1. Introduction
Nowadays, with the fast advance in technology and medicine, new chemicals arise and become extensively used worldwide; these are called emerging pollutants. These compounds are recalcitrant and systematically enter into the environment via conventional treatment processes, in which their degradation is frequently incomplete [
1,
2,
3]. Much remains to be explored about the characteristics of these pollutants and their behaviour, real toxicity, and adverse/hazardous effects in the environment, which are still unknown and are also not covered by legislation.
This study is focused on 17α-ethinylestradiol (EE2), an emerging pollutant considered one of the most hazardous compounds for aquatic and terrestrial organisms [
3]. EE2, an artificial hormone derived from the natural estrogen 17β-estradiol, is commonly utilized as a constituent of oral contraceptives [
4]. Recognized as an endocrine-disrupting agent, it has been newly added to the EU Watch List under the Water Framework Directive [
5], addressing emerging contaminants in aquatic environments.
Tang and colleagues [
6] investigated the presence of EE2 in several municipal Waste Water Treatment Plants (WWTPs) across 29 countries and reported that its mean concentrations were 78.4 and 12.3 ng/L in the influents and effluents, respectively. The authors observed that the mean effluent concentration of EE2 exceeded the registered lowest-observed-effect concentration by over 61 times, emphasizing the critical necessity of eliminating EE2 from WWTPs [
6].
The highest levels of pollution with EE2 are observed in areas with high human population density, hospital and pharmaceutical industrial activities, and agricultural and aquaculture practices [
6]. Data from several studies indicated that EE2 is not completely removed by conventional wastewater treatment processes due to inadequate systems, leading to surface waters contamination [
6,
7,
8,
9,
10]. Tang et al. [
6] found evidence that EE2 concentrations in surface waters were notably higher in developing countries compared to developed ones. Bhandari et al. [
11] reported concentrations of EE2 in similar aquatic surface resources ranging from 0.2 to 1.5 ng/L [
11].
More recent research studies have further demonstrated the presence of EE2 in surface waters, particularly in drinking water. Tang and colleagues [
6] ranked the top 10 countries based on the average concentration of EE2 in surface water. Notably, they found that Vietnam, Cambodia, and China were the three most polluted countries, with concentrations of 27.7 ng/L, 22.1 ng/L, and 21.5 ng/L of EE2, respectively. On the other hand, Portugal, with a concentration of 6.6 ng/L of EE2, had the lowest among the listed countries. In Europe, Tang and collaborators [
6] observed that France has fewer contaminated surface waters, while Portugal has the most contamination with EE2. However, both countries had an average concentration of over 1 ng/L of EE2 [
6]. For example, Rocha and co-authors [
12] detected an approximate EE2eq concentration of 50 ng/L in the Ria de Aveiro lagoon, recognized for its ecological and economic significance in Portugal. Additionally, Ribeiro and team [
13] identified EE2 concentrations reaching as high as 101.9 ng/L in the Douro River estuary. Sodré and Sampaio [
14] found 4.47 ng/L of EE2 in the Paranoa Lake, Brazil, which is an artificial reservoir, while Bradley and collaborators [
15] reported a concentration of 1.4 ng/L in Chicago, IL, USA.
EE2’s environmental contamination of hydric resources is escalating each day, posing an increasingly severe threat to both aquatic and terrestrial organisms. To safeguard the natural aquatic ecosystems, the European Commission has advocated a significantly more stringent average quality standard of 0.035 ng/L for aquatic surfaces [
6]. It is imperative to pursue more efficient and economical methods for its removal [
16], particularly through the utilization of biological approaches.
Several studies have reported that environmental contamination with EE2, even at concentrations as low as ng/L, exerts significant hazardous (ecotoxicological) acute and chronic effects, at the cellular, whole-organism, and population level, on organisms, particularly on animals. Additionally, this drug may bioaccumulate. The endocrine-disrupting effects caused by EE2 encompass behavioural alterations, a decrease in growth rate, and reproductive dysregulation, culminating in feminization phenomena. Moreover, EE2 exposure has been associated with oxidative stress, lipid peroxidation disruptions in the regulation of both proapoptotic and antiapoptotic processes, and there is a potential for inducing neoplastic mechanisms and genotoxic damage [
17,
18,
19].
According to EE’s kinetic reaction rate (k
biol) of 7–9 L/(gSS.day) and its octanol–water partition coefficient (logK
ow) of 2.8–4.2, it could be predicted that the removal of the drug may occur mainly by biodegradation and moderately by adsorption [
6,
9,
20].
Biodegradation is a sustainable, low-cost solution and one of the main processes by which bacteria can convert/metabolize these hazardous organic pollutants, generally leading to their mineralization [
21].
Numerous metabolic products were detected from the breakdown of EE2; an example is
Sphingobacterium sp. JCR5, a bacterium that utilizes EE2, converting it, in a first step, into ketone, estrone (E1) by the oxidization of C-17 of EE2. Subsequently, the hydroxylation and ketonization of C-9 of E1 occurs, with further cleavage of the B ring of E1. After the hydroxylation of the A ring of E1, it yields the compounds 3,4-dihydroxy-9,10-secoandrosta-1,3,5(10)-triene-9,17-dione, 2-hydroxy-2,4-dienevaleric acid, and 2-hydroxy-2,4-diene-1,6-dioic acid. Some successive breakdown reactions, involving the meta-cleavage of the A ring through dioxygenase, result in the generation of final byproducts, CO
2, and water [
22]. The mass spectrum interpretation during the breakdown of EE2 revealed that, in the initial stage, the drug undergoes oxygenation to form E1, 2-hydroxy-2,4-dienevaleric acid and 2-hydroxy-2,4-diene-1,6-dioic acid, which are key metabolic breakdown intermediates. The initial pathway was similar to one formerly documented for a bacterium,
Comamonas testosteroni TA441, which is known for degrading testosterone. The latter represents a metabolic byproduct with an evident cleavage position of 3-hydroxy-4,5-9,10-disecoestrane-1(10),2-diene-5,9,17-trione-4-oic acid, differing from the previous pathway.
Bioremediation processes are crucial in wastewater treatment. Understanding the composition, diversity, and degrading abilities of specialized bacterial communities is essential [
23]. Hence, the importance of finding effective bioremediation processes, which can be obtained through the use of organisms with excellent capabilities to degrade these recalcitrant compounds, is highlighted. With this goal in mind, this study focuses on the search for bacteria with unique capabilities and enzymatic activities, particularly those found in extreme environments, such as mining sites.
Mine caves and their acid mine drainage (AMD) wastes are considered extreme environments/habitats since it is very difficult for organisms to survive under their severe conditions, such as for example, very low pH, high/low temperatures, high concentrations of toxic metals, lack of light, oxygen or water, radiation, and accessibility to different energy sources [
24,
25,
26].
In the case of AMD, this is an extreme form of water pollution that is the consequence of the natural action of mining activities in which sulphur-bearing minerals (mostly iron sulphides) are exposed to air (oxygen), humidity, and acidophilic iron-oxidizing bacteria. This exposure leads to the generation of sulfuric acid, iron dissolution, and the precipitation of ferric ions. The sulfuric acid originates an acidic pH solution which dissolves heavy metals from mined materials, producing a water with high concentrations of metals such as aluminium, zinc, copper, arsenic, cadmium, lead, etc. [
27,
28]. Therefore, AMD is characterized by special hydro chemical and ecological features in which a low biodiversity, mainly comprising acidophilic organisms, is able to survive.
The bacteria isolated in mining extreme environments can be important for the biodegradation of emerging pollutants, for example, in bioaugmentation processes, at low pH and increased metals and sulphate concentrations, but also at neutral pH. For example, polycyclic aromatic hydrocarbons (PAH) contamination often occurs on acidic environments which occasionally have high temperatures, such as AMD basins [
29].
The aim of this study is focused on the importance of finding bacterial consortia and isolates from extreme environments, such as mines and their AMD, to be used in bioremediation processes, in this case focused on the degradation of EE2. Although contamination with pharmaceuticals, namely EE2, is not expected in the mine samples studied here, the microorganisms recovered from these less explored environments may have adapted metabolic enzymes/pathways/mechanisms allowing them to completely degrade these compounds into less harmful ones or even to mineralize them.
Therefore, the putative products of EE2 bacterial biodegradation and their respective putative metabolic mechanisms were also analysed. For this purpose, bacterial consortia were retrieved from samples obtained from mines and AMD.
The bacterial communities under study were pre-selected through a screening process, identifying those with the capacity to degrade paracetamol. Paracetamol was considered a model pharmaceutical due to its chemical composition and structure, comprising a benzene-aromatic ring, an amide group, and a hydroxyl group. It was chosen for its considerable solubility in water (14 g/L at 25 °C) and its widespread usage and detection as one of the most extensively found drugs in wastewater and all hydric resources [
30].
Two strains of Aeromonas sp. MLN-TP5 and Aeromonas salmonicida MLN-TP7 were isolated from the consortia recovered from AMD, and they were found to efficiently degrade EE2 and its metabolites. As far as is known, Aeromonas salmonicida was recovered and identified for the first time in this study in AMD, a harsh environment. It displays the ability to degrade EE2, making this bacterium an excellent candidate for bioaugmentation processes, potentially even in low pH environments.
2. Materials and Methods
2.1. Sampling (Inoculum Source)
Mine samples were collected in December 2019 from two inactive mines: Lousal mine, located in Grândola, Alentejo, Portugal (38°2′5″ N, 8°25′23″ W) belonging to the Iberian Pyrite Belt’s in the northwest (NW) sector, within a region confined to the north (N), east (E), and south (S) by the Tertiary sediments of the Alvalade basin; and Poderosa mine, located in El Campillo, Huelva at approximately 8 km NW of the well-known Rio Tinto mining region located in the western part of Andalusia, Spain (
Figure 1).
A scientific staff collected samples of mine rocks, sediments, and acid mine drainage (AMD) of the Lousal mine in Grândola (Portugal) and the Poderosa mine in Huelva (Spain). Sampling was carried out by scraping approximately 6 and 10 g of sediments and rocks and approximately 40 mL of AMD. Samples were stored in 50 mL falcon tubes. After collection, samples were shipped from Lousal and Poderosa to the Gambelas campus (University of Algarve) in a cool box to keep a constant temperature. The procedure of enriching bacterial consortia was initiated rapidly upon the arrival of the samples. Sediments and AMD samples were stored in a refrigerator at 4 °C, while bacterial consortia samples were stored at −80 °C.
2.2. Bacterial Consortia Recovery from Mine Sediments and AMD
Bacterial communities were recovered from samples collected in the walls and floor sediments of the diverse sites of Poderosa and Lousal mines, as well as from acidic lagoon water, denominated acid mine drainage (AMD), from the Lousal mine.
The mineralogic characterisation of the solid (wall and floor sediments) samples was performed by X-ray diffraction (XRD) employing a PANalytical X’Pert Pro powder diffractometer, following the procedures outlined in the protocol detailed by Palma et al. [
31].
For AMD characterisation, both redox potential (Eh) and pH were determined immediately after sample harvesting by employing a pH/Eh Meter (GLP 21, Crison Hach Lange, Barcelona, Spain). Subsequently, an aliquot of the AMD sample was centrifuged at 4000 rpm for 6 min; the liquid fraction was subsequently moved to a new container and acidified with HNO3 at 6 M. Sulphate concentration was determined utilising the sulfaVer4 method (Hach-Lange, Sköndal, Sweden) at 450 nm, employing a UV–visible spectrophotometer (DR 2800, Hach-Lange).
The metals concentration (Zn, Fe, Cu, Mn and Na) was analysed using flame atomic absorption spectroscopy with an Analytic Jena NovAA 350 model spectrometer. The aluminium concentration in selected samples was determined via UV/Visible spectroscopy at 522 nm (Hach-Lange, Sköndal, Sweden) utilising the AluVer3 procedure (Hach-Lange, Sköndal, Sweden). Three measurements were recorded for each sample.
For bacterial communities’ recovery, the samples collected from Lousal and Poderosa mines were washed with 9 mL of Ringer’s solution in 50 mL glass flasks containing 1 g of sample, subjected to orbital stirring at 150 rpm for 4 h. After, 500 µL of the previous solution was transferred to a 50 mL glass bottle with 5 mL of Nutrient Broth and underwent incubation at a temperature of 25 °C for a period ranging from 24 to 48 h. Following the incubation, bacterial cultures were maintained in glycerol (20% v/v) kept at −80 °C.
2.3. Bacterial Inocula Enrichment
The inoculum enrichments were carried out using the bacterial cultures previously prepared from the samples collected from mines.
The recovered bacterial consortia’s initial cultures were reactivated by utilising 100 μL of glycerol cultures in 1 mL of Luria Bertani Broth (LB) for 24 h at ambient temperature (23 °C–28 °C), subjected to 150 rpm in the absence of light conditions. To enhance the growth of biomass, 1 mL of the preceding culture was extracted and introduced into 9 mL of LB to finalise the activation procedure. To attain sufficient growth, the samples were placed in the absence of light and incubated at 28 °C for 24 h.
Nineteen bacterial communities recovered from samples collected at Lousal mine and six bacterial consortia obtained from samples at Poderosa mine were used for the screening procedure in the presence of paracetamol, serving as the model compound. For the enrichment process, subcultures were set up in sterile glass bottles with 90 mL of LB and 10 mL (10% v/v) of the original culture, both with and without the addition of 50 and 100 mg/L paracetamol. The incubation took place at an ambient temperature (23 °C–28 °C), during which subcultures were subjected to 150 rpm in darkness for 48 h.
The bacterial consortia that displayed the best growth performance in the presence of paracetamol were named as LF, LN, LAC, and PB, PF, PDE, starting with L of Lousal and P of Poderosa mines as a reference to the place where the samples were collected, followed by letters of the alphabet, respectively.
In all assays, bacterial growth was assessed by monitoring the optical density at 600 nm (OD600) at the beginning of the experiment and throughout the incubation period, using a Hach-Lange spectrophotometer DR-2800 (Sköndal, Sweden).
2.4. EE2 Bioremoval Studies with Bacterial Communities Obtained from Mines
Following the enrichment in LB in the presence of paracetamol, the bioremoval experiments were carried out utilising as inoculum 10% (
v/
v) of every single bacterial community in a Mineral Salt Medium (MSM) made as described in Palma et al. [
30], with 60 mg/L of paracetamol as the unique carbon source. A bacterial consortium sourced from a wastewater treatment plant sludge from a WWTP was used as a positive control. To assess the ability of bacterial consortia to biodegrade EE2, three communities (LF, LN, LAC) recovered from Lousal mine, and three communities (PB, PF, PDE) from Poderosa mine, each exhibiting good paracetamol removal capabilities, were individually tested, and then combined to form two new consortia designated as LACFN and PBDEF, respectively.
A new inoculum enrichment in LB in the presence of 10 mg/L of EE2 was performed.
To prepare the culture medium the following steps were carried out. Stock solutions of 1000 mg/L of EE2 were prepared in volumetric flasks of 100 mL, as follows: 0.100 g of 17α-ethinylestradiol was dissolved in methanol and the volume was completed with it. For example, to achieve a concentration of 9 mg/L of EE2 in 500 mL of MSM, 4.5 mL of the stock solution was added to 495.5 mL of MSM culture medium. This process occurred in the laminar flow chamber and the solution to be added was filtered through a 0.2 μm pore size PES syringe filter obtained from VWR (Leuven, Belgium). To compensate for filtration losses, an additional volume was added (approximately 4.6 mL). The concentration of this initial culture medium was analysed by HPLC, and the real concentration was determined.
The inoculum from the enrichment, which attained an optical density (OD) at 600 nm within the range of 0.8 to 1, was centrifuged and washed twice with MSM. Cultures were performed using 10% (
v/
v) of this inoculum (which corresponds to an initial OD of approximately 1.5 × 10
7 CFUs to 8.5 × 10
8 CFUs) resuspended with MSM plus 11.5 mg/L EE2 (inoculated 5 mL of the culture into 45 mL of MSM containing the drug). The bacterial cultures were incubated at 150 rpm and 28 °C in the absence of light to prevent the photodegradation of the drug. Sampling was conducted after 48, 92, and 168 h. Positive controls were carried out with inoculum without EE2 and negative controls were performed in the absence of inoculum, only in the presence of the drug. The tests were carried out in three replicates. Bacterial growth was assessed following the procedure outlined above in
Section 2,
Section 2.3 of the Material and Methods. The culture media, after inoculation, underwent filtration using a 0.2 μm syringe filter to remove bacterial biomass, and the negative control assays underwent the same sample treatment conditions before being subjected to analysis by HPLC and GC-MS.
The biodegradation removal efficiencies and kinetics were assessed by using (Equation (1)):
in which C
0 represents the initial concentration of EE2 at t = 0 and C
t indicates the EE2 concentration during the experiment, assessed at 280 nm.
Presuming that the bioremoval of EE2 conforms to first-order kinetics [
32], the experimental results were incorporated into the equation
where k
app represents the kinetic constant for removal, C
0 and C are the concentrations of EE2 at the initial time and at a specific time, respectively, and t corresponds to the experimental time measured in days. The estimation of the half-life (t
1/2) of EE2, following a pseudo-first-order degradation, was calculated as described in Palma et al. [
33].
2.4.1. Preparation of Bacterial Isolates
The isolates were obtained from the communities which were able to degrade EE2.
All the processes for bacterial isolation were carried out as detailed in Palma et al. [
30], with the exception that the general medium used in the present study was LB instead of Marine Broth.
Bacterial isolation was carried out using 1 mL of inoculum from bacterial consortia LN, LACFN and PF, PBDEF. Isolation protocol was performed as described by Palma et al. [
20,
30,
34,
35].
Genomic DNA of every single bacterial isolate were obtained utilizing NZY Microbial gDNA Isolation kit (NZYTech, Lisboa, Portugal). The 16S rRNA gene was subjected to amplification, and the resulting PCR products underwent direct sequencing through the Sanger method, as detailed in Palma et al. [
20,
30,
34,
35], resulting in a partial 16S rRNA gene of about 1484 bp. Taxonomic identification was determined by conducting a Blast search against the Gene Bank database of NCBI and utilising the RDP Classifier. Multiple sequence alignment was carried out in MEGA-X version 10.0.5 using the ClustalW package, as explained in Palma et al. [
30].
The comparison between 16S rRNA gene sequences of the identified bacterial isolates and their corresponding type strains supported the phylogenetic analysis. Information concerning the type strains, registered in BacDive—the Bacterial Diversity Metadatabase, followed the guidelines outlined by the DSMZ (
https://bacdive.dsmz.de/ (accessed on 7 February 2023): species:
Aeromonas salmonicida subsp.
salmonicida CECT 894
T with BacDive ID:275;
Aeromonas piscicola strain CECT7443
T with BacDive ID:282;
Aeromonas bestiarum CIP 7430
T with BacDive ID:260;
Rhizobium altiplani strain BR 10423
T with BacDive ID:133176;
Agrobacterium rhizogenes strain ATCC 11325
T with BacDive ID:13571;
Rhizobium lusitanum strain ZP1-1
T with BacDive ID:137407;
Rhizobium freirei strain PRF 81
T with BacDive ID:134045;
Burkholderia fungorum strain CIP 107096
T with BacDive ID:1954;
Burkholderia insulsa strain PNG-April
T with BacDive ID:130232.
The GenBank database has deposited the nucleotide sequence accession numbers for the 16S rRNA partial sequences of the six isolated bacteria that demonstrated the ability to growth in the presence of EE2. These accession numbers are as follows: Isolate 1_Aeromonas sp. strain MLN-TP5 (OQ186609), Isolate 2_Aeromonas salmonicida strain MLN-TP7 (OQ186687) and Isolate 3_Aeromonas bestiarum strain MLACFN-TC2 (OQ186610) were isolated from consortia LN and LACFN from Lousal mine and Isolate 4_Rhizobium lusitanum strain MPF-TL2 (OQ186611), Isolate 5_Rhizobium lusitanum strain MPF-TL1 (OQ186613) and Isolate 6_Paraburkholderia fungorum strain MPBDEF-TCP1 (OQ186612) were isolated from consortia PF and PBDEF from Poderosa mine.
2.4.2. Experiments on the Removal Efficacy Using Bacterial Isolates
Following the identification of bacterial isolates, bioremoval assays were conducted in liquid cultures of MSM with approximately 9 mg/L of EE2 as the sole carbon source. Cultures with 5% (2.5 mL) (
v/
v) of each isolate (inoculum) were sub-cultured into 50 mL of MSM containing EE2. Culture media for inoculation were prepared as outlined in
Section 2,
Section 2.4 of Material and Methods.
The negative controls were performed using culture medium containing EE2 without inoculum. The bacterial cultures were incubated at 150 rpm and 28 °C in the absence of light to prevent the drug’s photodegradation. Sampling was performed after 48, 168, and 216 h. A positive control was conducted without the presence of the drug. The experiments were carried out in three replicates. The bacterial growth was analysed as described above in
Section 2,
Section 2.3 of Material and Methods. The biodegradation removal efficiencies and kinetics were assessed as described above in
Section 2,
Section 2.4 of Material and Methods.
In this study, heightened concentrations of EE2, surpassing those typically found in the environment, were employed for various reasons elucidated in previous studies [
20,
30,
34,
35].
2.4.3. EE2 and Metabolites Analysis
EE2 was identified and quantified using High Performance Liquid Chromatography (HPLC), following the methodology outlined in Palma et al. [
30]. With the results obtained by HPLC, the removal efficiencies of EE2 were calculated. The promising results (in which the removal was significant) obtained by quantification in HPLC were further analysed by gas chromatography–mass spectrum (GC-MS). Putative metabolites resulting from the assays involving EE2 were detected and identified using a GC-MS procedure detailed in the study by Palma et al. [
20]. The GC-MS spectra acquired from experimental samples were interpreted based on a comparison with the National Institute and Technology (NIST) spectra database 2.0. The comparisons were made using the spectrum of standard or based on the most dominant peak (
m/
z) injected in analogous conditions, as detailed in Palma et al. [
30].
2.5. Statistical Analysis
The statistical analysis to compare the percentage removal efficiencies of EE2 by biotic experiments (bacterial consortia and isolates in the presence of the drug as sole carbon source) to the respective abiotic control (containing only the drug) was conducted as outlined in [
30].
For every single experimental condition, 3 independent assays were conducted. Sampling occurred at intervals of 48, 168, and 216 h. The observed effect in the samples was statistically significant at a significant level of 5% error (α = 0.05).
3. Results and Discussion
3.1. Mine Samples Characterisation
In the present study, bacterial communities were recovered from samples collected from the Lousal and Poderosa mines, two inactive copper mines which belong to the Iberian Pyrite Belt that comprise the major crustal sulphur anomaly and, therefore, have the greatest concentration of volcanogenic massive sulphide deposits worldwide [
36,
37,
38,
39]. The exploitation of these mines was mainly oriented towards extracting valuable metals such as gold (Au), silver (Ag), and base metals including copper (Cu), zinc (Zn), lead (Pb), and pyrite [
36,
37,
38,
39].
Lousal and Poderosa Mine Samples
Several samples were collected from different sampling points comprising floor sediments, wall sediments, and an acidic lagoon (AMD wastes) from the Lousal and Poderosa mines.
The designation, information, and characterisation of samples from which the bacterial consortia with the ability to biodegrade paracetamol and EE2 (used in the biodegradation experiments in the presence of EE2) were recovered are as described in
Figure 2.
The XRD characterisation of samples from the Lousal mine’s walls and floor revealed a predominant composition of various mineral categories: silicates (silicon dioxide, quartz, olivine), phyllosilicates (kaolinite, muscovite), oxides (hematite), carbonates (calcite), sulphide (arsenopyrite, octosulfur), and aluminium phosphate, while Poderosa mine’s samples were mainly composed of silicates (quartz), phyllosilicates (muscovite), sulphates (pickeringite, gypsum, natrojarosite, jarosite), and phosphate (brushite).
These findings are consistent with those reported by Inverno et al. [
38] that characterised the Phyllite–Quartzite Group of Iberian Pyrite Belt, which is constituted by dark shales, packets of quartz sandstones, quartzwacke siltstones, rare conglomerates, and carbonates (limestones) at the top. This Belt is also composed of a Volcanic Sedimentary Complex, which mainly comprises shales, thin-bedded siltstones, and minor volcanogenic sedimentary rocks such as felsic volcanic rocks (rhyolites, rhyodacites and dacites), mafic volcanic rocks (basalts and dolerites), volcanogenic enormous sulphide deposits, and minor andesites [
38].
The main constituents of the Lousal mine’s AMD are described in
Table 1.
The AMD collected in the Lousal mine has a very low pH and a high content of metals (e.g., 182.3 mg/L of iron, 65.3 mg/L of Al) and 3098 mg/L of sulphate. The pH measured at the time of sampling was 2.90; however, at the moment it was utilised as the inoculum source, it had slightly decreased to 2.54 (
Table 1). The redox potential (E
h) of +566 mV aligns with the characteristics of AMD. This positive Eh value is in accordance with the oxidative conditions typically observed in AMD-affected environments, reflecting the prevalence of sulphate and other oxidised species in the system [
40]. It should be noted that the AMD characteristics are also strongly influenced by the season of its collection, for example, the samples used/studied here were collected in December (winter) when rains are common, which dilutes the AMD, making it less concentrated in metals and sulphates; in summer, with the dry weather, the concentrations of metals and sulphates are expected to be higher. The AMD characterisation is similar to that obtained in other studies [
40,
41] and, as can be seen, AMD represents an extreme habitat, with a very low pH and high concentrations of metals, making the conditions of survival very challenging.
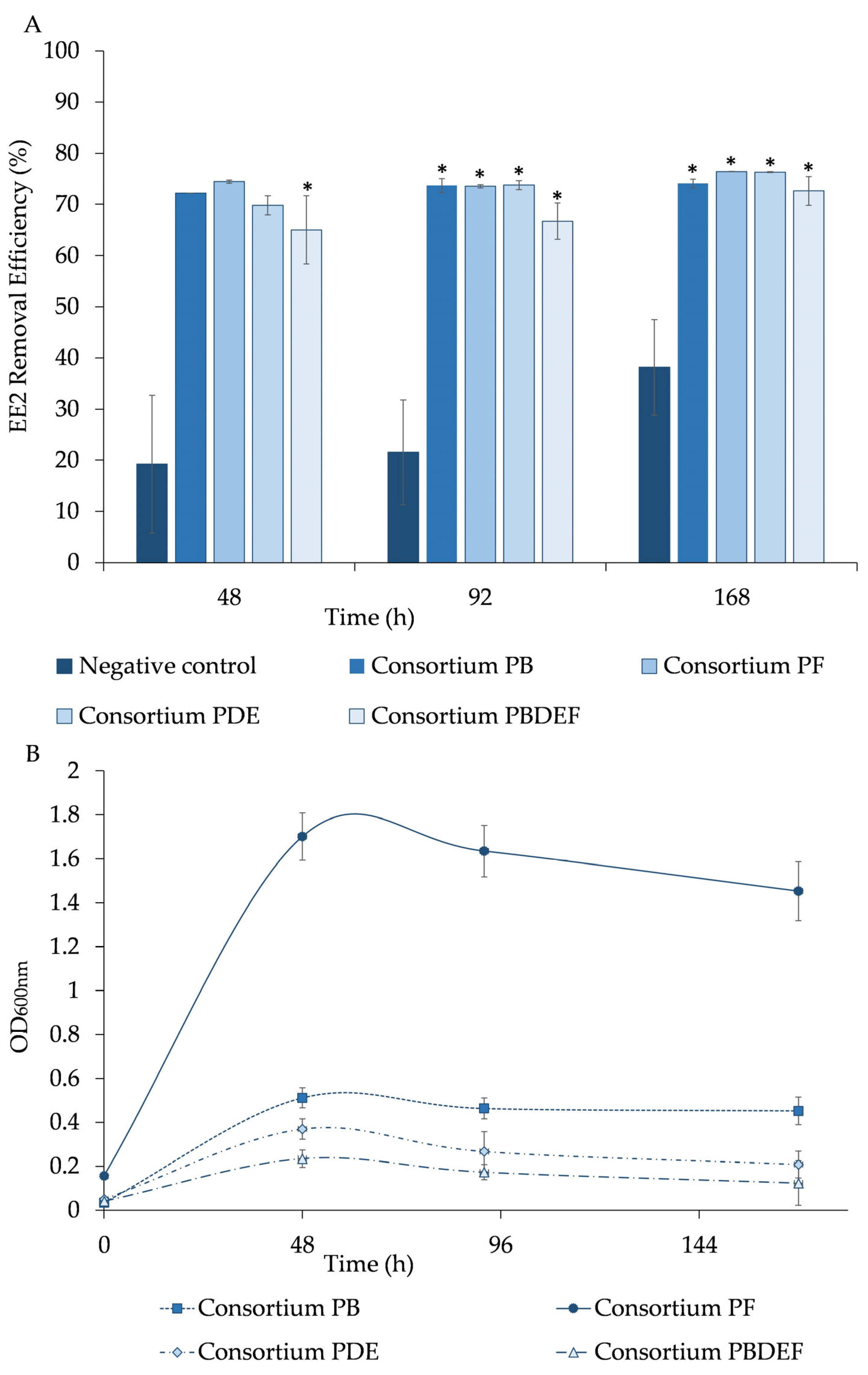
3.2. Bacterial Communities Isolated from Mine Samples with Ability to Remove EE2
The experiments on paracetamol removal revealed that the bacterial communities LF, LN, LAC and PB, PF, PDE recovered from the Lousal and Poderosa mines displayed the ability to degrade paracetamol; these efficient consortia were joined together and denominated as LACFN (Lousal) and PBDEF (Poderosa). The bacterial consortia obtained (10% (
v/
v) of inoculum) were tested at 11.5 mg/L of EE2, the bioremoval of which is the focus of the present study (
Figure 3 and
Figure 4).
The results revealed that the bacterial consortia LF, LN, LAC, and LACFN reduced to more than half (50.6 ± 0.1%, 76 ± 1%, 71 ± 2%, and 71 ± 10%) of 11.5 mg/L of EE2 after 48 h, respectively, using it as a sole carbon source (
Figure 3A). These three communities exhibited elevated microbial growth, consistently exceeding an OD
600nm value of 0.6 during the 48 h experimental period (
Figure 3B). The bacterial growth of the LN consortium is evidenced with OD600nm values of 1.2 ± 0.5, while LF consortium is the one which presented less growth with OD600nm values of 0.5 ± 0.2 (
Figure 3B). The bacterial consortia LF, LN, LAC, and LACFN removed (55 ± 4%, 77.59 ± 0.09%, 73.7 ± 0.5%, and 74 ± 2%), and (60 ± 1%, 78.33 ± 0.03%, 73.9 ± 0.4%, and 77 ± 1%), respectively, after 92 and 168 h (
Figure 3A). In terms of OD600nm, a decrease was evidenced in the bacterial growth for all consortia, after 92 h of experimentation (
Figure 3B). In the abiotic control, the EE2 removal was of 19 ± 13%, 22 ± 10%, and 38 ± 3%, after 48 h, 92 h, and 168 h, respectively.
During the 168 h experiment, the removal of EE2 in cultures with the LF consortium did not show a significant difference (p > 0.05) compared to the negative control (abiotic control). For LN and LAC consortia, the drug degradation was not significantly different when compared with the abiotic control after 48 h, while, during the rest of the assay, the drug’s removal was significantly higher (p < 0.05) in the presence of bacteria than in the abiotic control. In the assays in which a mixture of all the consortia (LACFN) was used, the removal of EE2 was always significantly higher (p < 0.05) throughout the 168 h assay. Thus, it can be inferred that both chemical interactions, as demonstrated in the abiotic control and biodegradation, may have a significant role in the removal of EE2. Therefore, approximately 30 to 50% of EE2 removal is estimated to be attributed to bacterial activity (biodegradation).
The best bacterial consortium, named LN, and its isolates were recovered from samples collected from AMD wastes of the Lousal mine (Portugal).
The results showed that the bacterial consortia PB, PF, PDE, and PBDEF removed 72 ± 1%, 74.4 ± 0.3%, 70 ± 2%, and 65 ± 7%, respectively, of 11.5 mg/L EE2 after 48 h (
Figure 4A). The bacterial growth of PB consortium culture stood out from the other consortia due to its high OD
600nm of 1.7 ± 0.1, while PF, PDE, and PBDEF showed lower values of OD
600nm between 0.5 and 0.3, after 48 h (
Figure 4B). The consortia PB, PF, PDE and PBDEF removed (74 ± 1%, 73.5 ± 0.3%, 73.8 ± 0.9% and 67 ± 4%, respectively), and (74.1 ± 0.9%, 76.41 ± 0.03%, 76.3 ± 0.1% and 73 ± 3%, respectively), after 92 and 168 h, respectively (
Figure 4A). Again after 92 h of experiment a decrease in the bacterial growth was observed for all the consortia (PB with a OD
600nm of 0.46 ± 0.05; PF a with OD
600nm of 1.6 ± 0.1; PDE with a OD
600nm of 0.27 ± 0.09; and PBDEF with a OD
600nm of 0.17 ± 0.03) (
Figure 4B). In the abiotic control, the EE2 removal was of 19 ± 13%, 22 ± 10%, and 38 ± 3%, after 48 h, 92 h, and 168 h, respectively.
Initially, the removal of the drug in the presence of the PB, PF, and PDE consortia was not significantly different (p > 0.05) when compared with the abiotic control, thus suggesting that both chemical interactions and bacterial activity could likely contribute to the drug’s removal. Only for the PBDEF consortium (a combination of consortia) did the depletion efficiency exhibit a statistically significant increase (p < 0.05) compared to the abiotic control throughout the entire experiment of 168 h. However, in the remaining duration of the assay, the removal of the drug by PB, PF, and PDE consortia was significantly higher (p < 0.05) in the presence of bacteria compared to the abiotic control.
3.3. Bacterial Isolation from Consortia with Ability for EE2 Bioremoval
The isolates were obtained from cultures LN, LACFN, PF, and PBDEF consortia in MSM supplemented with 50 mg/L of EE2 as an exclusive carbon source.
After an incubation of 48 h at 28 °C in the absence of light, the colony growth was generally good during the initial and first dilution stages. Isolates were obtained from the dilutions 10−1 for LN consortium, 10−2 for LACFN consortium, 10−4 for PF consortium, and 10−1 for PBDEF consortium. Six isolates capable of growing in the presence of 50 mg/L of EE2 as the sole carbon source were identified: two strains from LN consortium, one from LACFN, two from PF, and one from PBDEF, which were named as Isolate 1, 2, 3, 4, 5, and 6, respectively. In the presence of the EE2, every isolate exhibited the ability to undergo three generations of isolation.
The identification of isolates relied on the analysis of their 16S rRNA sequences, through a search in the NCBI BLAST database. The selection was based on the first entry that exhibited the highest similarity (>98%) and matched with the following species: Isolate 1_Aeromonas sp. strain MLN-TP5 (OQ186609) displayed 99.57% of similarity with Aeromonas salmonicida; Isolate 2_ Aeromonas salmonicida strain MLN-TP7 (OQ186687) showed a similarity of 99.72% with the same species; Isolate 3_Aeromonas bestiarum strain MLACFN-TC2 (OQ186610) revealed a similarity of 99% with Aeromonas bestiarum (these bacteria were isolated from consortia LN and LACFN, respectively); Isolate 4_Rhizobium lusitanum strain MPF-TL2 (OQ186611) and Isolate 5_Rhizobium lusitanum strain MPF-TL1 (OQ186613) displayed a similarity of 98.88% and 99.63%, respectively, with Rhizobium lusitanum; and Isolate 6_Paraburkholderia fungorum strain MPBDEF-TCP1 (OQ186612) showed a similarity of 99% with Paraburkholderia fungorum, which were isolated from the consortia PF and PBDEF, respectively.
To categorise and classify the bacteria accurately, identification was performed using BLAST, along with specific biochemical features obtained for each isolate and a phylogenetic tree. Isolate 1 and 2 present a very high similarity with the genus
Aeromonas salmonicida, which is corroborated with some biochemical tests; for example, both isolates and its type strain are catalase- and oxidase-positive (BacDive ID:275), as well as the Isolate 3 assigned to
Aeromonas bestiarum (BacDive ID:260). The species of this genus are facultative anaerobes which can be halotolerant and grow in 3% salt [
42], therefore having the capacity to survive in seawater; however, it is the first time that these species have been detected in mine extreme environments such as AMD. Isolate 4 and 5 are two strains of
Rhizobium lusitanum which are catalase- and oxidase-positive in their type strain (BacDive ID:137407).
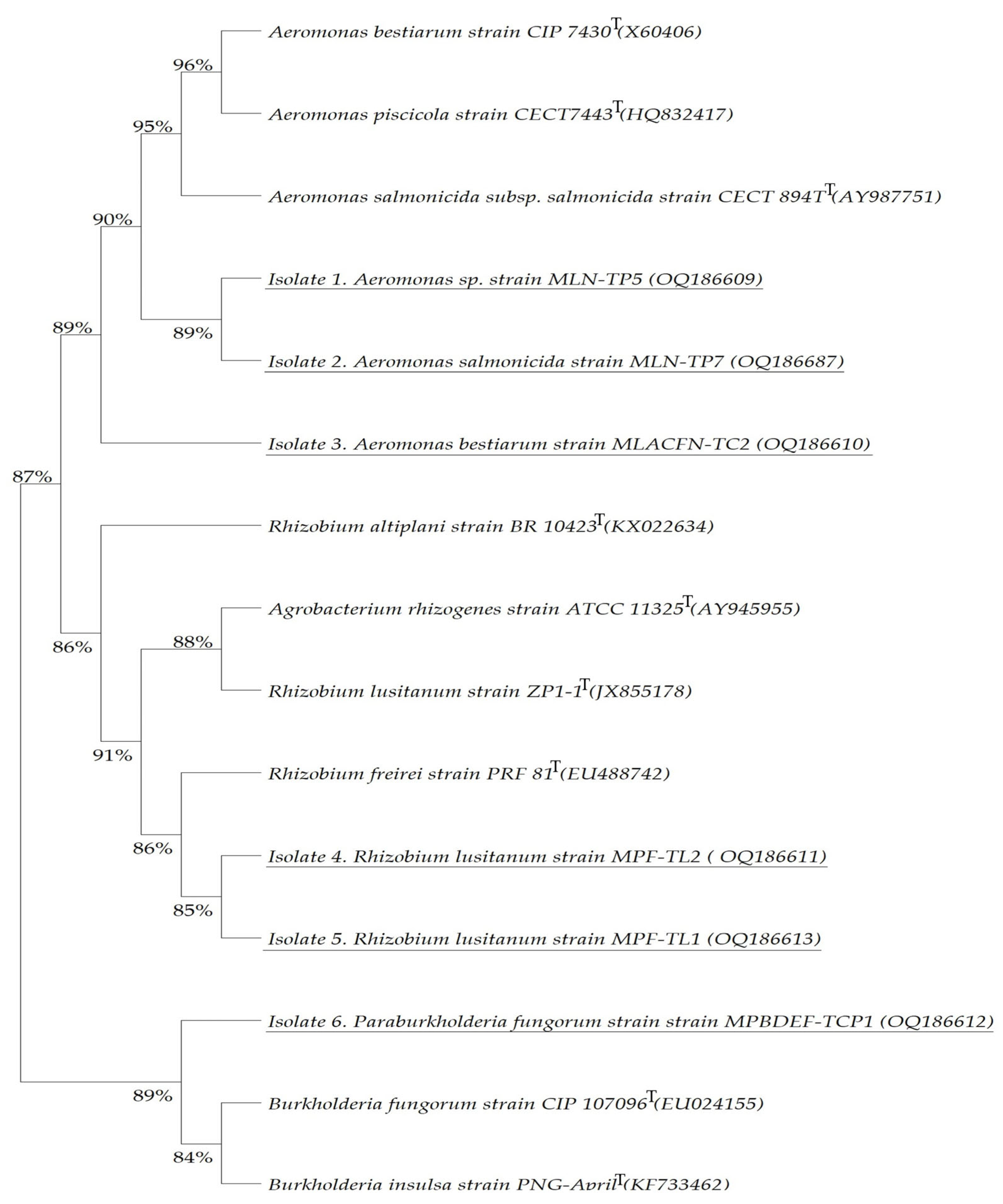
Paraburkholderia fungorum strain MPBDEF-TCP1 is catalase-positive and oxidase-negative as its type strain (BacDive ID:1954). All the obtained bacterial isolates are Gram-negative. To consistently associate the obtained bacteria with recognised species, a maximum likelihood phylogenetic tree was assembled utilising the nearly complete 16S rRNA gene sequences of the isolates. This comparison was made with the corresponding type strain for each isolate (
Figure 5).
The phylogenetic tree revealed that Isolates 1 and 2 isolated from AMD of the Lousal mine, which were assigned to
Aeromonas sp. strain MLN-TP5 and
Aeromonas salmonicida strain MLN-TP7, respectively, display a 89% similarity, and both presented a 95% similarity with
Aeromonas salmonicidaT,
Aeromonas piscicolaT, and
Aeromonas bestiarumT strains, defending their close relatedness with these species (
Figure 5). Isolate 3, assigned to
Aeromonas bestiarum strain MLACFN-TC2, has a 90% similarity with the Isolates 1 and 2,
Aeromonas salmonicidaT,
Aeromonas piscicolaT type strains, and with its own type strain
Aeromonas bestiarumT. Both Isolate 4 and 5 strains, assigned to
Rhizobium lusitanum MPF-TL2 and MPF-TL1 species, share a 85% similarity, an 86% with
Rhizobium freireiT type strain, and an 88% with its type strain
Rhizobacterium lusitanumT and also
Agrobacterium rhizogenesT; they displayed the highest similarity with
Rhizobium altiplaniT, revealing that these isolates are closely related to these species (
Figure 5). Isolate 6_
Paraburkholderia fungorum MPBDEF-TCP1 shows a close relation of 89% with
Burkholderia fungorumT and
Burkholderia insulsaT and, more interestingly, shares an 87% similarity with the
Aeromonas genus group and
Rhizobium genus group; however, it is more closely related to the Isolates 1, 2, and 3 and their type strains with an 89% similarity, while displaying a similarity of 86% with Isolates 4 and 5 (
Figure 5). The results are supported by high bootstrap values, revealing that these species are closely related (
Figure 5).
Aeromonas genus is environmentally ubiquitous and mainly found in several aquatic ecosystems.
Aeromonas species were isolated from surface, underground, drinking, bottled, waste, sea, and irrigation waters [
43]. However, as far as we know, this is the first instance where
Aeromonas salmonicida and
Aeromonas bestiarum were found in AMD. Very few works were found regarding the use of
Aeromonas salmonicida as biodegrading microorganisms. For example, the authors of reference [
44] found that the
Aeromonas salmonicida subsp.
salmonicida strain ETH-3 recovered from activated sludge was able to completely degrade 10,500 mg/L of ethyl formate within 36 h at 30 °C.
Paraburkholderia genus was mainly isolated from soil and plant roots or nodules, while some species were identified in aquatic ecosystems [
45]. In the present study,
Paraburkholderia fungorum was isolated from a consortium recovered from a mixture of wall, soil sediments, and water collected from the Poderosa mine. These species are normally known to have a lot of functions, for example, in producing phytohormones, antibiotics, and lytic enzymes, in solubilising soil minerals, etc., and it also displays bioremediation potential. For example,
Burkholderia fungorum strain FM-2 displayed the ability to degrade 300 mg/L of phenanthrene (fungicide) as an exclusive carbon source over a wide range of pH levels (between pH 3 and 9) and to tolerate high concentrations of toxic metals (Cd (II) and Zn (II)) while degrading phenanthrene [
46].
Rhizobium lusitanum was found in root nodule bacteria, specifically nodulating
Phaseolus vulgaris [
47]. Massot et al. [
48] found that
Rhizobium sp. strain was able to metabolise almost 50% of 50 mg/L glyphosate in 9 days.
3.4. Bioremoval of EE2 by Mine-Recovered Bacterial Isolates
Experiments to explore the potential bioremoval profile of EE2 by the obtained bacterial isolates,
Aeromonas sp. strain MLN-TP5,
Aeromonas salmonicida strain MLN-TP7,
Aeromonas bestiarum strain MLACFN-TC2,
Rhizobium lusitanum MPF-TL2, and
Paraburkholderia fungorum MPBDEF-TCP1, were performed (
Figure 6).
The cultures with 5% (
v/
v) of
Aeromonas sp. strain MLN-TP5 as inoculum spiked with 9 mg/L EE2 as the sole carbon source presented a removal of 75 ± 8%, 89 ± 7%, and 97 ± 5% after 48, 168, and 216 h, respectively (
Figure 6A). The bioremoval rate of EE2 was 0.2548 d
−1, the coefficient of determination (R
2) was 0.9992, and t
1/2 was 2.7 days. The OD
600nm of the initial population was 0.333, with a slight decrease of 0.252 ± 0.002 after 48 h of incubation; however, the OD
600nm remained practically constant during all the experiments (0.236 ± 0.002 and 0.233 ± 0.001 of 168 and 216 h, respectively) (
Figure 6B).
The bioremoval of 9 mg/L of EE2 in association with
Aeromonas salmonicida strain MLN-TP7 was 84 ± 6%, 95 ± 8%, and below the limits of detection after 48, 168, and 216 h, respectively, (
Figure 6A) with a reaction rate of 0.8593 d
−1, R
2 of 1, and a t
1/2 of 0.81 days (19.4 h). This value is lower than the t
1/2 value reported by Menashe and coworkers [
49] for EE2 in WWTPs (24 h), suggesting that the introduction of this strain in the sludge may improve the rate of EE2 biodegradation. The OD
600nm of the initial population was 0.430, with a very slight decrease of 0.410 ± 0.01 after 48 h incubation. For this isolate, OD
600nm remained constant during all the experiments (0.399 ± 0.009 and 0.394 ± 0.008 of 168 and 216 h, respectively), revealing that the drug was apparently not toxic to this strain (
Figure 6B). These bacterial growth profiles may have several explanations, such as, that bacteria take time to adapt to the drug as a sole carbon source and start to use it as readily degradable source. Drug consumption may be necessary to keep the bacteria alive, but this is not enough to noticeably increase its biomass, which was already high from the beginning. Also, EE2 can be co-metabolically degraded by these bacterial strains.
In the case of
Aeromonas bestiarum MLACFN-TC2, the removal of the drug was 56 ± 20%, 73 ± 2%, and 90 ± 8% considering 48, 168, and 216 h of assay, respectively (
Figure 6A), and a reaction rate of 0.2375 d
−1, R
2 of 0.9309, and a t
1/2 of 2.9 days. In terms of bacterial growth, the OD
600nm of the initial population was 0.364 and, as with the previous species of the
Aeromonas genus, the optical density did not suffer significant variations (0.294 ± 0.007, 0.283 ± 0.001, and 0.278 ± 0.004 of 48, 168, and 216 h, respectively) (
Figure 6B).
In the case of
Rhizobium lusitanum MPF-TL2, the removal of the drug was 48 ± 10%, 65 ± 7%, and 75 ± 2% considering 48, 168, and 216 h of assay, respectively (
Figure 6A), and a rate of 0.1055 d
−1, R
2 of 0.9982, and t
1/2 = 6.6 days. The OD
600nm of the initial population was 0.304, suffering a decrease as the assay progressed (0.28 ± 0.01, 0.25 ± 0.01, and 0.22 ± 0.01 of 48, 168, and 216 h, respectively) (
Figure 6B).
In
Paraburkholderia fungorum MPBDEF-TC1 cultures, the removal of EE2 was 48 ± 27%, 77 ± 8%, and 78 ± 3% considering 48, 168, and 216 h of assay, respectively (
Figure 6A), and a rate of 0.1102 d
−1, R
2 of 0.8449, and t
1/2 = 6.3 days (if following a pseudo-first-order kinetics; t
1/2 = 40.8 days if following a pseudo-zero-order kinetics). Interestingly, the removal rate of EE2 by this bacterium did not exhibit behaviour consistent with a first-order kinetics, since the R
2 from the linear regression is 0.8449, thus less than 0.95. It is likely that the drug removal may follow a pseudo-zero-order reaction in which the reaction rate is not dependent on the EE2 concentration. The OD
600nm of the initial population was 0.378. However, this bacterium showed the most evidenced decrease in OD
600nm (0.314 ± 0.007, 0.29 ± 0.01, and 0.229 ± 0.009 of 48, 168, and 216 h, respectively) throughout the assay (
Figure 6B).
In the abiotic control performed to evaluate the chemical interactions of the medium with the drug, the removal was 9 ± 11%, 26 ± 8%, and 28 ± 8% considering 48, 168, and 216 h of assay, respectively (
Figure 6A), and a removal rate of 0.0208 d
−1, R
2 of 0.9402, and t
1/2 = 33 days.
In cultures with isolates of Aeromonas sp. strain MLN-TP5 and Aeromonas salmonicida strain MLN-TP7, the EE2 removal was greater (p < 0.05) than in the abiotic/negative control throughout the whole 216 h experiment. In cultures in which Rhizobium lusitanum MPF-TL2 and Paraburkholderia fungorum MPBDEF-TCP1 were the inoculum, the EE2 removal efficiency was not significantly different (p > 0.05) than in the abiotic/negative culture, considering 48 h of assay. However, during the rest of the assay, the drug removal showed a more statistically significant increase (p < 0.05) in the presence of the inoculum than in the negative control.
The contribution of the chemical interactions in the presence of inoculum was estimated to be in the range of approximately 13 to 37%.
The presence of methanol in the culture medium seemed to have no adverse or toxic effects on the bacteria. Furthermore, methanol may potentially serve as an additional carbon source, stimulating bacterial activity and facilitating the subsequent utilisation of EE2. This is supported by the observed degradation of the drug by bacteria over time.
These results suggest that Aeromonas sp. strain MLN-TP5 and Aeromonas salmonicida strain MLN-TP7 are multifunctional bacterial isolates which may integrate cost-effective bioremediation strategies for soils, sediments, and waters polluted with multiple contaminants, namely in acidic environments undergoing long-term contamination. Furthermore, its ability to thrive in harsh environments suggests its resistance, potentially enabling it to prosper under demanding environmental conditions. This includes soils, sediments, and water bodies contaminated with multiple pollutants, as well as acidic environments. This adaptability extends to WWTPs, where conditions may undergo wide variations, such as low influent pH or low water alkalinity due to nitrification.
Identification of EE2 Metabolic Products
EE2 biodegradation was investigated by HPLC and GC-MS analysis. The putative metabolic products that originated during EE2 removal were analysed via GC-MS.
17α-ethinylestradiol was identified with a retention time of 19.433 min for GC-MS and 8.38 min for HPLC. Various intermediates were chosen due to their detection solely in the presence of bacteria, eliminating those found in the negative controls (abiotic) samples.
For the detection/identification of EE2’s putative metabolites, the cultures of the genus
Aeromonas strains MLN-TP5 and MLN-TP7 with the best removal results were studied (
Figure S1, Supplementary Material). The GC-MS spectra acquired from the samples throughout the experiments were analysed and [
30] important peaks, which may elucidate the putative metabolites and subsequent pathways involved in EE2 degradation by these bacteria, were observed (
Table 2,
Figure S1).
Some of the evident peaks detected may correspond to compounds that could be part of, or a result of, alternative or different metabolic pathways for these bacterial isolates. These compounds are not discussed in this context.
In all samples, including the abiotic ones, a huge peak was detected at 5.522 min that is likely a medium compound or other impurity. A contamination with bisphenol A was also found at 9.672 min.
For both
Aeromonas strains MLN-TP5 and MLN-TP7, the degradation of EE2 seems to occur via the metabolization of the drug into estra-1,3,5(10)-trien-17-one,3-hydroxy-(estrone, E1), since a major peak compatible with this compound appeared at 9.968 min (
m/
z of 270), after 48 h of incubation. This result supports that the degradation of EE2 may start by the oxidation of the D ring’s C-17 into a ketone, resulting in E1. Other peaks detected after 48 h of experiment match with compounds that may be putative intermediates or dead end products of EE2 degradation, such as 2-hydroxyandrosta-1,4, diene-3,6,17-trione at 11.848 min, and also D-mannonic acid γ-lactone (lactone structure) and 2-ketoglutaric acid (oxo dicarboxylic acid) at 6.458 min, which are simple compounds found in this early stage (
Table 2). Those former ones are intermediate metabolites that may enter into the citric acid cycle (TCA), indicating that EE2 may be readily used as a sole carbon source by these isolates. The isolates also seem to be able to degrade its recalcitrant metabolites, such as E1. The lactone structures (D-mannonic acid γ-lactone and Gluconic acid γ-lactone) identified here may already result from the cleavage of the D ring from the C ring of E1.
A significant peak consistent with E1, with a retention time of 11.190 min (
m/
z of 270), was identified and persisted even after 168 h of the assay. Other putative intermediates, such as norepinephrine at 11.393 min, 2,4-dihydroxy-4,6-dimethoxy-3-methylchalcone at 11.848 min, butenedioic acid or fumaric acid at 7.929 min, and citric acid at 9.512 min, were identified after 168 h of experiment. Butenedioic acid and citric acid are dicarboxylic acids and intermediate metabolites in the TCA cycle (
Table 2).
However, neither E1 nor its parent compound (EE2) were detected after 216 h of experiment. In this period, the most evidenced peaks belong to compounds such as gluconic acid γ-lactone (lactone structure) detected at 7.903 min, pentenedioic acid (dicarboxylic acid) at 5.288 min, and β-hydroxypyruvic acid (2-oxo monocarboxylic acid) at 5.541 min (
Table 2).
Several mechanisms were proposed to explain EE2 degradation by bacteria. However, the compounds identified here are in agreement with the ones already reported by Lee and Liu [
50] and Palma et al. [
20]. Lee and Liu [
50] studied estradiol (E2) degradation by bacteria from activated sludge. They proposed that, in the initial stages, the drug suffered ring cleavage and oxidation, leading to the formation of E1 at the D ring, resulting in a lactone structure. Lactones are structures which are easily produced by water removal from γ-hydroxy acids. The authors and colleagues [
50] found the 5-hydroxy-15-methyl-13-oxatetracyclo [8.7.0.0.0.]-heptadeca-2(7),3,5-trien-14-one, that was formed by the further oxidation of estrone, which may suffer oxidative fission (meta-cleavage) at C-4 and C-5 in ring A of E1, resulting in the generation of dicarboxylic acid intermediates. Subsequently, its metabolic intermediates may experience ring cleavage and enter the TCA or citric acid cycle, ultimately leading to EE2 complete degradation (mineralisation) (
Table 2).
Another putative intermediate metabolite compatible with prost-13-en-1-oic acid was detected at 20.396 min, likely resulting from the aperture of the A, B, and C rings. This metabolite may have undergone cleavage and subsequent mineralisation into CO
2 and H
2O. This metabolite was also detected in the degradation of EE2 by
Pantoea agglomerans [
20].
Additionally, a noteworthy compound detected was 2-amino-adipic acid at 6.477 min. It is important to note that the pathway for adipic acid generation is derived from glucose. In this context, its generation likely arises from EE2, as it serves as the exclusive carbon source. Additionally, it is formed utilising intermediates, wherein the intermediate 2-oxoadipate is produced from 2-oxoglutarate (a TCA cycle intermediate) during the initial enzymatic phase of the α-aminoadipate pathway for L-lysine biosynthesis [
51]. Furthermore, adipic acid is generated as an intermediate in the microbial degradation of cyclohexane, cyclohexanol, cyclohexanone, or caprolactam by certain bacteria, including
Arthrobacter sp.,
Rhodococcus sp.,
Acinetobacter sp., and
Pseudomonas strains [
51].
One of the major peaks, which appeared at 10.796 min into the assay in the presence of the isolate, is compatible with N-α-acetyl-L-lysine, an acetylated di-amino acid and 5-aminovaleric acid at 10.758 min. This compound may be important as a clue for proving the extremophile nature/capacity of the
Aeromonas strains isolated here, which have, for the first time, been found in AMD, since this compound is a rarely occurring osmolyte that can be synthesised and accumulated by certain halophiles under osmotic stress [
52]. The accumulation of N-α-acetyl-L-lysine has the potential to function as a thermolyte, a compound that enhances heat stress tolerance and survival in
S. halodurans. A thermolyte is a solute that assists microorganisms in coping with high-temperature environments by stabilizing their proteins and membranes, thus promoting adaptation under these conditions [
53]. In the case of these
Aeromonas strains, they may also confer resistance to acidic conditions and to the presence of high concentrations of heavy metals and toxic compounds, e.g., pharmaceuticals.
3.5. Hypothetical Potential of Aeromonas salmonicida for EE2 Biodegradation Based on Its Phenotypical Characteristics
The results obtained indicate that the Aeromonas strains MLN-TP5 and MLN-TP7 studied here may possess enzymatic tools for EE2 degradation, as they demonstrate the ability to efficiently remove the drug.
The genetic potential of Aeromonas strains can be studied by comparing genes encoding similar functional enzymes by sequence homology.
This particular investigation was conducted by comparing the phenotypic characteristics of members belonging to the same species, specifically,
Aeromonas salmonicida strains from the 52 genomes deposited in BV-BRC 3.28.21 database [
54]. Investigating specific genes associated with enzymes that play a role in EE2 removal allowed us to make inferences about the bacterium’s biodegradation capabilities and the metabolic pathways it employs.
Some authors have reported that the aerobic degradation of oestrogens follows a mechanism known as the 4,5-seco pathway. The first step proposed involves the cleavage of the central ring, taking place between the fourth (C-4) and fifth (C-5) carbon atoms in the A-ring. Therefore, several particular enzymes are reported for EE2 degradation [
55,
56]. In the process described by Chen et al. [
56], estrone undergoes hydroxylation at C-4 catalyzed by an enzyme known as estrone 4-hydroxylase. Subsequently, a 17β-estradiol dehydrogenase and 4-hydroxyestrone 4,5-dioxygenase are involved in the 17-dehydrogenation and meta-cleavage of the estrogen A ring in the subsequent steps. These specific enzymes are not listed in the PATRIC database for
Aeromonas salmonicida strains. However, similar to that which occurs with androgens degradation, some actinobacteria display the capability of utilising similar or homologous enzymes for the aerobic degradation of these compounds via an analogous pathway [
55], so the same could be happening in this case. Also, Chen et al. [
56] stated that other extradiol dioxygenases are not completely excluded and may also contribute to the meta-cleavage of 4-hydroxyestrone. The authors report that some meta-cleavage pathways include a [2Fe-2S] ferredoxin that allows the reactivation of the metal ions of extradiol dioxygenases that become oxidised through catalytic reactions [
56]. Also, Chen et al. [
56] stated that the involvement of another extradiol dioxygenases in the meta-cleavage of 4-hydroxyestrone cannot be entirely ruled out. The authors report that certain meta-cleavage pathways incorporate a [2Fe-2S] ferredoxin, facilitating the reactivation of metal ions within extradiol dioxygenases that undergo oxidation during catalytic reactions [
56].
Therefore, some examples of enzymes that
Aeromonas salmonicida possesses, and that may play a role in EE2 degradation, are suggested here, such as, for example, biphenyl-2,3-diol 1,2-dioxygenase III-related protein (PATRIC local family PLF_642_00002657; RefSeq locus tag BHR46_02575; location complement (205678..206046); accession MIIS01000002) [
54]. Playing a crucial role in the degradation pathway of biphenyl and poly-chlorinated biphenyls (PCBs), this enzyme facilitates the initial ring cleavage step. It catalyses the reaction by introducing two oxygen atoms into the catechol ring, which is produced by cis-2,3-dihydrobiphenyl-2,3-diol dehydrogenase.
Also, 2-polyprenylphenol hydroxylase and gene
visC (PATRIC local family PLF_642_00000363; RefSeq locus tag ASA_2634; location complement (2817442..2818677); accession NC_009348) [
54] play a role in the central meta-cleavage pathway of aromatic compound degradation [
57].
The studied genomes contain [2Fe-2S] ferredoxin enzyme (PATRIC local family PLF_642_00001905; RefSeq locus tag IYQ_01572; Protein ID EHI54426.1; location 251808..252146; accession AGVO01000002) [
46,
54]; apart from the aforementioned dioxygenases, other enzymes may also play a role in the metabolism of these types of compounds. Another enzyme found which may display a role in EE2 degradation is the Beta-ketoadipate enol-lactone hydrolase (PATRIC local family PLF_642_00000103; RefSeq locus tag BHR46_01780; location complement (24823..25614); accession MIIS01000002) [
54]. Beta-Ketoadipate enol-lactone hydrolase plays a role in the chloroaromatic degradation pathway and catalyses a conventional step in the utilisation of protocatechuate and cis, cis-muconate by bacteria [
57]. For example, aromatic compound dissimilation is initiated by
A. calcoaceticus, generating catechol or protocatechuate from various substrates. The degradation process follows the beta-ketoadipate pathway, with two separate branches that enzymatically convert catechol or protocatechuate through aromatic ring fission into intermediates of the tricarboxylic acid cycle [
58].
The phenotypical features were compared with strains that were likely not exposed to EE2. Although members of this species may possess the genes coding for enzymes described in scientific publications for EE2 bioconversion, these genes remained inactive, and the corresponding proteins were not expressed/produced. Much remains to be discovered, as the biodegradation potential of
Aeromonas salmonicida is not extensively studied, and many gene-encoding proteins with unknown roles exist [
30].
4. Conclusions
The consortium that displayed the highest removal efficiency was LN, which was recovered from the AMD of the Lousal mine. It was able to remove approximately 78.33 ± 0.03% of 11.5 mg/L EE2 after 168 h (7 days) of incubation, showing the best removal efficiency among consortia recovered from the Lousal and Poderosa mines. From this consortium, strains identified as Aeromonas sp. and Aeromonas salmonicida (MLN-TP5 and MLN-TP7) were isolated for the first time in acid mine drainage waters. These strains efficiently removed EE2 as the sole carbon source. Aeromonas strain MLN-TP5 removed 75 ± 8%, 89 ± 7%, and 97 ± 5% of 9 mg/L of EE2, while strain MLN-TP7 efficiently removed 84 ± 6% and 95 ± 8%, reaching concentrations of this drug in the medium below the limits of detection after 48, 168, and 216 h (9 days). Notably, these two Aeromonas strains also demonstrated the ability to degrade EE2’s toxic metabolites via E1, lactone compounds, and dicarboxylic acids, which enter the TCA cycle. Aeromonas strain MLN-TP7, in particular, exhibits remarkable potential in the removal EE2, making it a highly promising candidate for bioremediation processes. Therefore, for future perspectives, it would be interesting to employ these bacterial strains in bioaugmentation. One strategy could involve enriching flocculant or granular sludge from WWTP systems with these isolates, namely in conditions where pH variations may occur. This approach aims to establish a process capable of generating a highly efficient system, potentially leading to a more efficient anoxic/aerobic system in the latter case. Such a system could simultaneously reduce COD, nitrates, nitrites, phosphorus, and pharmaceuticals in a single step. In addition to bioaugmentation, the identification of bacteria present in acidic environments and capable of degrading pharmaceuticals may stimulate research into co-treatment strategies, namely aimed at the effective treatment of both AMD (deficient in carbon sources) and other types of wastewater (e.g., domestic or industrial, rich in pharmaceuticals, with high contents of carbon sources).




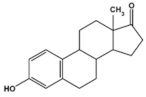 Estrone (E1)
Estrone (E1)

 Lactone (X1)
Lactone (X1)