Adeno-Associated Virus-Mediated CRISPR-Cas13 Knockdown of Papain-like Protease from SARS-CoV-2 Virus
Abstract
1. Introduction
2. Materials and Methods
2.1. SARS-CoV-2 Genome Bioinformatics Study
2.2. Cell Culture
2.3. Construction of PLpro Stable Cell Line
2.4. RNA Secondary Structures and Design of crRNA for CRISPR-Cas13 Systems
2.5. Compact CRISPR-Cas13 Plasmid Construction
2.6. Recombined Adeno-Associated Virus (rAAV) Production and Titration
2.7. rAAV Infection and Western Blotting
2.8. Total RNA Extraction and Reverse Transcriptase PCR (RT-PCR)
2.9. Real-Time Quantitative PCR (RT-qPCR)
2.10. Immunofluorescence (IF) Imaging
2.11. Statistical Analysis
3. Results
3.1. SARS-CoV-2 Antiviral Target Genome Mining
3.2. Structure-Based Design of crRNAs Simultaneously Targeting Three Distant Sites of PLpro mRNA
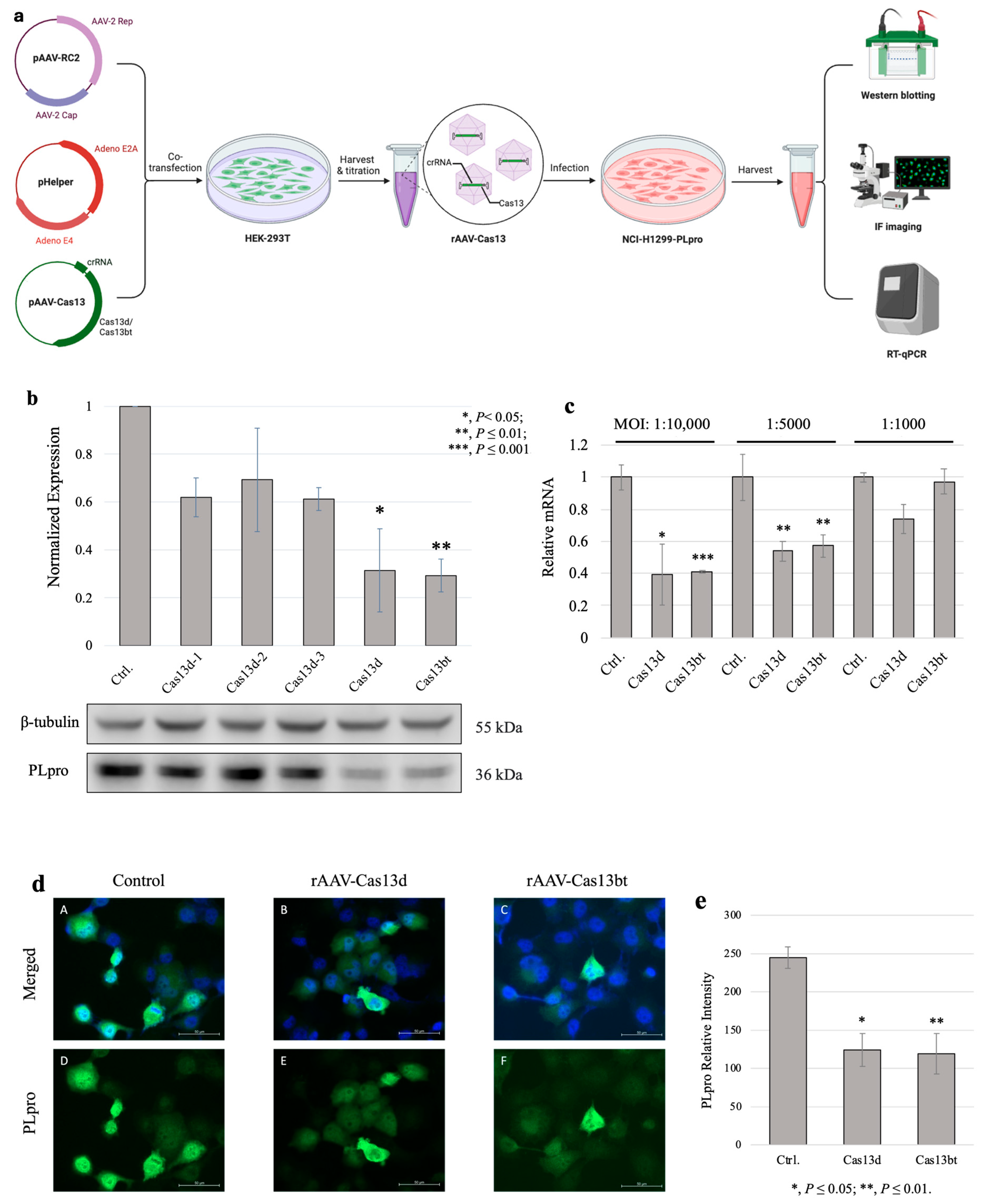
3.3. Compact CRISPR-Cas13 Knockdown PLpro with rAAV Vehicles
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Ciotti, M.; Ciccozzi, M.; Terrinoni, A.; Jiang, W.-C.; Wang, C.-B.; Bernardini, S. The COVID-19 pandemic. Crit. Rev. Clin. Lab. Sci. 2020, 57, 365–388. [Google Scholar] [CrossRef] [PubMed]
- Huang, C.; Wang, Y.; Li, X.; Ren, L.; Zhao, J.; Hu, Y.; Zhang, L.; Fan, G.; Xu, J.; Gu, X.; et al. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet 2020, 395, 497–506. [Google Scholar] [CrossRef]
- COVID-19 Dashboard. Available online: https://coronavirus.jhu.edu/map.html (accessed on 19 January 2023).
- Coronavirus Disease 2019 (COVID-19) Treatment Guidelines. Available online: https://www.ncbi.nlm.nih.gov/books/NBK570371/ (accessed on 19 January 2023).
- Wang, C.; Horby, P.W.; Hayden, F.G.; Gao, G.F. A novel coronavirus outbreak of global health concern. Lancet 2020, 395, 470–473. [Google Scholar] [CrossRef]
- Duchene, S.; Featherstone, L.; Haritopoulou-Sinanidou, M.; Rambaut, A.; Lemey, P.; Baele, G. Temporal signal and the phylodynamic threshold of SARS-CoV-2. Virus Evol. 2020, 6, veaa061. [Google Scholar] [CrossRef] [PubMed]
- van Dorp, L.; Acman, M.; Richard, D.; Shaw, L.P.; Ford, C.E.; Ormond, L.; Owen, C.J.; Pang, J.; Tan, C.C.S.; Boshier, F.A.T.; et al. Emergence of genomic diversity and recurrent mutations in SARS-CoV-2. J. Mol. Epidemiol. Evol. Genet. Infect. Dis. 2020, 83, 104351. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Liu, J.; Johnson, B.A.; Xia, H.; Ku, Z.; Schindewolf, C.; Widen, S.G.; An, Z.; Weaver, S.C.; Menachery, V.D.; et al. Delta spike P681R mutation enhances SARS-CoV-2 fitness over Alpha variant. bioRxiv 2022, 39, 110829. [Google Scholar] [CrossRef]
- Baez-Santos, Y.M.; St John, S.E.; Mesecar, A.D. The SARS-coronavirus papain-like protease: Structure, function and inhibition by designed antiviral compounds. Antivir. Res. 2015, 115, 21–38. [Google Scholar] [CrossRef]
- Amin, S.A.; Banerjee, S.; Ghosh, K.; Gayen, S.; Jha, T. Protease targeted COVID-19 drug discovery and its challenges: Insight into viral main protease (Mpro) and papain-like protease (PLpro) inhibitors. Bioorganic Med. Chem. 2021, 29, 115860. [Google Scholar] [CrossRef]
- Shin, D.; Mukherjee, R.; Grewe, D.; Bojkova, D.; Baek, K.; Bhattacharya, A.; Schulz, L.; Widera, M.; Mehdipour, A.R.; Tascher, G.; et al. Papain-like protease regulates SARS-CoV-2 viral spread and innate immunity. Nature 2020, 587, 657–662. [Google Scholar] [CrossRef]
- Fu, Z.; Huang, B.; Tang, J.; Liu, S.; Liu, M.; Ye, Y.; Liu, Z.; Xiong, Y.; Zhu, W.; Cao, D.; et al. The complex structure of GRL0617 and SARS-CoV-2 PLpro reveals a hot spot for antiviral drug discovery. Nat. Commun. 2021, 12, 488. [Google Scholar] [CrossRef]
- Kannan, S.; Altae-Tran, H.; Jin, X.; Madigan, V.J.; Oshiro, R.; Makarova, K.S.; Koonin, E.V.; Zhang, F. Compact RNA editors with small Cas13 proteins. Nat. Biotechnol. 2022, 40, 194–197. [Google Scholar] [CrossRef] [PubMed]
- Li, C.; Samulski, R.J. Engineering adeno-associated virus vectors for gene therapy. Nat. Rev. Genet. 2020, 21, 255–272. [Google Scholar] [CrossRef] [PubMed]
- Abudayyeh, O.O.; Gootenberg, J.S.; Essletzbichler, P.; Han, S.; Joung, J.; Belanto, J.J.; Verdine, V.; Cox, D.B.T.; Kellner, M.J.; Regev, A.; et al. RNA targeting with CRISPR–Cas13. Nature 2017, 550, 280–284. [Google Scholar] [CrossRef] [PubMed]
- Lotfi, M.; Rezaei, N. CRISPR/Cas13: A potential therapeutic option of COVID-19. Biomed. Pharmacother. 2020, 131, 110738. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, H.; Wilson, H.; Jayakumar, S.; Kulkarni, V.; Kulkarni, S. Efficient Inhibition of HIV Using CRISPR/Cas13d Nuclease System. Viruses 2021, 13, 1850. [Google Scholar] [CrossRef]
- Yin, L.; Zhao, F.; Sun, H.; Wang, Z.; Huang, Y.; Zhu, W.; Xu, F.; Mei, S.; Liu, X.; Zhang, D.; et al. CRISPR-Cas13a Inhibits HIV-1 Infection. Mol. Ther. Nucleic Acids 2020, 21, 147–155. [Google Scholar] [CrossRef]
- Hadfield, J.; Megill, C.; Bell, S.M.; Huddleston, J.; Potter, B.; Callender, C.; Sagulenko, P.; Bedford, T.; Neher, R.A. Nextstrain: Real-time tracking of pathogen evolution. Bioinformatics 2018, 34, 4121–4123. [Google Scholar] [CrossRef]
- Wessels, H.-H.; Méndez-Mancilla, A.; Guo, X.; Legut, M.; Daniloski, Z.; Sanjana, N.E. Massively parallel Cas13 screens reveal principles for guide RNA design. Nat. Biotechnol. 2020, 38, 722–727. [Google Scholar] [CrossRef]
- Pfaffl, M.W. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 2001, 29, e45. [Google Scholar] [CrossRef]
- Chuang, Y.F.; Wang, P.Y.; Kumar, S.; Lama, S.; Lin, F.L.; Liu, G.S. Methods for in vitro CRISPR/CasRx-Mediated RNA Editing. Front. Cell Dev. Biol. 2021, 9, 667879. [Google Scholar] [CrossRef]
- Tong, Y.; Whitford, C.M.; Blin, K.; Jørgensen, T.S.; Weber, T.; Lee, S.Y. CRISPR–Cas9, CRISPRi and CRISPR-BEST-mediated genetic manipulation in streptomycetes. Nat. Protoc. 2020, 15, 2470–2502. [Google Scholar] [CrossRef] [PubMed]
- qPCR Primer & Probe Design Tool. Available online: https://eurofinsgenomics.eu/en/ecom/tools/qpcr-assay-design/ (accessed on 29 January 2023).
- McClain, C.B.; Vabret, N. SARS-CoV-2: The many pros of targeting PLpro. Signal Transduct. Target. Ther. 2020, 5, 223. [Google Scholar] [CrossRef] [PubMed]
- Abudayyeh, O.O.; Gootenberg, J.S.; Konermann, S.; Joung, J.; Slaymaker, I.M.; Cox, D.B.T.; Shmakov, S.; Makarova, K.S.; Semenova, E.; Minakhin, L.; et al. C2c2 is a single-component programmable RNA-guided RNA-targeting CRISPR effector. Science 2016, 353, aaf5573. [Google Scholar] [CrossRef]
- Flotte, T.R. Size does matter: Overcoming the adeno-associated virus packaging limit. Respir. Res. 2000, 1, 16–18. [Google Scholar] [CrossRef]
- Bandaru, S.; Tsuji, M.H.; Shimizu, Y.; Usami, K.; Lee, S.; Takei, N.K.; Yoshitome, K.; Nishimura, Y.; Otsuki, T.; Ito, T. Structure-based design of gRNA for Cas13. Sci. Rep. 2020, 10, 11610. [Google Scholar] [CrossRef] [PubMed]
- Harvey, W.T.; Carabelli, A.M.; Jackson, B.; Gupta, R.K.; Thomson, E.C.; Harrison, E.M.; Ludden, C.; Reeve, R.; Rambaut, A.; Peacock, S.J.; et al. SARS-CoV-2 variants, spike mutations and immune escape. Nat. Rev. Microbiol. 2021, 19, 409–424. [Google Scholar] [CrossRef]
- Zawbaa, H.M.; Osama, H.; El-Gendy, A.; Saeed, H.; Harb, H.S.; Madney, Y.M.; Abdelrahman, M.; Mohsen, M.; Ali, A.M.A.; Nicola, M.; et al. Effect of mutation and vaccination on spread, severity, and mortality of COVID-19 disease. J. Med. Virol. 2022, 94, 197–204. [Google Scholar] [CrossRef] [PubMed]
- Roe, M.K.; Junod, N.A.; Young, A.R.; Beachboard, D.C.; Stobart, C.C. Targeting novel structural and functional features of coronavirus protease nsp5 (3CL(pro), M(pro)) in the age of COVID-19. J. Gen. Virol. 2021, 102, 001558. [Google Scholar] [CrossRef]
- de Wit, E.; van Doremalen, N.; Falzarano, D.; Munster, V.J. SARS and MERS: Recent insights into emerging coronaviruses. Nat. Reviews. Microbiol. 2016, 14, 523–534. [Google Scholar] [CrossRef]
- Lamers, M.M.; Haagmans, B.L. SARS-CoV-2 pathogenesis. Nat. Rev. Microbiol. 2022, 20, 270–284. [Google Scholar] [CrossRef]
- Lei, J.; Hilgenfeld, R. Structural and mutational analysis of the interaction between the Middle-East respiratory syndrome coronavirus (MERS-CoV) papain-like protease and human ubiquitin. Virol. Sin. 2016, 31, 288–299. [Google Scholar] [CrossRef] [PubMed]
- Klemm, T.; Ebert, G.; Calleja, D.J.; Allison, C.C.; Richardson, L.W.; Bernardini, J.P.; Lu, B.G.; Kuchel, N.W.; Grohmann, C.; Shibata, Y.; et al. Mechanism and inhibition of the papain-like protease, PLpro, of SARS-CoV-2. EMBO J. 2020, 39, e106275. [Google Scholar] [CrossRef] [PubMed]
- Patchsung, M.; Jantarug, K.; Pattama, A.; Aphicho, K.; Suraritdechachai, S.; Meesawat, P.; Sappakhaw, K.; Leelahakorn, N.; Ruenkam, T.; Wongsatit, T.; et al. Clinical validation of a Cas13-based assay for the detection of SARS-CoV-2 RNA. Nat. Biomed. Eng. 2020, 4, 1140–1149. [Google Scholar] [CrossRef] [PubMed]
- Kellner, M.J.; Koob, J.G.; Gootenberg, J.S.; Abudayyeh, O.O.; Zhang, F. Sherlock: Nucleic acid detection with CRISPR nucleases. Nat. Protoc. 2019, 14, 2986–3012. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, T.M.; Zhang, Y.; Pandolfi, P.P. Virus against virus: A potential treatment for 2019-nCov (SARS-CoV-2) and other RNA viruses. Cell Res. 2020, 30, 189–190. [Google Scholar] [CrossRef] [PubMed]
- Moreno, A.M.; Palmer, N.; Alemán, F.; Chen, G.; Pla, A.; Jiang, N.; Leong Chew, W.; Law, M.; Mali, P. Immune-orthogonal orthologues of AAV capsids and of Cas9 circumvent the immune response to the administration of gene therapy. Nat. Biomed. Eng. 2019, 3, 806–816. [Google Scholar] [CrossRef]
- Wagner, D.L.; Peter, L.; Schmueck-Henneresse, M. Cas9-directed immune tolerance in humans—A model to evaluate regulatory T cells in gene therapy? Gene Ther. 2021, 28, 549–559. [Google Scholar] [CrossRef]
- Manghwar, H.; Li, B.; Ding, X.; Hussain, A.; Lindsey, K.; Zhang, X.; Jin, S. CRISPR/Cas Systems in Genome Editing: Methodologies and Tools for sgRNA Design, Off-Target Evaluation, and Strategies to Mitigate Off-Target Effects. Adv. Sci. 2020, 7, 1902312. [Google Scholar] [CrossRef]
- Naeem, M.; Majeed, S.; Hoque, M.Z.; Ahmad, I. Latest Developed Strategies to Minimize the Off-Target Effects in CRISPR-Cas-Mediated Genome Editing. Cells 2020, 9, 1608. [Google Scholar] [CrossRef]
- Boulikas, T. Nuclear localization signals (NLS). Crit. Rev. Eukaryot. Gene Expr. 1993, 3, 193–227. [Google Scholar]
- Lange, A.; Mills, R.E.; Lange, C.J.; Stewart, M.; Devine, S.E.; Corbett, A.H. Classical Nuclear Localization Signals: Definition, Function, and Interaction with Importin α. J. Biol. Chem. 2007, 282, 5101–5105. [Google Scholar] [CrossRef] [PubMed]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yang, Y.; Kessler, M.G.C.; Marchán-Rivadeneira, M.R.; Zhou, Y.; Han, Y. Adeno-Associated Virus-Mediated CRISPR-Cas13 Knockdown of Papain-like Protease from SARS-CoV-2 Virus. J 2024, 7, 393-405. https://doi.org/10.3390/j7030023
Yang Y, Kessler MGC, Marchán-Rivadeneira MR, Zhou Y, Han Y. Adeno-Associated Virus-Mediated CRISPR-Cas13 Knockdown of Papain-like Protease from SARS-CoV-2 Virus. J. 2024; 7(3):393-405. https://doi.org/10.3390/j7030023
Chicago/Turabian StyleYang, Yuehan, Mara Grace C. Kessler, M. Raquel Marchán-Rivadeneira, Yuxi Zhou, and Yong Han. 2024. "Adeno-Associated Virus-Mediated CRISPR-Cas13 Knockdown of Papain-like Protease from SARS-CoV-2 Virus" J 7, no. 3: 393-405. https://doi.org/10.3390/j7030023
APA StyleYang, Y., Kessler, M. G. C., Marchán-Rivadeneira, M. R., Zhou, Y., & Han, Y. (2024). Adeno-Associated Virus-Mediated CRISPR-Cas13 Knockdown of Papain-like Protease from SARS-CoV-2 Virus. J, 7(3), 393-405. https://doi.org/10.3390/j7030023





