in fact were based on the logistic regression model (
43)–(
44). If this is a fluke it’s an intriguing one.
There is one special case where the covariance penalty formula (
76) can be unbiasedly estimated without recourse to resampling: if
is the normal model
, and
—so
—then
Stein’s unbiased risk estimate (SURE) is defined to be
where the partial derivatives are calculated directly from the functional form of
. Section 2 of [
4] gives an example comparing
with
. Each term
measures the influence of
on its own estimate.
4.2. Degrees of Freedom
The OLS model
yields the familiar estimate
of
, where
is the projection matrix
as in (
70);
has
where
. Mallows’
formula
becomes
in this case. In other words, the covariance penalty that must be added to the apparent error
is directly proportional to
p, the degrees of freedom of the OLS model.
Suppose that
with matrix
not necessarily a projection. It has become common to define
’s degrees of freedom as
playing the role of
p in the
Formula (
92). In this way,
becomes a
lingua franca for comparing linear estimators
of different forms. (The referee points out that formulas such as (
92) are more often used for model selection rather than error rate prediction. Zhang and Yang [
13] consider model selection applications, as does Remark B of [
4].)
A nice example is the
ridge regression estimator
a fixed non-negative constant;
is the usual OLS estimator while
“shrinks”
toward
, more so as
increases. Some linear algebra gives the degrees of freedom for (
94) as
where the
are the eigenvalues of
.
The generalization of Mallows’ Formula (
68) in Theorem 1 (
76) has penalty term
which again measures the self-influence of each
on its own estimate
. The choice of
in
Figure 3 results in
, squared error, and
, in which case
equals
, and (
96) becomes
, as in Mallows’ Formula (
68). This suggests using
as a measure of degrees of freedom (or its estimate
from (
81)) for a general estimator
.
Some support comes from the following special situation: suppose
is obtained from a
p-parameter generalized linear model, with prediction error measured by the appropriate deviance function. (Binomial deviance for logistic regression as in the transplant example.) Theorem 2 of [
14], Section 6, then gives the asymptotic approximation
as in (
91)–(
92), the intuitively correct answer.
Approximation (
99) leads directly to
Akaike’s information criterion (AIC). In a generalized linear model, the total deviance from the MLE
is
denoting the density function ([
2], Hoeffding’s Lemma). Suppose we have glm’s of different sizes
p we wish to compare. Minimizing
over the choice of model is then equivalent to maximizing the total log likelihood
minus a dimensionality penalty,
which is the AIC.
Approximation (
99) is not razor-sharp:
is the transplant simulation logistic regression but the 100 estimates
averaged 23.08 with standard error 0.40. Degrees of freedom play a crucial role in model selection algorithms. Resampling methods allow us to assess
(
98) even for very complicated fitting algorithms
.
5. Conformal Inference
If there is a challenger to cross-validation for “oldest resampling method” it is permutation testing, going back to Fisher in the 1930s. The newest prediction error technique, conformal inference, turns out to have permutation roots, as briefly reviewed next.
A clinical trial of an experimental drug has yielded independent real-valued responses for control and treatment groups:
Student’s
t-test could be used to see if the new drug was giving genuinely larger responses but Fisher, reacting to criticism of normality assumptions, proposed what we would now call a nonparametric two-sample test.
Let
be the combined data set,
and choose some score function
that contrasts the last
m z-values with the first
n, for example the difference of means,
Define
as the set of scores for all permutations of
,
ranging over the
permutations.
The permutation
p-value for the treatment’s efficacy in producing larger responses is defined to be
the proportion of
having scores
exceeding the observed score
. Fisher’s key idea was that if in fact all the observations came from the same distribution
F,
(implying that Treatment is the same as Control), then all
permutations would be equally likely. Rejecting the null hypothesis of No Treatment Effect if
has null probability (nearly)
.
Usually
is too many for practical use. This is where the sampling part of resampling comes in. Instead of all possible permutations, a randomly drawn subset of
B of them, perhaps
, is selected for scoring, giving an estimated permutation
p-value
In 1963, Hodges and Lehmann considered an extension of the null hypothesis (
107) to cover location shifts; in terms of cumulative distribution functions (cdfs), they assumed
where
is a fixed but unknown constant that translates the
v’s distribution by
units to the right of the
u’s.
For a given trial value of
let
and compute its permutation
p-value
A 0.95 two-sided nonparametric confidence interval for
is then
The only assumption is that, for the true value of
, the
components of
are i.i.d., or more generally exchangeable, from some distribution
F.
Vovk has proposed an ingenious extension of this argument applying to prediction error estimation, a much cited reference being [
15]; see also [
16]. Returning to the statement of the prediction problem at the beginning of
Section 2,
is the observed data and
a new (predictor, response) pair, all
pairs assumed to be random draws from the same distribution
F,
is observed but not
, and it is desired to predict
. Vovk’s proposal,
conformal inference, produces an exact nonparametric distribution of the unseen
.
Let
be a proposed trial value for
, and define
as the data set
augmented with
,
A prediction rule
gives estimates
(It is required that
be invariant under reordering of
’s elements.)
For some score function
let
where
measures disagreement between
y and
, larger values of
indicating less conformity between observation and prediction. (More generally
can be any function
.)
If the proposed trial value
were in fact the unobserved
then
and
would be exchangeable random variables, because of the i.i.d. assumption (
113). Let
be the ordered values of
. Assuming no ties, the
n values
partition the line into
intervals, the first and last of which are semi-infinite. Exchangeability implies that
has probability
of falling into any one of the intervals.
A conformal interval for the unseen
consists of those values of
for which
“conforms” to the distribution (
117). To be specific, for a chosen miscoverage level
, say 0.05, let
and
be integers approximately proportion
from the endpoints of
,
The conservative two-sided level
conformal prediction interval
for
is
The argument is the same as for the Hodges–Lehmann interval (
112), now with
and
.
Interval (
119) is computationally expensive since all of the
, not just
, change with each choice of trial value
. The
jackknife conformal interval begins with the jackknife estimates
where
, that is
(
10) with
deleted. The scores
(
116) are taken to be
for some function
s, for example
. These are compared with
, and
is computed as at (
119). Now the score distribution (
117) does not depend on
(nor does
), greatly reducing the computational burden.
The jackknife conformal interval at
was calculated for the small example of
Figure 2 using
for
. For this choice of scoring function, interval (
119) is
with conformal probability
. The square dots in
Figure 8 are the values
for
, with
having probability
of falling into each of the 21 intervals. Conformal probability for the full range
is
.