Affinity and Correlation in DNA
Abstract
1. Introduction
- (a)
- There is a statistical difference between the coding and non-coding areas of DNA. The different statistical behavior of these two kinds of sequences can be used to build methods for identifying these areas of the DNA. For example, a fractal method to distinguish coding and non-coding sequences in a complete genome has been proposed [22]. This method shows that, following the thermodynamics formulation of multifractal measures, in a graphic of the analogous of specific heat, points of coding and non-coding sequences of many prokaryote genomes are roughly distributed in different regions.
- (b)
- This statistical difference is not due to the biological function of DNA. In general, any sequence of letters is quasi-random, and the regularities have nothing to do with the message conveyed, but only with specific properties of the used language. The same letter translated from English in Italian or in French will have other regularities (departures from randomness) due to the different letter usage of Latin origin languages with respect to English language. This happens every time when in the presence of a code.
2. Method
- DKL(P,Q) > 0, DKL(P,Q) ≠ DKL(Q,P) and DKL(Pi,Qi) = 0 if, and only if, Pi = Qi.
- The propagation of short-range correlations (affinities);
- The large amount of specific sequences in DNA, for example, the Alu sequence, which we will consider later; and
- The hydrogen or disulfide bridges in proteins that directly bind two distant sites in these sequences.
3. Results and Discussion
- Evolutionary reasons could provide information on past DNA and allow us to identify the laws of its evolution. The correlations, therefore, would be a consequence of such laws of genome evolution, and current genomes could be seen as a picture of the ongoing genome evolution process and, assuming that the symmetries on its composition derive from past genomes, could shed some light on the origin of life [46,47,48]. In this case, however, we must take into account that genomes show a great variability of constituent elements as consequence of their rates of mutation, genetic recombination events, horizontal gene transfers and gene losses or gains [49]. For example, two random DNA sequences can show up to 50% identical sequences when gaps are allowed [50,51,52]. As a result, many evolutionary analysis tools work well at short evolutionary distances, but only a few of them have worked well over longer time distances [53].
- The structural reasons explain how DNA is made. In this case, given that the structure of DNA is determined by physical and system constraints, the latter is dependent on its information content. These authors show that the composition of these codes lie at or around the local minima of the information function. The fact that codes do not evolve towards maximizing the information function leads these authors to assume that there is a mechanism that induces genetic codes to minimize information and that this mechanism is driving the evolution of this code. At some point in the evolution of living organisms, the number of constituents was blocked, and the cell began to develop a genetic code with non-random information content that corresponds to a trend towards one of the lows of the information function, or at least close to such state. This image agrees with most of the considerations about the correlation between the complex biological systems and non-random information content. The authors of [54] also suggest that based on the complexity of DNA sequences, a model for duplications of DNA sequences can be a fruitful approach to understanding long-range correlations.
- (a)
- All stacking energies of the dimers are negative, i.e., the dimers are, in all cases, more stable than the two isolated base pairs.
- (b)
- There are 11 dimers (ApA, ApC, ApG, CpC, CpT, GpA, GpG, GpT, TpA, TpC and TpT) with equal or similar stacking energy. For these dimers, in fact, we have an average stacking energy of 13.56 kcal/mol with a small dispersion in the range [13.22 ÷ 13.96].
- (c)
- There is only one dimer, ApT, with a stacking energy value lower than that of group (b) and, in particular, with a stacking energy of 11.66 kcal/mol.
- (d)
- The other four dimers, ApT, CpA (and TpG equal to it) and CpG, have stacking energies higher than that of group (b), with an average value of 16.62 kcal/mol, a dispersion in the range [15.95 ÷ 18.44 and limit values of CpA = TpG = 15.95 kcal/mol and CpG = 18.44 kcal/mol.
Second Chargaff Rule (SCR)
4. Conclusions
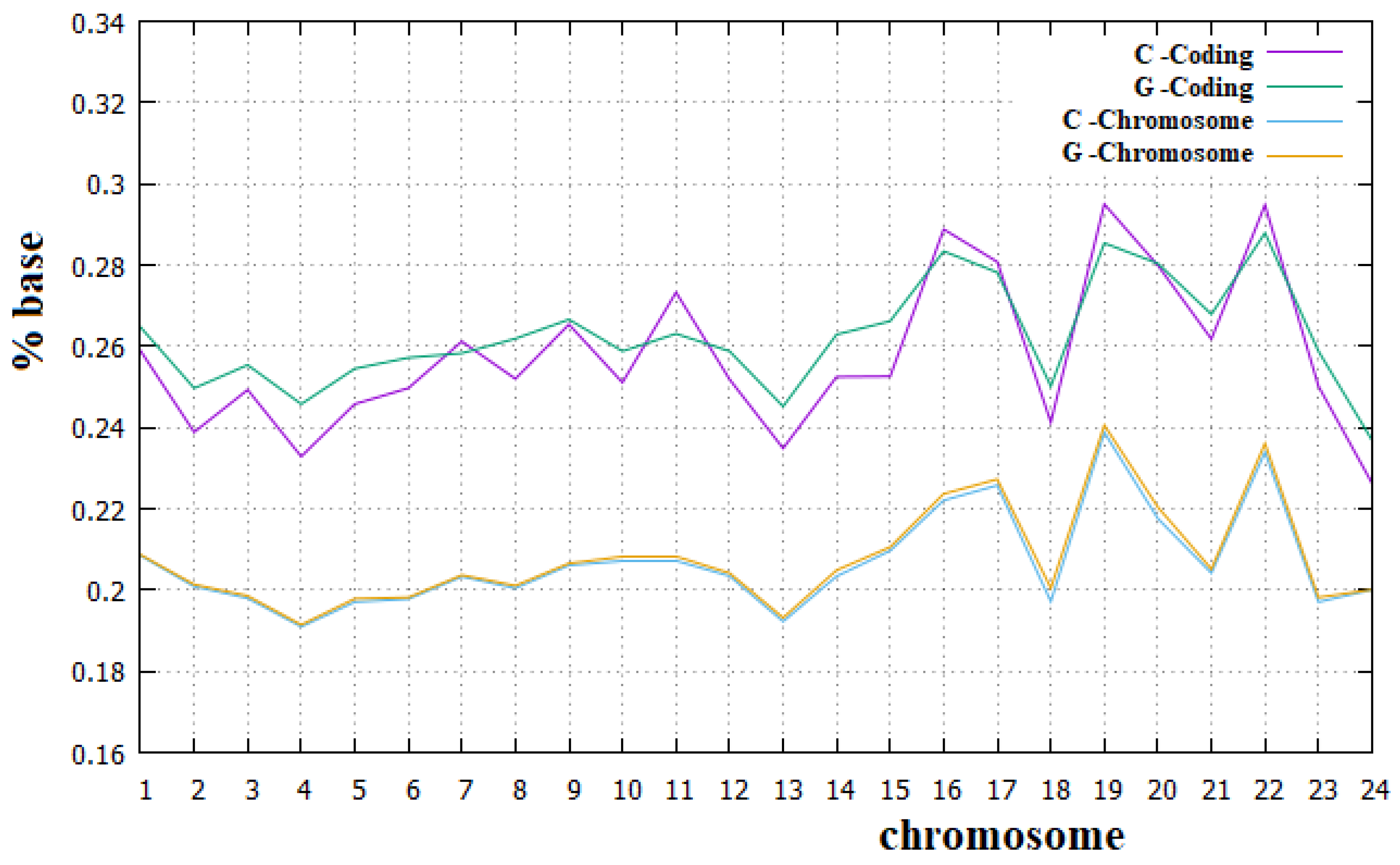
- The difference between human chromosome sequences and their coding parts is relevant both in the relationships between sites and in some specific compositional rules, such as the second Chargaff rule.
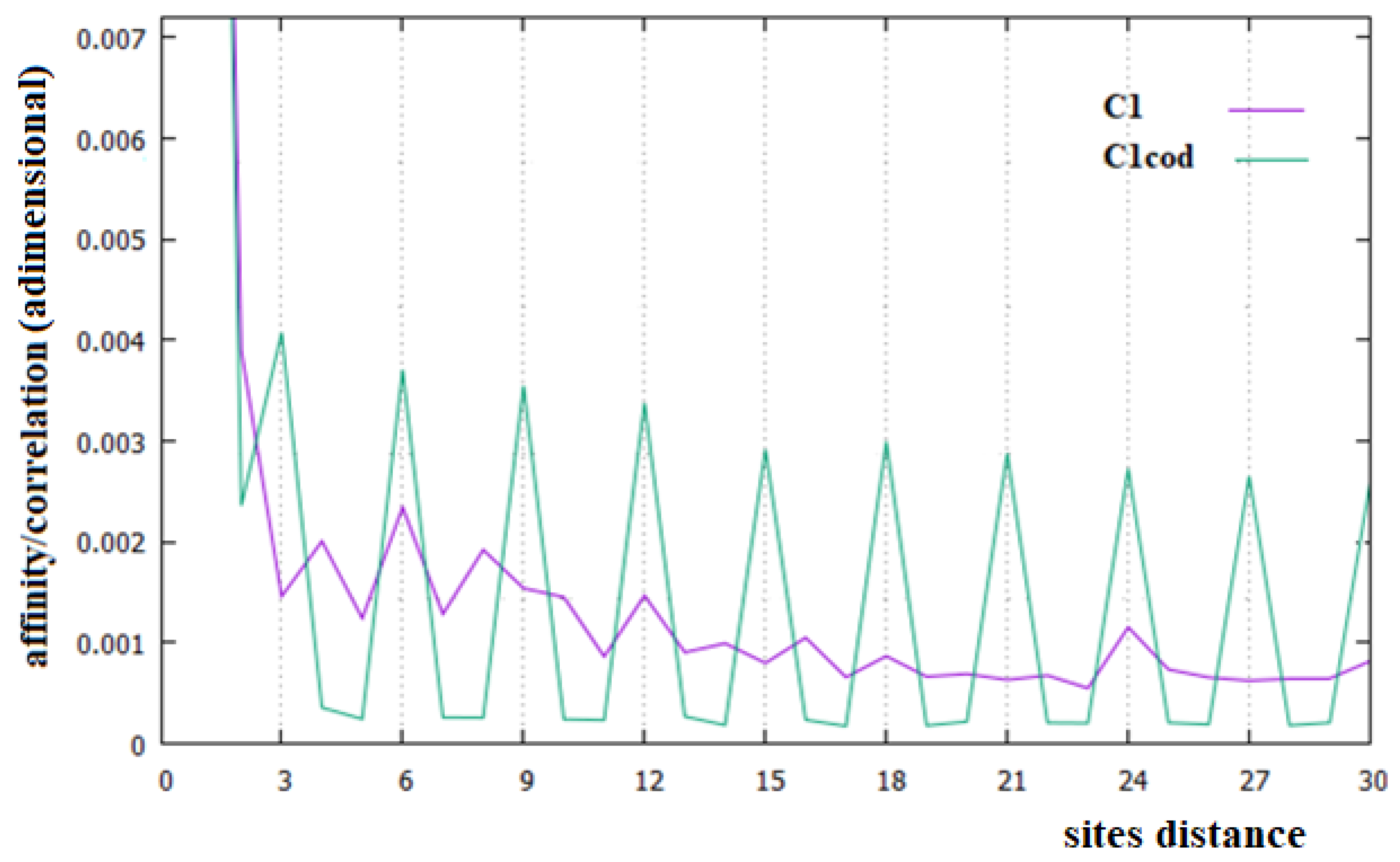
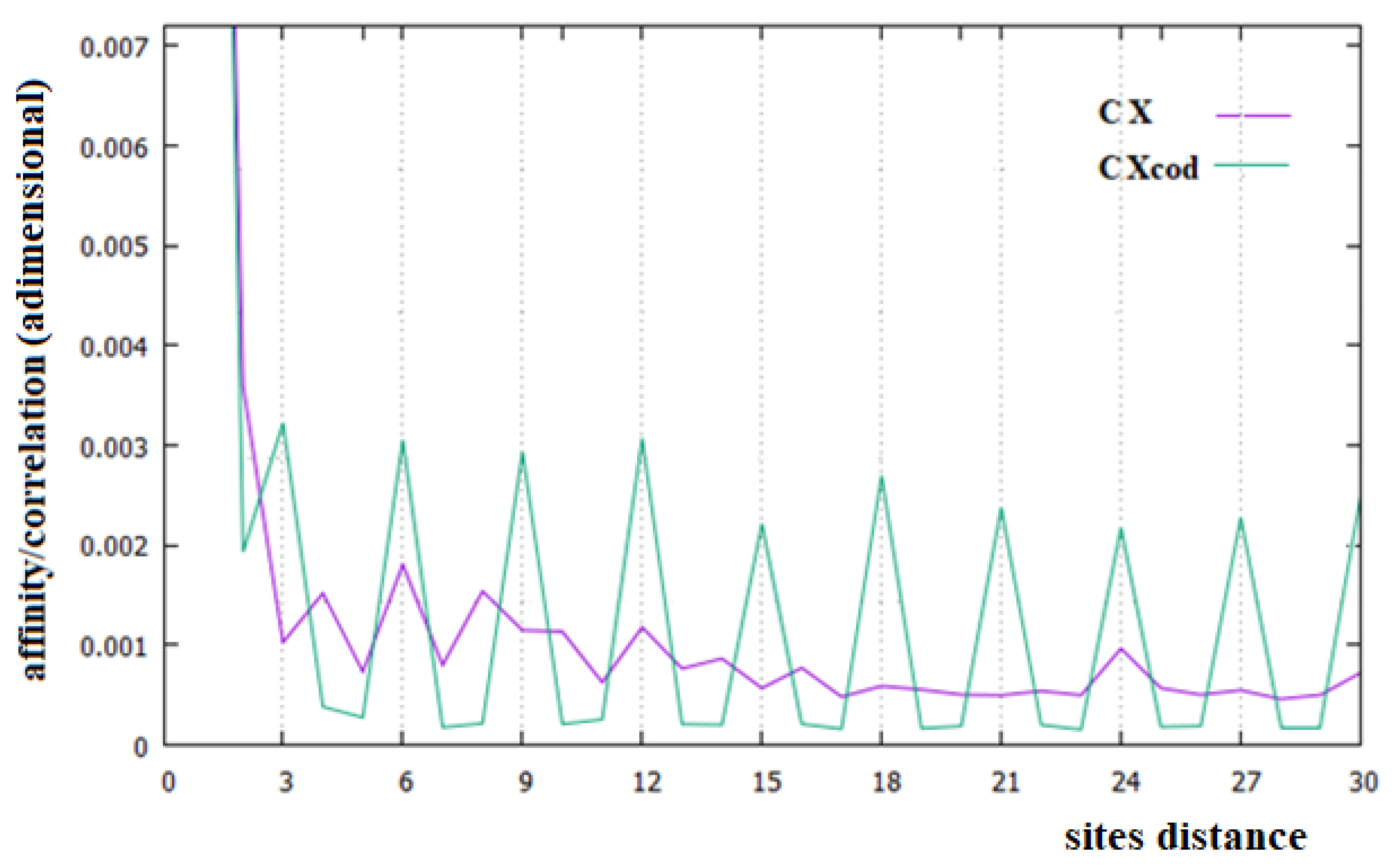
- The most important relationship between sites in all the DNA sequences examined is that between two consecutive base pairs, which indicates an energetic stabilization of these couples of base pairs due to the stacking interaction.
- The evidence of the relationship in two successive triplets of DNA-coding sequences demonstrates the existence of a relationship between two successive amino acids in proteins. This is obviously impossible if all the relationships between the sites of a macromolecule are statistical evidence and do not involve causes; in this article, due to stacking interactions and this relationship in coding sequences, we divided the concept of a relationship between sites into two concepts: affinity and correlation, the first with physical causes and the second without.
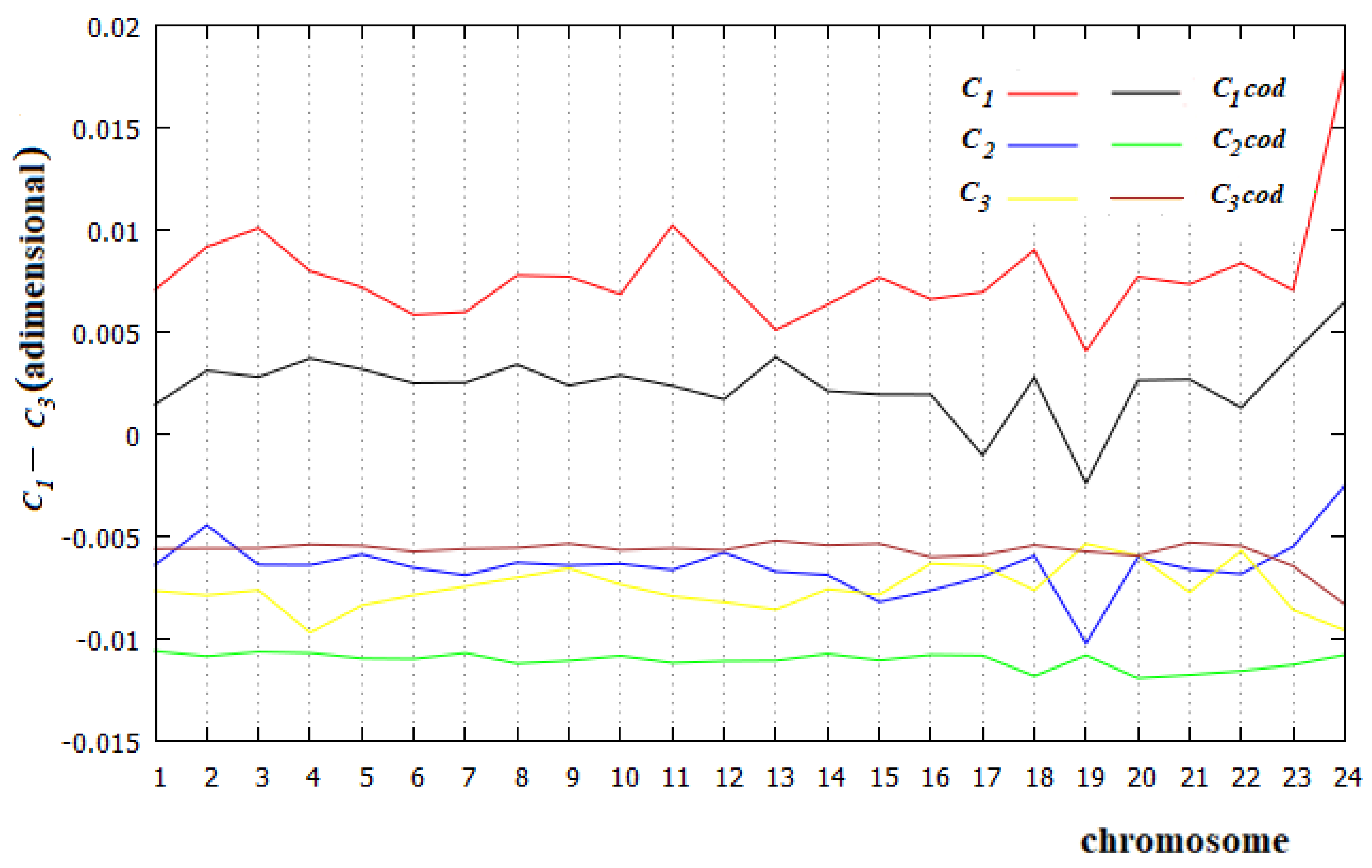
- The causal relationships, named in this paper affinity, of C1 in all chromosomes and of C3 in the related coding parts, are the most important relationships in DNA. This is in contrast to the current idea that all the relationships between sites of these macromolecules are only statistical evidence, but in perfect agreement with the results of [22,23]. These affinities may be due to different processes, but it is certainly also necessary to consider the energetic interaction between pairs of monomers, which is an interaction not considered in the literature.
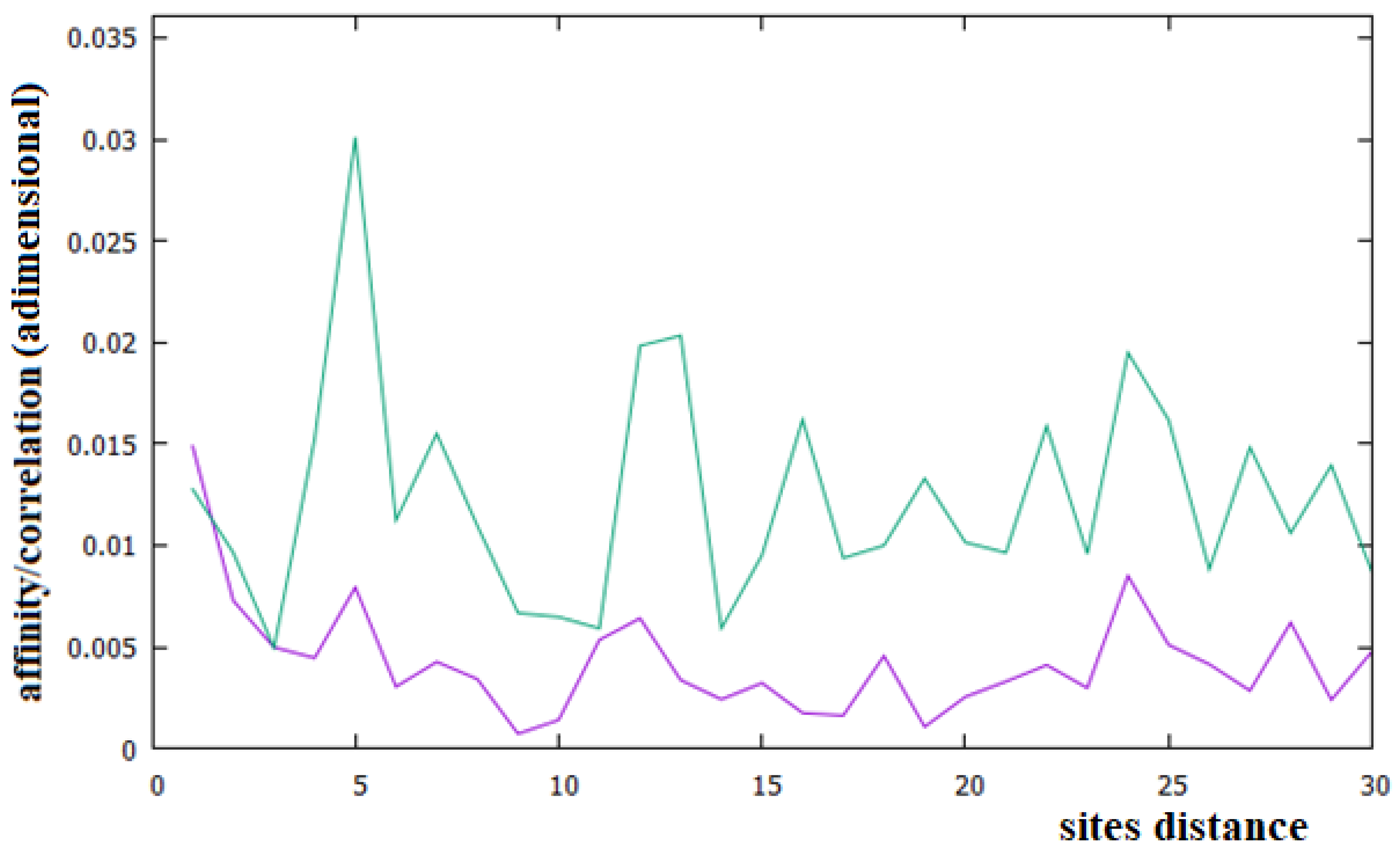
- In some large sequences with several Mbp, such as those of some non-coding parts of chromosomes, there is evidence of specific, long-rage correlations that may be related to several large replicate sequences (for example, the Alu sequence).
- The second Chargaff rule is substantially valid for all human chromosomes, but not for their coding parts.
Funding
Informed Consent Statement
Conflicts of Interest
Appendix A
| 1 | gccgggcgcg | gtggcgcgtg | cctgtagtcc | cagctactcg | ggaggctgag | gctggaggat |
| 61 | cgcttgagtc | caggagttct | gggctgtagt | gcgctatgcc | gatcgggtgt | ccgcactaag |
| 121 | ttcggcatca | atatggtgac | ctcccgggag | cgggggacca | ccaggttgcc | taaggagggg |
| 181 | tgaaccggcc | caggtcggaa | acggagcagg | tcaaaactcc | cgtgctgatc | agtagtggga |
| 241 | tcgcgcctgt | gaatagccac | tgcactccag | cctgggcaac | atagcgagac | cccgtctct |
References
- Ohno, S. So Much ‘Junk DNA’ in our Genome in Evolution of genetic systems. In Brookhaven Symposia in Biology; Smith, H.H., Ed.; Gordon and Breach: New York, NY, USA, 1972; Volume 23, pp. 366–370. [Google Scholar]
- Rowe, G.W.; Trainor, L.E.H. On the informational content of viral DNA. J. Theor. Biol. 1983, 101, 151–170. [Google Scholar] [CrossRef]
- Li, W.; Kaneko, K. Long-range correlation and partial 1=f spectrum in a noncoding DNA sequence. Europhys. Lett. 1992, 17, 655–660. [Google Scholar] [CrossRef]
- Peng, C.K.; Buldyrev, S.V.; Goldberger, A.L.; Havlin, S.; Sciortino, F.; Simons, M.; Stanley, H.E. Long-range correlations in nucleotide sequences. Nature 1992, 356, 168–170. [Google Scholar] [CrossRef] [PubMed]
- Nee, S. Uncorrelated DNA walks. Nature 1992, 357, 450. [Google Scholar] [CrossRef] [PubMed]
- Maddox, J. Long range correlations within DNA. Nature 1992, 358, 103–105. [Google Scholar] [CrossRef] [PubMed]
- Prabhu, V.V.; Claverie, J.M. Correlations in intronless DNA. Nature 1992, 359, 782–786. [Google Scholar] [CrossRef] [PubMed]
- Munson, P.J.; Taylor, R.C.; Michaels, G.S. DNA correlations. Nature 1992, 360, 636–640. [Google Scholar] [CrossRef]
- Lio, P.; Ruffo, S.; Buiatti, M. Third codon G+C periodicity as a possible signal for an internal selective constraint. J. Theor. Biol. 1992, 171, 215–223. [Google Scholar] [CrossRef]
- Buldyrev, S.V.; Goldberger, A.L.; Havlin, S.; Mantegna, R.N.; Matsa, M.E.; Peng, C.K.; Simons, M.; Stanley, H.E. Long range correlation properties of coding and noncoding DNA sequences. Phys. Rev. E 1995, 51, 5084–5091. [Google Scholar] [CrossRef]
- Arneodo, A.; Bacry, E.; Graves, P.V.; Muzy, J.F. Characterizing long-range correlations in DNA sequences from wavelet analysis. Phys. Rev. Lett. 1995, 74, 3293–3296. [Google Scholar] [CrossRef]
- Luo, L.F.; Tsai, L.; Zhou, Y.M. Informational parameters of nucleic acid and molecular evolution. J. Theor. Biol. 1988, 130, 351–361. [Google Scholar] [CrossRef]
- Luo, L.F.; Tsai, L. DNA walk and fractal analysis of nucleotide sequences. Acta Sci. Natur. Univ. Intramongolicae 1996, 27, 781–789. [Google Scholar]
- Voss, R.F. Evolution of long-range fractal correlations and 1/f noise in DNA base sequences. Phys. Rev. Lett. 1992, 68, 3805–3808. [Google Scholar] [CrossRef] [PubMed]
- Chechetkin, V.R.; Turygin, A.Y. The spectral criteria of disorder in nonperiodic sequences. J. Phys. A 1994, 27, 4875–4898. [Google Scholar] [CrossRef]
- Chechetkin, V.R.; Turygin, A.Y. Study of correlation in DNA sequence. J. Theor. Biol. 1996, 178, 205–217. [Google Scholar] [CrossRef]
- Luo, L.F.; Ji, F.M. The correlation spectrum of nucleotide sequences. How to extract signals from background noise? Acta Sci. Natur. Univ. Intramongolicae 1995, 26, 419–426. [Google Scholar]
- Luo, L.F.; Tsai, L. Fractal Dimension of Nucleic Acid Sequences and its Relation to Evolutionary Level. Chinese Phys. Lett. 1988, 5, 421–424. [Google Scholar]
- Gatlin, L. Information Theory and Living System; Columbia Univ. Press: New York, NY, USA, 1972. [Google Scholar]
- Luo, L.F.; Li, H. The statistical correlation of nucleotides in protein coding DNA sequences. Bull. Math. Biol. 1991, 53, 345–353. [Google Scholar] [CrossRef]
- Herzel, H.; Grosse, I. Measuring correlations in symbol sequences. Phys. A 1995, 216, 518–542. [Google Scholar] [CrossRef]
- Zhou, L.Q.; Yu, Z.G.; Deng, J.Q.; Anh, V.; Long, S.C. A fractal method to distinguish coding and non-coding sequences in a complete genome based on a number sequence representation. J. Theor. Biol. 2005, 232, 559–567. [Google Scholar] [CrossRef]
- Li, J.; Zhang, L.; Li, H.; Ping, Y.; Xu, Q.; Wang, R.; Tan, R.; Wang, Z.; Liu, B.; Wang, Y. Integrated entropy-based approach for analyzing exons and introns in DNA sequences. BMC Bioinform. 2019, 20, 283. [Google Scholar] [CrossRef] [PubMed]
- Šponer, J.; Leszczynski, J.; Hobza, P. Nature of Nucleic Acid-Base Stacking: Nonempirical Ab Initio and Empirical Potential Characterization of 10 Stacked Base Dimers. Comparison of Stacked and H-Bonded Base Pairs. J. Phys. Chem. 1996, 100, 5590–5596. [Google Scholar] [CrossRef]
- Hobza, P.; Šponer, J.; Polasek, M. H-Bonded and Stacked DNA-Base Pairs—Cytosine Dimer—An Ab-Initio 2nd-Order Møller-Plesset Study. J. Am. Chem. Soc. 1995, 117, 792–798. [Google Scholar] [CrossRef]
- Šponer, J.; Gabb, H.A.; Leszczynski, J.; Hobza, P. Base-Base and Deoxyribose-Base Stacking Interactions in B-DNA and Z-DNA: A Quantum-Chemical Study. Biophys. J. 1997, 73, 76–87. [Google Scholar] [CrossRef]
- Hobza, P.; Šponer, J. Structure, Energetics, and Dynamics of the Nucleic Acid Base Pairs: Nonempirical Ab Initio Calculations. Chem. Rev. 1999, 99, 3247–3276. [Google Scholar] [CrossRef]
- Parker, T.M.; Sherrill, C.D. Assessment of Empirical Models versus High-Accuracy Ab Initio Methods for Nucleobase Stacking: Evaluating the Importance of Charge Penetration. J. Chem. Theory Comput. 2015, 11, 4197–4204. [Google Scholar] [CrossRef]
- Poater, J.; Swart, M.; Bickelhaupt, F.M.; Fonseca Guerra, C. BDNA Structure and Stability: The Role of Hydrogen Bonding, π-π Stacking Interactions, Twist-Angle, and Solvation. Org. Biomol. Chem. 2014, 12, 4691–4700. [Google Scholar] [CrossRef]
- Hamlin, T.A.; Fonseca Guerra, J.; Bickelhaupt, F.M. B-DNA Model Systems in Non-Terran Bio-Solvents: Implications for Structure, Stability and Replication. Phys. Chem. Chem. Phys. 2017, 19, 16969–16978. [Google Scholar] [CrossRef]
- Řezáč, J.; Huang, Y.; Hobza, P.; Beran, G.J.O. Benchmark Calculations of Three-Body Intermolecular Interactions and the Performance of Low-Cost Electronic Structure Methods. J. Chem. Theory Comput. 2015, 11, 3065–3079. [Google Scholar] [CrossRef]
- Mahadevi, A.S.; Sastry, G.N. Cooperativity in Noncovalent Interactions. Chem. Rev. 2016, 116, 2775–2825. [Google Scholar] [CrossRef]
- Parker, T.M.; Hohenstein, E.G.; Parrish, R.M.; Hud, N.V.; Sherrill, C.D. Quantum-Mechanical Analysis of the Energetic Contributions to π Stacking in Nucleic Acids versus Rise, Twist, and Slide. J. Am. Chem. Soc. 2013, 135, 1306–1316. [Google Scholar] [CrossRef] [PubMed]
- Villani, G. Theoretical Investigation of the Coupling between Hydrogen-Atom Transfer and Stacking Interaction in Adenine–Thymine Dimers. Chem. Phys. Chem. 2013, 14, 1256–1263. [Google Scholar] [CrossRef] [PubMed]
- Villani, G. Theoretical investigation of the coupling between hydrogen atoms transfer and stacking interaction in guanine–cytosine dimers. Phys.Chem. Chem. Phys. 2013, 15, 19242–19252. [Google Scholar] [CrossRef] [PubMed]
- Villani, G. Coupling Between Hydrogen Atoms Transfer and Stacking Interaction in Adenine-Thymine/Guanine-Cytosine Complexes: A Theoretical Study. J. Phys. Chem. B 2014, 118, 5439–5452. [Google Scholar] [CrossRef] [PubMed]
- Kruse, H.; Banás, P.; Šponer, J. Investigations of Stacked DNA Base-Pair Steps: Highly Accurate Stacking Interaction Energies, Energy Decomposition, and Many-Body Stacking Effects. J. Chem. Theory Comput. 2019, 15, 95–115. [Google Scholar] [CrossRef] [PubMed]
- Kullback, S.; Leibler, R.A. On Information and Sufficiency. Ann. Math. Stat. 1951, 22, 79–86. [Google Scholar] [CrossRef]
- Chang, C.H.; Hsieh, L.C.; Chen, T.Y.; Chen, H.D.; Luo, L.; Lee, H.C. Shannon information in complete genomes. J. Bioinform. Comput. Biol. 2005, 3, 587–608. [Google Scholar] [CrossRef]
- Bouaynaya, N.; Schonfeld, D. Nonstationary Analysis of Coding and Noncoding Regions in Nucleotide Sequences. IEEE J. Sel. Top. Signal Processing 2008, 2, 357–364. [Google Scholar] [CrossRef]
- Tsallis, C. Possible generalization of Boltzmann-Gibbs statistics. J. Stat. Phys. 1988, 52, 479–487. [Google Scholar] [CrossRef]
- Guerra, J.C.O.; Licinio, P.; Andrade, P.C.P. Comparison of period-3 correlation amplitudes in genomic DNA sequences. Genet. Mol. Res. 2013, 12, 4459–4467. [Google Scholar] [CrossRef]
- Batzer, M.A.; Deininger, P.L. Alu repeats and human genomic diversity. Nat. Rev. Genet. 2002, 3, 370–379. [Google Scholar] [CrossRef] [PubMed]
- Deininger, P. Alu elements: Know the SINEs. Genome Biol. 2011, 12, 236. [Google Scholar] [CrossRef] [PubMed]
- Perez, J.C. Codon Populations in Single-stranded Whole Human Genome DNA. Are Fractal and Fine-tuned by the Golden Ratio 1.618. Interdiscip. Sci. Comput. Life Sci. 2010, 2, 228–240. [Google Scholar] [CrossRef] [PubMed]
- Sobottka, M.; Hart, A.G. A model capturing novel strand symmetries in bacterial DNA. Biochem. Biophys. Res. Commun. 2011, 410, 823–828. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Zhang, S.-H.; Huang, Y.-Z. Limited contribution of stem-loop potential to symmetry of single-stranded genomic DNA. Bioinformatics 2010, 26, 478–485. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Y.; Mishra, B. Models of Genome Evolution. In Modeling in Molecular Biology; Ciobanu, G., Rozenberg, G., Eds.; Springer: Berlin, Germany, 2004; pp. 287–304. [Google Scholar]
- Zielezinski, A.; Vinga, S.; Almeida, J.; Karlowski, W.M. Alignment-free sequence comparison: Benefits, Applications, and Tools. Genome Biol. 2017, 18, 186. [Google Scholar] [CrossRef]
- Eddy, S.R. Where did the BLOSUM62 alignment score matrix come from? Nat. Biotechnol. 2004, 22, 1035–1036. [Google Scholar] [CrossRef]
- Gardner, P.P.; Wilm, A.; Washietl, S. A benchmark of multiple sequence alignment programs upon structural RNAs. Nucleic Acids Res. 2005, 33, 2433–2439. [Google Scholar] [CrossRef]
- Capriotti, E.; Marti-Renom, M.A. Quantifying the relationship between sequence and three-dimensional structure conservation in RNA. BMC Bioinform. 2010, 11, 322. [Google Scholar] [CrossRef]
- Earl, D.; Nguyen, N.; Hickey, G.; Harris, R.S.; Fitzgerald, S.; Beal, K.; Seledtsov, I.; Molodtsov, V.; Raney, B.J.; Clawson, H.; et al. Alignathon: A Competitive Assessment of Whole-Genome Alignment Methods. Genome Res. 2014, 24, 2077–2089. [Google Scholar] [CrossRef]
- Li, W.; Marr, T.G.; Kaneko, K. Understanding long-range correlations in DNA sequences. Phys. D 1994, 75, 392–416. [Google Scholar] [CrossRef]
- Chargaff, E. Structure and function of nucleic acids as cell constituents. Fed. Proc. 1951, 10, 654–659. [Google Scholar] [PubMed]
- Chargaff, E.; Lipshitz, R.; Green, C. Composition of the deoxypentose nucleic acids of four genera of seaurchin. J. Biol. Chem. 1952, 195, 155–160. [Google Scholar] [CrossRef]
- Rudner, R.; Karkas, J.D.; Chargaff, E. Separation of B. subtilis DNA into complementary strands, III. Proc. Natl. Acad. Sci. USA 1968, 60, 921–922. [Google Scholar] [CrossRef] [PubMed]
- Chargaff, E. Preface to a Grammar of Biology: A Hundred Years of Nucleic Acid Research. Science 1971, 172, 637–642. [Google Scholar] [CrossRef]
- Mitchell, D.; Bridge, R. A test of Chargaff’s second rule. Biochem. Biophys. Res. Commun. 2006, 340, 90–94. [Google Scholar] [CrossRef]
- Albrecht-Buehler, G. Asymptotically increasing compliance of genomes with Chargaff’s second parity rules through inversions and inverted transpositions. Proc. Natl. Acad. Sci. USA 2006, 103, 17828–17833. [Google Scholar] [CrossRef]
- Albrecht-Buehler, G. Inversions and inverted transpositions as the basis for an almost universal “format” of genome sequences. Genomics 2007, 90, 297–305. [Google Scholar] [CrossRef]
- Baisnee, P.F.; Hampson, S.; Baldi, P. Why are complementary DNA strands symmetric? Bioinformatics 2002, 18, 1021–1033. [Google Scholar] [CrossRef]
- Bell, S.J.; Forsdyke, D.R. Deviations from Chargaff’s second parity rule correlate with direction of transcription. J. Theor. Biol. 1999, 197, 63–76. [Google Scholar] [CrossRef]
- Chen, L.; Zhao, H. Negative correlation between compositional symmetries and local recombination rates. Bioinformatics 2005, 21, 3951–3958. [Google Scholar] [CrossRef] [PubMed]
- Fickett, J.W.; Torney, D.C.; Wolf, D.R. Base compositional structure of genomes. Genomics 1992, 13, 1056–1064. [Google Scholar] [CrossRef]
- Forsdyke, D.R. Relative roles of primary sequence and (G+C)% in determining the hierarchy of frequencies of complementary trinucleotide pairs in DNAs of different species. J. Mol. Evol. 1995, 41, 573–581. [Google Scholar] [CrossRef] [PubMed]
- Forsdyke, D.R.; Bell, S.J. Purine loading, stemloops and Chargaff’s second parity rule. Appl. Bioinform. 2004, 3, 3–8. [Google Scholar] [CrossRef]
- Hart, A.; Martínez, S. Statistical testing of Chargaff’s second parity rule in bacterial genome sequences. Stoch. Models 2011, 27, 272–317. [Google Scholar] [CrossRef]
- Hart, A.; Martínez, S.; Olmos, F. A Gibbs approach to Chargaff’s second parity rule. J. Stat. Phys. 2012, 146, 408–422. [Google Scholar] [CrossRef]
- Kong, S.G.; Fan, W.L.; Chen, H.D.; Hsu, Z.T.; Zhou, N.; Zheng, B.; Lee, H.C. Inverse symmetry in complete genomes and whole-genome inverse duplication. PLoS ONE 2009, 4, e7553. [Google Scholar] [CrossRef]
- Lobry, J.R.; Lobry, C. Evolution of DNA base composition under no-strand-bias conditions when the substitution rates are not constant. Mol. Bio. Evol. 1999, 16, 719–723. [Google Scholar] [CrossRef]
- Lobry, J.R.; Sueoka, N. Asymmetric directional mutation pressures in bacteria. Genome Biol. 2002, 3, research0058.1. [Google Scholar] [CrossRef]
- Mascher, M.; Schubert, I.; Scholz, U.; Friedel, S. Patterns of nucleotide asymmetries in plant and animal genomes. Biosystems 2013, 111, 181–189. [Google Scholar] [CrossRef]
- Okamura, K.; Wei, J.; Scherer, S.W. Evolutionary implications of inversions that have caused intra-strand parity in DNA. BMC Genom. 2007, 8, 160–166. [Google Scholar] [CrossRef] [PubMed]
- Powdel, B.R.; Satapathy, S.S.; Kumar, A.; Jha, P.K.; Buragohain, A.K.; Borah, M.; Ray, S.K. A study in entire chromosomes of violations of the intra-strand parity of complementary nucleotides (Chargaff’s second parity rule). DNA Res. 2009, 16, 325–343. [Google Scholar] [CrossRef] [PubMed]
- Rapoport, A.E.; Trifonov, E.N. Compensatory nature of Chargaff’s second parity rule. J. Biomol. Struct. Dyn. 2013, 31, 1324–1336. [Google Scholar] [CrossRef] [PubMed]
- Sueoka, N. Intrastrand parity rules of DNA base composition and usage biases of synonymous codons. J. Mol. Evol. 1995, 40, 318–325. [Google Scholar] [CrossRef]
- Wang, S.; Tu, J.; Jia, Z.; Lu, Z. High order intrastrand partial symmetry increases with organismal complexity in animal evolution. Sci. Rep. 2014, 4, 6400. [Google Scholar] [CrossRef]
- Zhang, S.H.; Huang, Y.Z. Characteristics of oligonucleotide frequencies across genomes: Conservation versus variation, strand symmetry, and evolutionary implications. Nat. Preced. 2008. [Google Scholar] [CrossRef]
- Zhang, H.; Li, P.; Zhong, H.S.; Zhang, S.H. Conservation vs. variation of dinucleotide frequencies across bacterial and archaeal genomes: Evolutionary Implications. Front. Microbiol. 2013, 4, 269–276. [Google Scholar] [CrossRef]
- Rosandić, M.; Vlahović, I.; Glunčić, M.; Paar, V. Trinucleotide’s quadruplet symmetries and natural symmetry law of DNA creation ensuing Chargaff’s second parity rule. J. Biomol. Struct. Dyn. 2016, 34, 1383–1394. [Google Scholar] [CrossRef]



| A | C | G | T | |
|---|---|---|---|---|
| A | 0.0223 | −0.0406 | 0.0377 | −0.0194 |
| C | 0.0503 | 0.0406 | −0.1415 | 0.0506 |
| G | 0.0320 | 0.0031 | 0.0050 | −0.0401 |
| T | −0.1162 | 0.0049 | 0.0960 | 0.0153 |
| A | T | C | G | |
|---|---|---|---|---|
| A | 0.3005 | 0.2621 | 0.2120 | 0.2254 |
| T | 0.2841 | 0.2764 | 0.1929 | 0.2466 |
| C | 0.2200 | 0.1850 | 0.2548 | 0.3402 |
| G | 0.1915 | 0.1936 | 0.3314 | 0.2835 |
| A | T | C | G | |
|---|---|---|---|---|
| A | 0.3265 | 0.2553 | 0.1729 | 0.2452 |
| T | 0.2164 | 0.3279 | 0.2059 | 0.2498 |
| C | 0.3489 | 0.3422 | 0.2594 | 0.0494 |
| G | 0.2878 | 0.2417 | 0.2109 | 0.2596 |
| A | T | C | G | |
|---|---|---|---|---|
| A | 0.3045 | 0.2098 | 0.1983 | 0.2874 |
| T | 0.1661 | 0.2445 | 0.2438 | 0.3456 |
| C | 0.3325 | 0.2798 | 0.2795 | 0.1082 |
| G | 0.3142 | 0.1891 | 0.2420 | 0.2547 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Villani, G. Affinity and Correlation in DNA. J 2022, 5, 214-231. https://doi.org/10.3390/j5020016
Villani G. Affinity and Correlation in DNA. J. 2022; 5(2):214-231. https://doi.org/10.3390/j5020016
Chicago/Turabian StyleVillani, Giovanni. 2022. "Affinity and Correlation in DNA" J 5, no. 2: 214-231. https://doi.org/10.3390/j5020016
APA StyleVillani, G. (2022). Affinity and Correlation in DNA. J, 5(2), 214-231. https://doi.org/10.3390/j5020016






