Fungal and Prokaryotic Communities in Soil Samples of the Aral Sea Dry Bottom in Uzbekistan
Abstract
1. Introduction
2. Materials and Methods
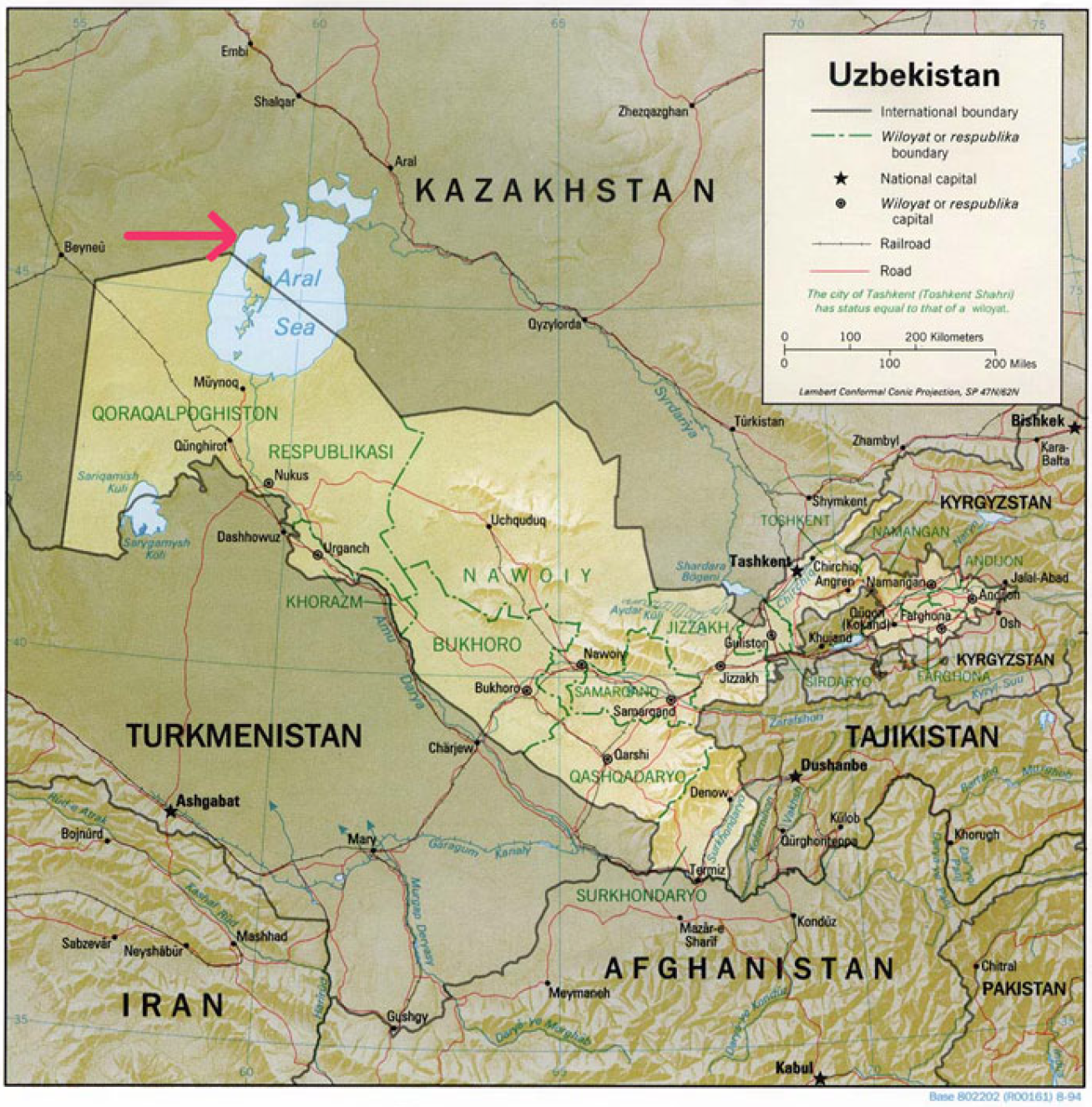
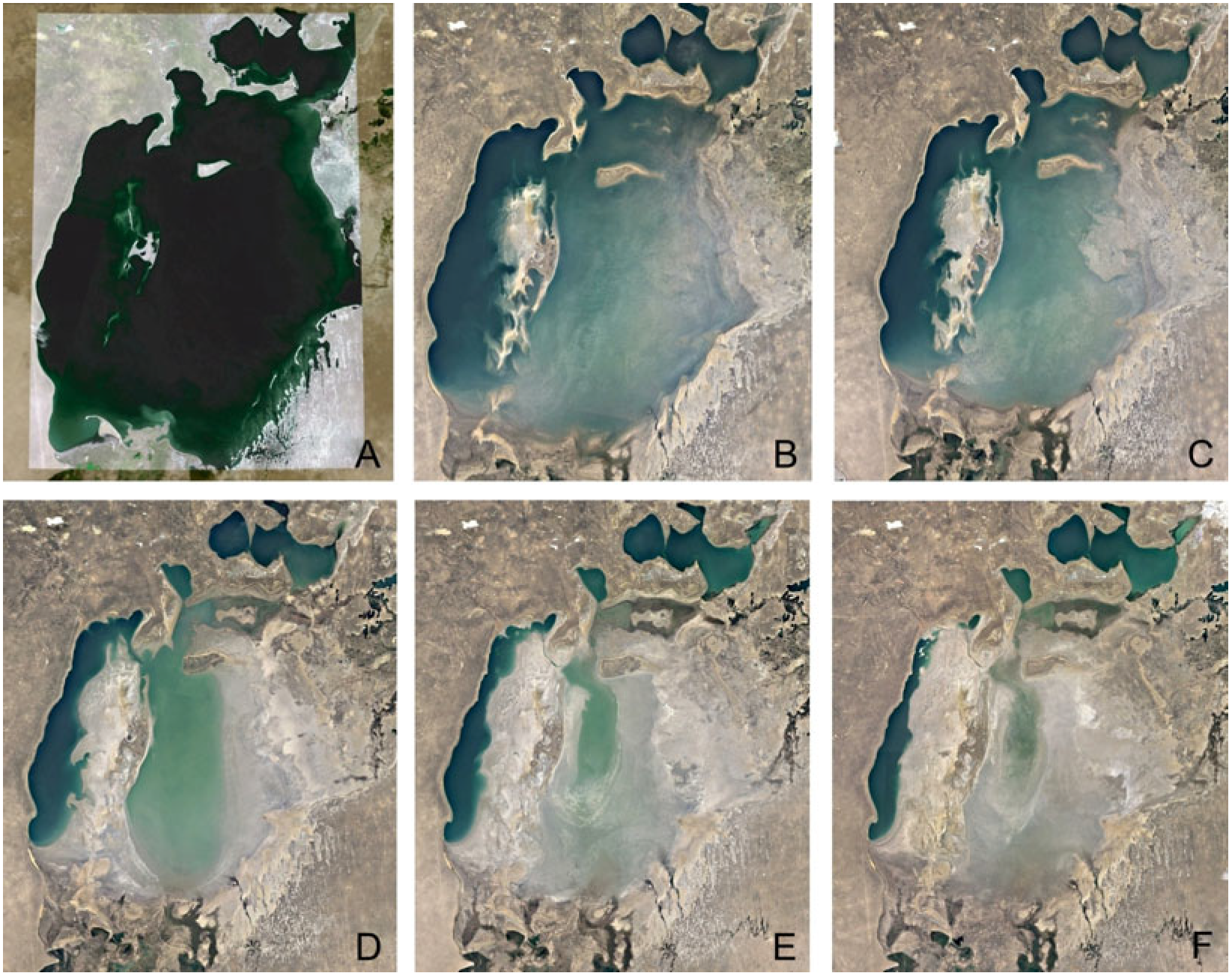
2.1. Study Area and Soil Sampling
2.2. Chemical Analyses
2.3. Mycological Analyses
2.4. Bacterial Analyses
2.5. Next-Generation Sequencing Analysis
2.5.1. DNA Amplification of Bacterial, Archaeal and Fungal Communities
2.5.2. High-Throughput Sequencing and Data Analysis
3. Results and Discussion
3.1. Soil Characteristic
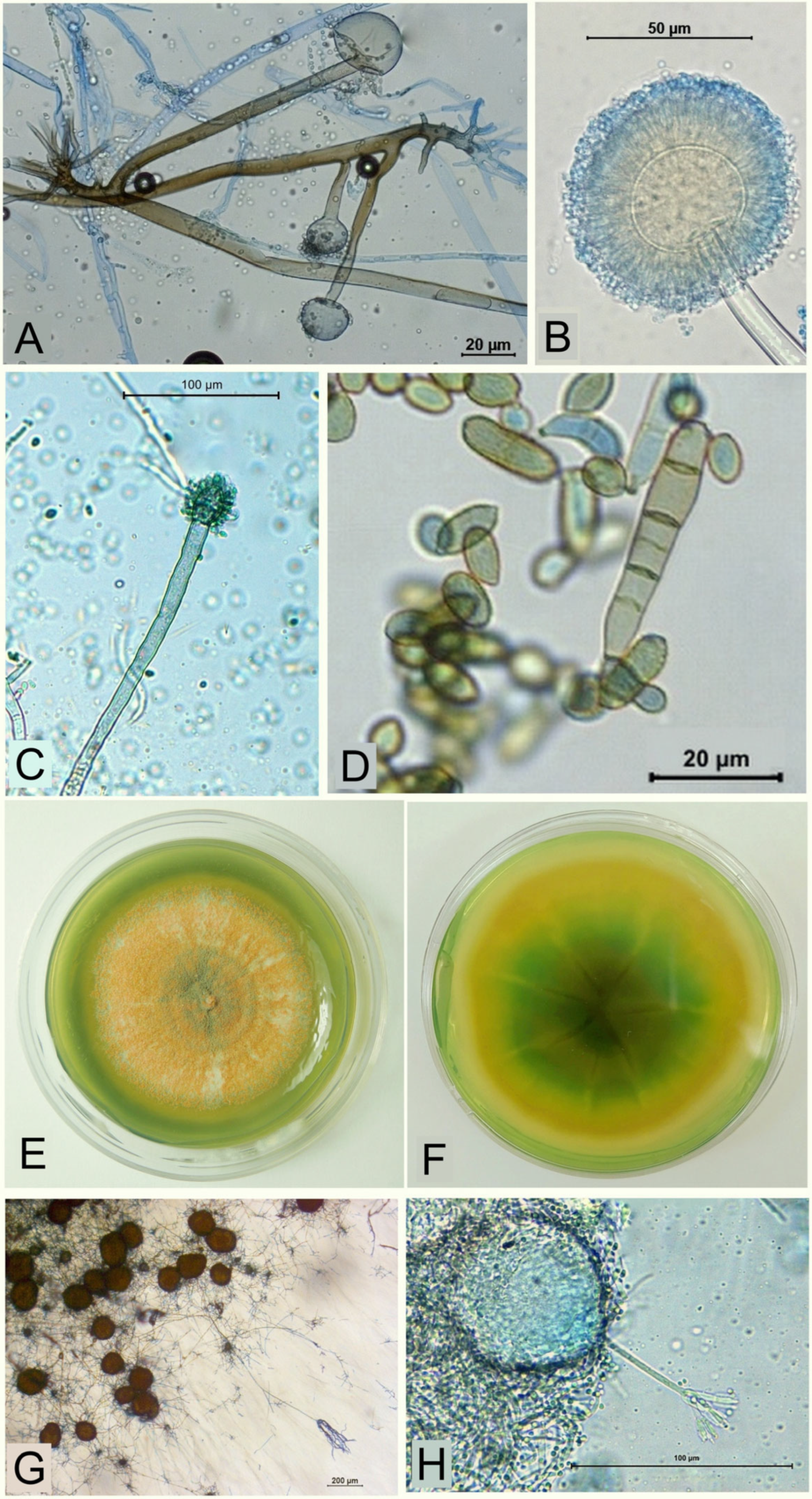
3.2. Cultivable Soil Microscopic Filamentous Fungi
3.3. Bacterial Isolates
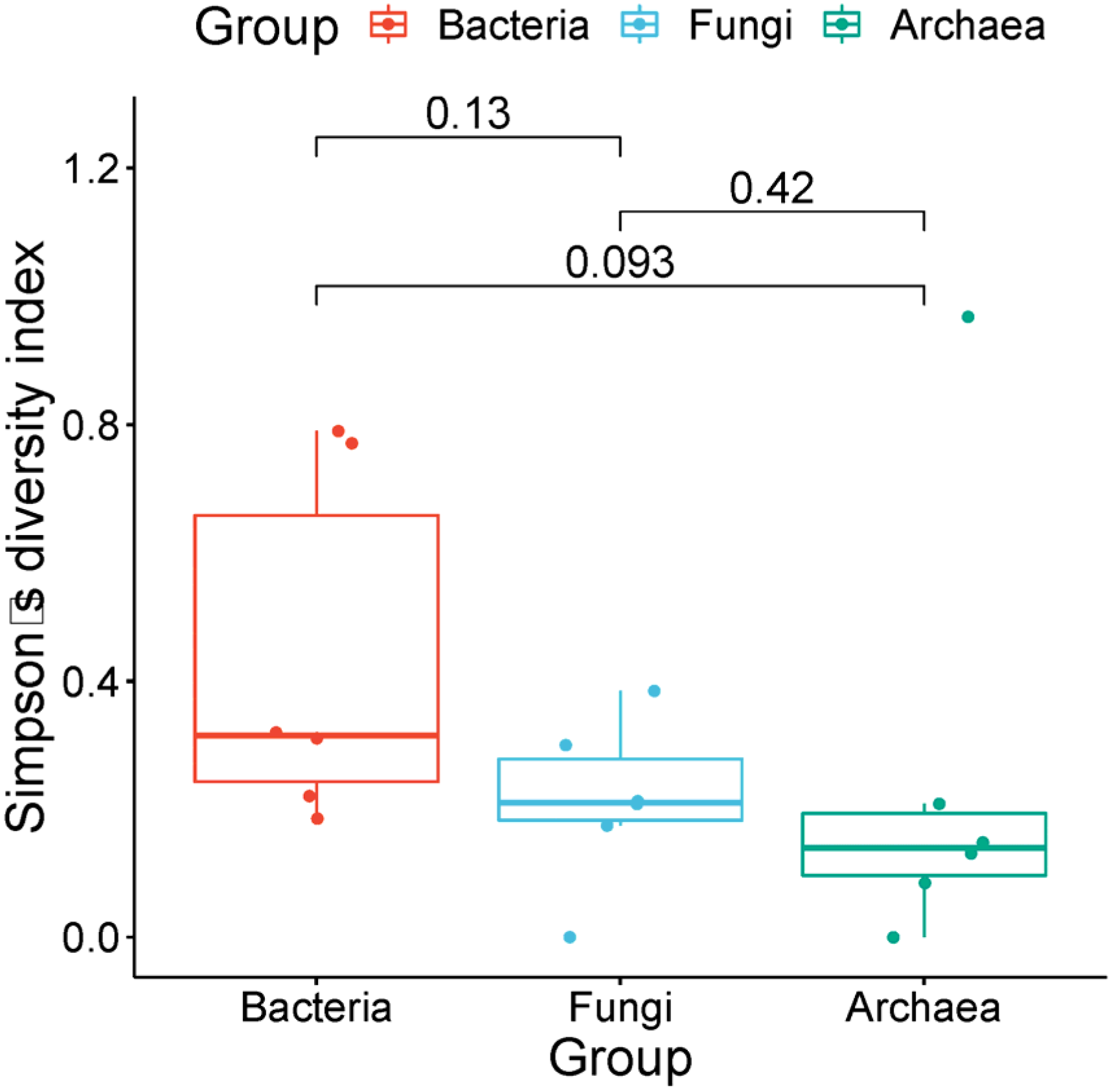
3.4. High-Throughput Sequencing Analysis
3.4.1. Fungi
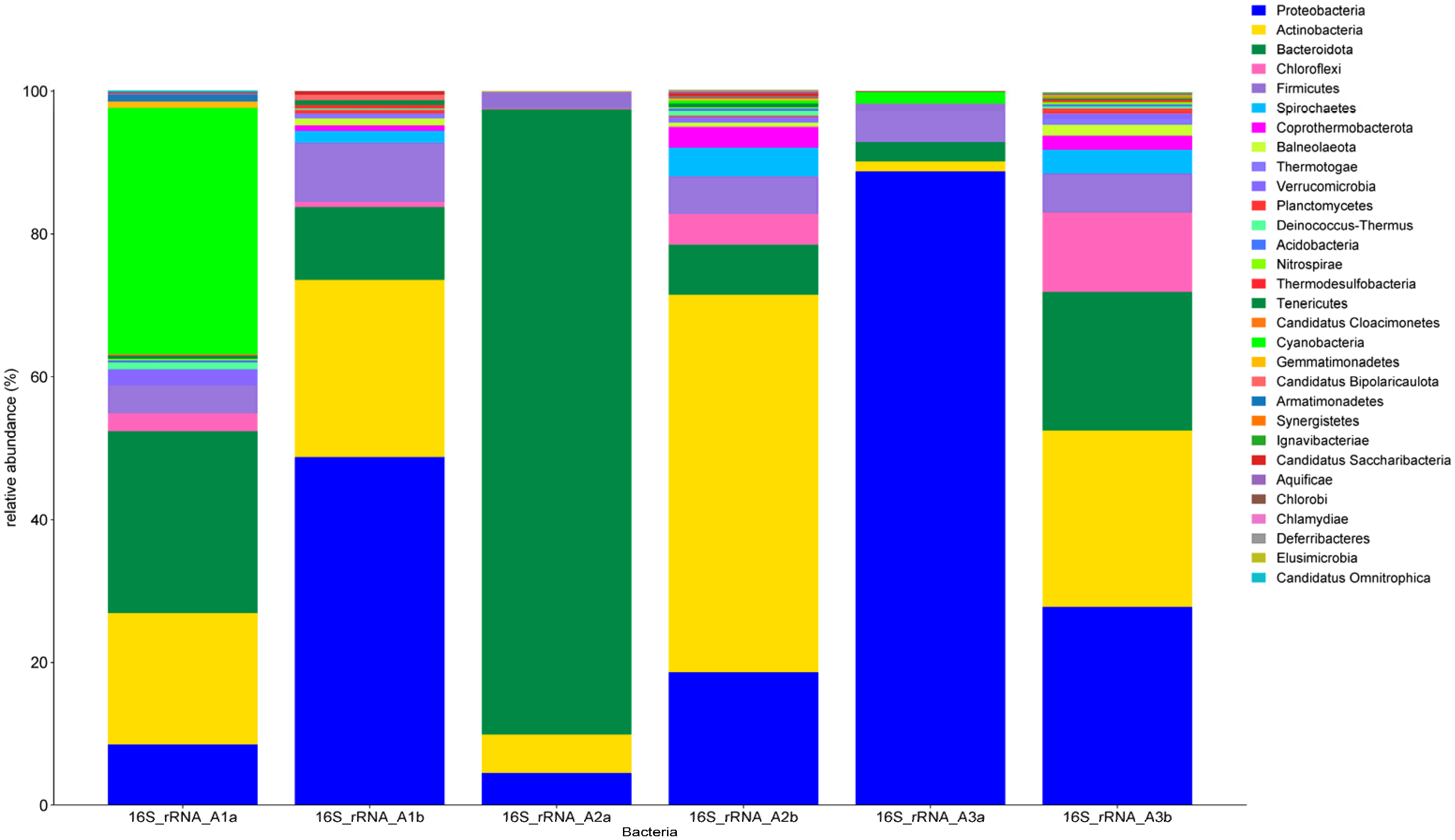
3.4.2. Bacteria
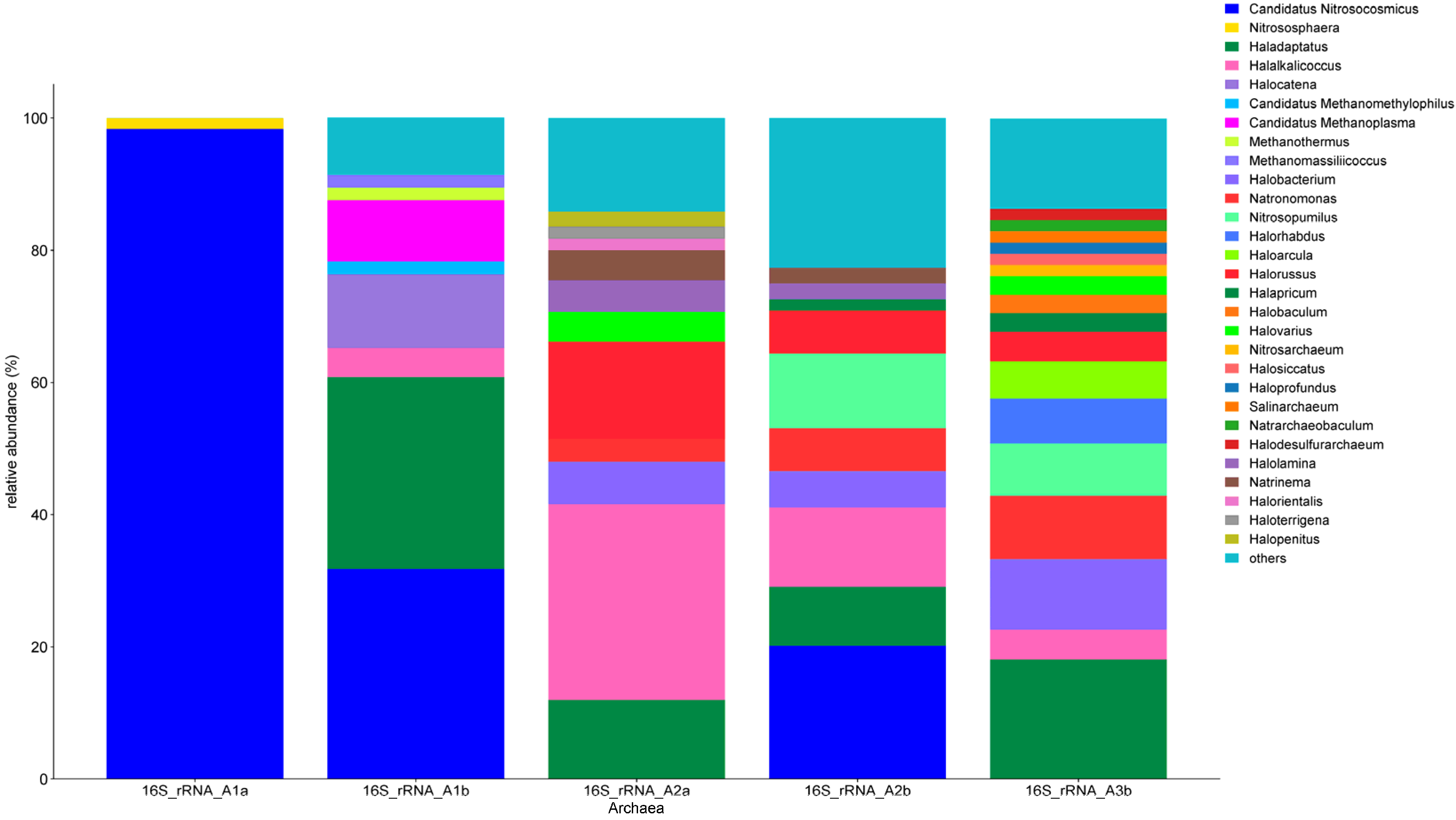
3.4.3. Archaea
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Matzhanova, K.K.; Orel, M.M.; Matzhanov, T.K. The Aral Sea crisis and its impact on the current state of water bodies and higher aquatic vegetation. ESJ 2021, 2, 82–86. Available online: https://scholarzest.com/index.php/esj/article/view/269 (accessed on 15 November 2023).
- Turdimambetov, I.R.; Madreymov, A.; Pauditsova, E.; Oteuliev, M.O.; Bekanov, K.K. Influence of harmful environmental factors on the rate of incidence of children in Karakalpakstan. Cent. Asian J. Geogr. Res. 2021, 3–4, 55–63. [Google Scholar]
- Rahi, K.A.; Halihan, T. Changes in the salinity of the Euphrates river system in Iraq. Reg. Environ. Chang. 2010, 10, 27–35. [Google Scholar] [CrossRef]
- Siyu, X.; Zhixun, X.; Pingwu, W. Soil salinity in the irrigated area of the Yellow river in Ningxia, China. Arid Soil Res. Rehabil. 1996, 10, 95–101. [Google Scholar] [CrossRef]
- Liu, W.; Ma, L.; Abuduwaili, J. Historical change and ecological risk of potentially toxic elements in the lake sediments from North Aral Sea, Central Asia. Appl. Sci. 2020, 10, 5623. [Google Scholar] [CrossRef]
- Matsapaeva, I.V.; Osinskaya, N.S.; Danilova, E.A. Concentrations of heavy metals in bottom sediments of Lake Dautkul as an indicator of anthropogenic impact in the area south of the Aral Sea. Water Res. 2010, 37, 586–590. [Google Scholar] [CrossRef]
- Ataniyazova, O.A.; Baumann, R.A.; Liem, A.K.D.; Mukhopadhyay, U.A.; Vogelaar, E.F.; Boersma, E.R. Levels of certain metals, organochlorine pesticides and dioxins in cord blood, maternal blood, human milk and some commonly used nutrients in the surroundings of the Aral Sea (Karakalpakstan, Republic of Uzbekistan). Acta Paediatr. 2001, 90, 801–808. [Google Scholar] [CrossRef] [PubMed]
- Oren, A. Halophilic microbial communities and their environments. Curr. Opin. Biotechnol. 2015, 33, 119–124. [Google Scholar] [CrossRef] [PubMed]
- Belozerskaya, T.; Aslanidi, K.; Ivanova, A.; Gessler, N.; Egorova, A.; Karpenko, Y.; Olishevskaya, S. Characteristics of extremophilic fungi from Chernobyl nuclear power plant. Curr. Res. Technol. Educ. Top. Appl. Microbiol. Microb. Biotechnol. 2010, 1, 88–94. [Google Scholar]
- Šimonovičová, A.; Kraková, L.; Pauditšová, D.; Pangallo, D. Occurrence and diversity of cultivable autochthonous microscopic fungi in substretes of old environmental loads from mining acitivities in Slovakia. Ecotoxicol. Environ. Saf. 2019, 172, 194–202. [Google Scholar] [CrossRef]
- Andronov, E.E.; Petrova, S.N.; Pinaev, A.G.; Pershina, E.V.; Rakhimgalieva, S.Z.; Akhmedenov, K.M.; Sergaliev, N.K. Analysis of the structure of microbial community in soils with different degrees of salinization using T-RFL Pandreal-time PCR techniques. Eurasian Soil Sci. 2012, 45, 147–156. [Google Scholar] [CrossRef]
- Yuan, B.C.; Li, Z.Z.; Liu, H.; Gao, M.; Zhang, Y.Y. Microbial biomass and activity in salt affected soils under arid conditions. Appl. Soil Ecol. 2007, 35, 319–328. [Google Scholar] [CrossRef]
- Yan, N.; Marschner, P.; Cao, W.H.; Zuo, C.Q.; Qin, W. Influence of salinity and water content on soil microorganisms. Int. Soil Water Conserv. Res. 2015, 3, 316–323. [Google Scholar] [CrossRef]
- Tavoosi, N.; Akhavan Sepahi, A.; Amoozegar, M.A.; Kiarostami, V. Toxic heavy metal/oxyanion tolerance in haloarchaea from some saline and hypersaline ecosystems. J. Basic Microbiol. 2023, 63, 558–569. [Google Scholar] [CrossRef] [PubMed]
- Naitam, M.G.; Kaushik, R. Archaea: An agro-ecological perspective. Curr. Microbiol. 2021, 78, 2510–2521. [Google Scholar] [CrossRef] [PubMed]
- Ayangbenro, A.S.; Babalola, O.O. Reclamation of arid and semi-arid soils: The role of plant growth-promoting archaea and bacteria. Curr. Plant Bio. 2021, 25, 100173. [Google Scholar] [CrossRef]
- Khan, S.A.; Akbar, A.; Permpornsakul, P.; Yanwisetpakdee, B.; Chen, X.; Anwar, M.; Ali, I. Molecular diversity of halophilic fungi isolated from mangroves ecosystem of Miani Hor, Balochistan, Pakistan. Pak. J. Bot. 2020, 52, 1823–1829. [Google Scholar] [CrossRef] [PubMed]
- Gunde-Cimerman, N.; Plemenitaš, A.; Oren, A. Strategies of adaptation of microorganisms of the three domains of life to high salt concentrations. FEMS Microbiol. Rev. 2018, 42, 353–375. [Google Scholar] [CrossRef] [PubMed]
- Fotedar, R.; Kolecka, A.; Boekhout, T.; Fell, J.W.; Al-Malki, A.; Zeyara, A.; Al Mari, M. Fungal diversity of the hypersaline inland Sea in Qatar. Bot. Mar. 2018, 61, 595–609. [Google Scholar] [CrossRef]
- Kladwang, W.; Bhumirattana, A.; Hywel-Jones, N. Alkaline-tolerant fungi from Thailand. Fungal Div. 2003, 13, 69–83. [Google Scholar]
- Edbeib, M.F.; Wahab, R.A.; Huyop, F. Halophiles. Biology, adaptation, and their role in decontamination of hypersaline environments. World J. Microbiol. Biotechnol. 2016, 32, 135. [Google Scholar] [CrossRef] [PubMed]
- Gostinčar, C.; Grube, M.; de Hoog, S.; Zalar, P.; Gunde-Cimerman, N. Extremotolerance in fungi: Evolution on the edge. FEMS Microbiol. Ecol. 2009, 71, 2–11. [Google Scholar] [CrossRef] [PubMed]
- Gonḉalves, M.F.M.; Vicente, T.L.F.; Esteves, A.C.; Alves, A. Novel halotolerant species of Emericellopsis and Parasarocladium associated with macroalgae in an estuarine environment. Mycologia 2020, 112, 154–171. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Li, S.J.; Li, J.J.; Liang, Z.Z.; Zhao, C.Q. Novel natural products from extremophilic fungi. Mar. Drugs 2018, 16, 194. [Google Scholar] [CrossRef] [PubMed]
- Musa, H.; Kasim, F.H.; Gunny, A.A.N.; Gopinath, S.C.B. Salt-adapted moulds and yeasts: Potentials in industrial and environmental biotechnology. Process. Biochem. 2018, 69, 33–41. [Google Scholar] [CrossRef]
- Bano, A.; Hussain, J.; Akbar, A.; Mehmood, K.; Anwar, M.; Hasn, M.S.; Ullah, S.; Sajid, S.; Ali, I. Biosorption of heavy metals by obligate halophilic fungi. Chemosphere 2018, 199, 218–222. [Google Scholar] [CrossRef] [PubMed]
- Kreuzberg-Mukhina, E.A. The Aral Sea basin: Changes in migratory and breeding waterbird populations due to major human-induced changes to the region’s hydrology. In Waterbirds around the World; Boere, G.C., Galbraith, C.A., Stroud, D.A., Eds.; The Stationery Office: Edinburgh, UK, 2006; pp. 283–284. [Google Scholar]
- Aladin, N.V.; Plotnikov, I.S.; Micklin, P.; Ballatore, T. Aral Sea: Water level, salinity and long-term changes in biological communities of an endangered ecosystem-past, present and future. NREI 2009, 15, 36. Available online: https://digitalcommons.usu.edu/nrei/vol15/iss1/36 (accessed on 20 January 2024).
- de Hoog, G.S.; Guarro, J.; Gené, J.; Ahmed, S.A.; Al-Hatmi, A.M.S.; Figueras, M.J.; Vitale, R.G. Atlas of Clinical Fungi The ultimate benchtool for diagnostics. In Part α: Introductions, Lower Fungui, Basidiomycetes, Yeasts, Filamentous Ascomycetes A-B, 4th ed.; Foundation Atlas of Clinical Fungi: Hilversum, The Netherlands, 2020; ISBN 978-94-93226-12-8. [Google Scholar]
- Pavlović, J.; Puškárová, A.; Planý, M.; Farkas, Z.; Rusková, M.; Kvalová, K.; Kraková, L.; Bučková, M.; Pangallo, D. Colored stains: Microbial survey of cellulose-based and lignin rich papers. Int. J. Biol. Macromol. 2023, 241, 124456. [Google Scholar] [CrossRef] [PubMed]
- Hubka, V.; Kolařík, M.; Kubátová, A.; Peterson, S.W. Taxonomic revision of Eurotium and transfer of species to Aspergillus. Mycologia 2013, 105, 912–937. [Google Scholar] [CrossRef]
- Visagie, C.M.; Houbraken, J.; Frisvad, J.C.; Hong, S.B.; Klaassen, C.H.W.; Perrone, G.; Seifert, K.A.; Varga, J.; Yaguchi, T.; Samson, R.A. Identification and nomenclature of the genus Penicillium. Stud. Mycol. 2014, 78, 343–371. [Google Scholar] [CrossRef]
- Grivalský, T.; Bučková, M.; Puškárová, A.; Kraková, L.; Pangallo, D. Water-related environments: A multistep procedure to assess the diversity and enzymatic properties of cultivable bacteria. World J. Microbiol. Biotechnol. 2016, 32, 42. [Google Scholar] [CrossRef] [PubMed]
- Ramírez, J.S.; Hoyos, V.C.; Guido Plaza, T. Phytosociology of weeds associated with rice crops in the department of Tolima, Colombia. Agron. Colomb. 2015, 33, 64–73. [Google Scholar] [CrossRef]
- Kraková, L.; Šoltys, K.; Budiš, J.; Grivalský, T.; Ďuriš, F.; Pangallo, D.; Szemes, T. Investigation of bacterial and archaeal communities: Novel protocols using modern sequencing by Illumina MiSeq and traditional DGGE-cloning. Extremophiles 2016, 20, 795–808. [Google Scholar] [CrossRef] [PubMed]
- Šimonovičová, A.; Kraková, L.; Piecková, E.; Planý, M.; Globanová, M.; Pauditšová, E.; Šoltys, K.; Budiš, J.; Szemes, T.; Gáfriková, J.; et al. Soil Microbiota of Dystric Cambisol in the High Tatra Mountains (Slovakia) after Windthrow. Sustainability 2019, 11, 6851. [Google Scholar] [CrossRef]
- Wood, D.E.; Lu, J.; Langmead, B. Improved metagenomic analysis with Kraken 2. Genome Biol. 2019, 20, 257. [Google Scholar] [CrossRef] [PubMed]
- Čurlík, J.; Bedrna, Z.; Hanes, J.; Holobradý, K.; Hrtánek, B.; Kotvas, F.; Masaryk, Š.; Paulen, J. Soil Reaction and Its Adjustment; Bratislava, Jaroslav Suchoň, Soma print Bratislava: Slovak, NY, USA, 2003; 249p. [Google Scholar]
- Nishikawa, J.; Nakashima, C. Japanese species of Alternaria and their species boundaries based on host range. Fungal Syst. Evol. 2020, 5, 197–281. [Google Scholar] [CrossRef] [PubMed]
- Durán, P.; Barra, P.J.; Jorquera, M.A.; Viscardi, S.; Fernandez, C.; Paz, C.; de la Luz Mora, M.; Bol, R. Occurrence of soil fungi in Antarctic Pristine environments. Front. Bioeng. Biotechnol. 2019, 7, 28. [Google Scholar] [CrossRef] [PubMed]
- Günyar, O.A.; Kirac, S.; Aldi, B.; Ergin, C. Isolation and identification of keratinophilic fungi in soil samples from excavation area of ancient city of Stratonikeia, Turkey and determination of its enzyme potentials. J. Environ. Biol. 2020, 41, 1521–1525. [Google Scholar] [CrossRef]
- Klapec, T.; Cholewa, G.; Cholewa, A.; Dutkiewicz, J.; Wójcik-Fatla, A. Fungal diversity of root vegetables and soil rhizosphere collected from organic and conventional farms in Eastern Poland. Ann. Agric. Environ. Med. 2018, 25, 374–381. [Google Scholar] [CrossRef]
- Jaouani, A.; Neifar, M.; Prigione, V.; Ayari, A.; Sbissi, I.; Amor, S.B.; Tekaya, S.B.; Varese, G.C.; Cherif, A.; Gtari, M. Diversity and enzymatic profiling of halotolerant micromycetes from Sebkha El Melah, a Saharan salt flat in Southern Tunisia. Bio. Med. Res. Int. 2014, 2014, 439197. [Google Scholar] [CrossRef]
- Hashimoto, K.; Oda, H.; Saito, Y.; Akimoto, M.; Nojiri, T.; Kawakami, Y. Isolation od Simplicillium sympodiophorum and Toxicocladosporium irritans from the blowout air of houshold air conditioners. Biocontrol. Sci. 2021, 26, 105–111. [Google Scholar] [CrossRef]
- Pethybridge, S.J.; Jones, S.J.; Shivas, R.G.; Hay, F.S.; Wilson, C.R.; Groom, T. Tan spot: A new disease of pyrethrum caused by Microsphaeropsis tanaceti sp. nov. Plant Pathol. 2008, 57, 1058–1065. [Google Scholar] [CrossRef]
- Soytong, K.; Kahonokmedhakul, S.; Song, J.; Tongon, R. Chaetomium Application in Agriculture. In Technology in Agriculture; IntechOpen Limited: London, UK, 2021; Open access peer. reviewed chapter. [Google Scholar] [CrossRef]
- Chen, A.J.; Hubka, V.; Frisvad, J.C.; Visagie, C.M.; Houbraken, J.; Meijer, M.; Varga, J.; Demirel, R.; Jurjević, Ž.; Kubátová, A.; et al. Polyphasic taxonomy of Aspergillus section Aspergillus (formerly Eurotium), and its occurrence in indoor environments and food. Stud. Mycol. 2017, 88, 37–135. [Google Scholar] [CrossRef] [PubMed]
- López, A.L.; Amador, G.B.T.; Tovar, L.J.M.; Ramírez, B.S.H.; Hernández, F.H. Aspergillus proliferans Onychomycossis forming fungal masses. Dermatol. CMQ 2021, 19, 174–179. [Google Scholar]
- Rodriguez-Andrade, E.; Cano-Lira, J.F.C.; Wiederhold, N.; Pérez-Cantero, A. A revision of amlbranchea-like fungi from clinical specimens in the United States of America reveals unexpected novelty. IMA Fungus 2021, 12, 25. [Google Scholar] [CrossRef] [PubMed]
- Shubert, K.; Groenewald, J.Z.; Braun, U.; Dijksteruis, J.; Starink, M.; Hill, C.F.; Zalar, P.; de Hoog, G.S.; Crous, P.W. Biodiversity in the Cladospoium herbarum complex (Davidiellaceae, Capnodiales), with standartisation of methods for Cladosporium taxonomy and diagnostics. Stud. Mycol. 2007, 58, 105–156. [Google Scholar] [CrossRef] [PubMed]
- Kochhar, S.; Gupta, V.S.; Sethi, H.S.; Capoor, M.R. A rare case of keratinomycosis due to Myriodontium keratinophilum. DJO 2018, 29, 54–56. [Google Scholar] [CrossRef]
- Deshmukh, S.K.; Verekar, S.A. prevalence of keratinophilic fungi in public park soils of Mumbai, India. Microbiol. Res. 2012, 3, e6. [Google Scholar] [CrossRef]
- Pangallo, D.; Kraková, L.; Chovanová, K.; Bučková, M.; Puškárová, A.; Šimonovičová, A. Disclosing a crypt. Microbial diversity and degradatiobn activity of the microflora isolated from funeral clothes of Cardinal Peter Pázmany. Microbiol. Res. 2013, 168, 289–299. [Google Scholar] [CrossRef]
- Kirichuk, N.N.; Pivkin, M.V.; Matveeva, T.V. Three new Penicillium species from marine subaqueous soils. Mycol. Prog. 2017, 16, 15–26. [Google Scholar] [CrossRef]
- Takahashi, K.; Sakai, K.; Nagano, Y.; Sakaguchi, S.O.; Lima, A.O.; Pellizari, V.H.; Iwatsuki, M.; Takishita, K.; Nonaka, K.; Fujikura, K.; et al. Cladomarine, a new anti-saprolegniasis compound isolared from the deep-sea fungus, Penicillium coralligerum YK-247. J. Antibiot. 2017, 70, 911–914. [Google Scholar] [CrossRef] [PubMed]
- Trabelsi, H.; Neji, S.; Hadrich, I.; Sellami, M.; Khemakhem, N.; Sellami, H.; Makni, F.; Hammami, B.; Ayadi, A. Unusual case of otomycosis caused by Saksenaea vasiformis. Med. Mycol. Case. Rep. 2020, 27, 68–71. [Google Scholar] [CrossRef] [PubMed]
- Jurjevic, Z.; Peterson, S.W.; Horn, B.W. Aspergillus section Versicolores: Nine new species and multilocus DNA sequences based phylogeny. IMA Fungus 2012, 3, 59–79. [Google Scholar] [CrossRef] [PubMed]
- Oetari, A.; Susetyo-Salim, T.; Sjamsuridzal, W.; Suherman, E.A.; Monica, M.; Wongso, R.; Fitri, R.; Nurlaili, D.G.; Ayu, D.C.; Teja, T.P. Occurrence of fungi on deriororated old dluwang manuscript from Indonesia. Int. Biodeter. Biodegr. 2016, 114, 94–103. [Google Scholar] [CrossRef]
- Haas, D.; Lesch, S.; Buzina, W.; Galler, H.; Gutschi, A.M.; Habib, J.; Pfeifer, B.; Luxner, J.; Reinthaler, F.F. Culturable fungi in potting soils and compost. Medical Mycol. 2016, 54, 825–834. [Google Scholar] [CrossRef] [PubMed]
- Parsi, F. Entomopathogenic fungi collected from Sunn Pest, Eurygaster integriceps Puton (Hemiptera: Scutelleridae), overwintering sites in central Iran. J. Biol. Today’s World 2019, 8, 210. [Google Scholar]
- Kondrasheva, K.V.; Umruzokov, A.A.; Kalenov, S.V.; Merkel, A.Y.; Chernyh, N.A.; Slobodkin, A.I.; Gavrilov, S.N.; Davranov, K.D. Calcinating Bacteria in Extreme Ecosystems of the Southern Aral Region. Microbiology 2023, 92, 473–480. [Google Scholar] [CrossRef]
- Gao, L.; Ma, J.; Liu, Y.; Huang, Y.; Mohamad, O.A.A.; Jiang, H.; Egamberdieva, D.; Li, W.; Li, L. Diversity and biocontrol potential of cultivable endophytic bacteria associated with halophytes from the West Aral Sea basin. Microorganisms 2021, 9, 1448. [Google Scholar] [CrossRef] [PubMed]
- Osman, J.R.; Fernandes, G.R.; Kamilova, E.; DuBow, M.S. Genomic microbiome analyses of surface sand samples from the Kyzyl-Kum Desert (Uzbekistan): Characterization and comparative study. Arch. Microbiol. 2023, 205, 90. [Google Scholar] [CrossRef]
- Manni, A.; Filali-Maltouf, A. Diversity and bioprospecting for industrial hydrolytic enzymes of microbial communities isolated from deserted areas of south-east Morocco. AIMS Microbiol. 2022, 8, 5. [Google Scholar] [CrossRef]
- Alexyuk, M.; Bogoyavlenskiy, A.; Alexyuk, P.; Moldakhanov, Y.; Berezin, V.; Digel, I. Epipelagic microbiome of the Small Aral Sea: Metagenomic structure and ecological diversity. MicrobiologyOpen 2021, 10, e1142. [Google Scholar] [CrossRef] [PubMed]
- Pátek, M.; Grulich, M.; Nešvera, J. Stress response in Rhodococcus strains. Biotechnol. Adv. 2021, 53, 107698. [Google Scholar] [CrossRef] [PubMed]
- Bennur, T.; Kumar, A.R.; Zinjarde, S.; Javdekar, V. Nocardiopsis species: Incidence, ecological roles and adaptations. Microbiol. Res. 2015, 174, 33–47. [Google Scholar] [CrossRef] [PubMed]
- Wicaksono, W.A.; Egamberdieva, D.; Berg, C.; Mora, M.; Kusstatscher, P.; Cernava, T.; Berg, G. Function-based rhizosphere assembly along a gradient of desiccation in the former Aral Sea. mSystems 2022, 20, e00739-22. [Google Scholar] [CrossRef] [PubMed]
- Murray, T.D.; Schroeder, B.K.; Schneider, W.L.; Luster, D.G.; Sechler, A.; Rogers, E.E.; Subbotin, S.A. Rathayibacter toxicus, other Rathayibacter species inducing bacterial head blight of grasses, and the potential for livestock poisonings. Phytopathol. 2017, 107, 804–815. [Google Scholar] [CrossRef] [PubMed]
- Malarczyk, D.G.; Panek, J.; Frąc, M. Triplex Real-Time PCR approach for the detection of crucial fungal berry pathogens—Botrytis spp., Colletotrichum spp. and Verticillium spp. Int. J. Mol. Sci. 2020, 21, 8469. [Google Scholar] [CrossRef] [PubMed]
- Moraes, D.; Rodrigues, J.G.C.; Silva, M.G.; Soares, L.W.; de Almeida Soares, C.M.; Bailão, A.M.; Silva-Bailão, M.G. Copper acquisition and detoxification machineries are conserved in dimorphic fungi. Fungal Biol. Rev. 2023, 44, 100296. [Google Scholar] [CrossRef]
- Jiang, H.; Huang, J.; Li, L.; Huang, L.; Manzoor, M.; Yang, J.; Wu, G.; Sun, X.; Wang, B.; Egamberdieva, D.; et al. Onshore soil microbes and endophytes respond differently to geochemical and mineralogical changes in the Aral Sea. Sci. Total Environ. 2021, 765, 142675. [Google Scholar] [CrossRef] [PubMed]
- Chen, D.D.; Fang, B.Z.; Manzoor, A.; Liu, Y.H.; Li, L.; Mohamad, O.A.A.; Shu, W.S.; Li, W.J. Revealing the salinity adaptation mechanism in halotolerant bacterium Egicoccus halophilus EGI 80432 T by physiological analysis and comparative transcriptomics. Appl. Microbiol. Biotechnol. 2021, 105, 2497–2511. [Google Scholar] [CrossRef]
- Shurigin, V.; Hakobyan, A.; Panosyan, H.; Egamberdieva, D.; Davranov, K.; Birkeland, N.K. A glimpse of the prokaryotic diversity of the Large Aral Sea reveals novel extremophilic bacterial and archaeal groups. Microbiol. Open 2019, 8, e00850. [Google Scholar] [CrossRef]



| Samples | pHH20 | % | ||||
|---|---|---|---|---|---|---|
| TC | TN | C Org. | Humus | CaCO3 | ||
| A1a | 8.4 | 1.6 | 0.03 | 0.1 | 0.17 | 14 |
| A1b | 8.6 | 1.0 | 0.01 | 0.1 | 0.17 | 8.5 |
| A2a | 8.1 | 2.1 | 0.04 | 0.2 | 0.34 | 16.5 |
| A2b | 8.5 | 2.6 | 0.04 | 0.2 | 0.34 | 19.3 |
| A3a | 8.2 | 1.6 | 0.02 | 0.5 | 0.86 | 8.6 |
| A3b | 8.6 | 3.2 | 0.04 | 0.6 | 1.03 | 22.3 |
| Samples | Ca2+ | Na2+ | Mg2+ | K+ | Al3+ | Mn2+ | Fe2+ | H+ | Salinity |
|---|---|---|---|---|---|---|---|---|---|
| g/kg | % | ||||||||
| A1a | 1.1 | 0.06 | 0.4 | 0.1 | 0.02 | 0.005 | 0.56 | 0.002 | 0.22 |
| A1b | 3.1 | 0.9 | 1.3 | 0.1 | 0.02 | 0.005 | 0.56 | 0.002 | 0.60 |
| A2a | 3.3 | 1.3 | 1.6 | 0.4 | 0.02 | 0.005 | 0.56 | 0.002 | 0.72 |
| A2b | 2.1 | 4.8 | 3.3 | 0.5 | 0.02 | 0.005 | 0.56 | 0.002 | 1.13 |
| A3a | 26.0 | 5.6 | 3.6 | 0.3 | 0.02 | 0.005 | 0.56 | 0.002 | 3.61 |
| A3b | 2.5 | 5.9 | 0.7 | 0.5 | 0.02 | 0.005 | 0.56 | 0.002 | 1.02 |
| Genera/Species/Sequence Similarity | Study Soil Samples | |||||
|---|---|---|---|---|---|---|
| A1a | A1b | A2a | A2b | A3a | A3b | |
| KF747365 Alternaria alternata 100% | + | − | − | − | − | − |
| MG780401 Alternaria atra 100% | + | − | + | − | − | − |
| MF462308 Alternaria japonica 100% | + | − | − | − | − | − |
| MH345962 Alternaria tenuissima 100% | + | − | − | − | − | − |
| MT645322 Aspergillus flavus 100% | − | − | − | + | + | − |
| MT487775 Aspergillus fumigatus 100% | + | − | − | + | − | + |
| KX258805 Aspergillus pseudoglaucus 100% | − | − | − | + | − | − |
| LN898704 Aspergillus jensenii 100% | − | − | − | − | + | − |
| KJ701548 Aspergillus niger 100% | − | − | − | + | + | + |
| MW058064 Aspergillus oryzae 99% | − | − | − | + | − | − |
| KX696377 Aspergillus proliferans 100% | − | − | − | + | − | − |
| MH857026 Auxarthron umbrinum 100% | − | − | − | + | − | − |
| MT529803 Chaetomium globosum 100% | − | − | + | − | − | − |
| MT131338 Cladosporium cladosporioides 100% | − | − | − | − | + | + |
| MF472979 Cladosporium floccosum 100% | − | − | + | − | − | − |
| OR243878 Cladosporium herbarum 100% | − | − | + | − | − | − |
| OQ608649 Cladosporium iridis 100% | − | − | − | + | − | + |
| MG787259 Cladosporium sphaerospermum 100% | + | − | − | − | − | |
| ON229430 Epicoccum nigrum 100% | − | − | − | − | − | + |
| MH191137 Isaria farinosa 100% | − | − | − | − | − | + |
| MK719910 Neomicrosphaeropsis italica 99% | − | − | + | − | − | − |
| MH861337 Myriodontium keratinophilum 100% | − | − | − | + | − | − |
| MN947607 Parengyodontium album 100% | − | + | − | − | − | |
| MT103060 Penicillium chrysogenum 100% | − | + | − | − | + | + |
| KP016836 Penicillium coralligerum 99% | − | − | − | + | − | − |
| KP411588 Penicillium echinulatum 100% | − | − | − | − | + | − |
| MN643064 Penicillium expansum 99% | − | + | − | − | − | − |
| GU566230 Penicillium rugulosum 100% | − | − | − | + | − | − |
| KC206538 Rhizopus microsporus 100% | + | − | + | − | − | − |
| FR687323 Saksenaea vasiformis 100% | − | − | − | + | − | − |
| NR_111027 Simplicillium sympodiophorum 100% | − | − | + | − | − | − |
| MK802874 Stachybotrys chlorohalonata 100% | − | + | − | − | − | − |
| ∑ 15 genera/32 species | 7 | 4 | 7 | 12 | 6 | 7 |
| A1a | A1b | A2a | A2b | A3a | A3b | |
|---|---|---|---|---|---|---|
| A1a | - | S/J = 0.64 | S/J = 0.47 | S/J = 0.35 | S/J = 0.54 | S/J = 0.47 |
| A1b | S/J = 0.64 | - | S/J = 0.36 | S/J = 0.25 | S/J = 0.36 | S/J = 0.33 |
| A2a | S/J = 0.36 | S/J = 0.36 | - | S/J = 0.37 | S/J = 0.54 | S/J = 0.5 |
| A2b | S/J = 0.35 | S/J = 0.25 | S/J = 0.37 | - | S/J = 0.5 | S/J = 0.57 |
| A3a | S/J = 0.47 | S/J = 0.36 | S/J = 0.54 | S/J = 0.63 | - | S/J = 0.37 |
| A3b | S/J = 0.47 | S/J = 0.33 | S/J = 0.5 | S/J = 0.57 | S/J = 0.37 | - |
| Genera/Species/Sequence Similarity | Study Soil Samples | |||||
|---|---|---|---|---|---|---|
| A1a | A1b | A2a | A2b | A3a | A3b | |
| KF747044 Massilia sp. 99% | + | − | − | − | − | − |
| MF077219 Massilia varians 99% | + | − | − | − | − | − |
| JQ977399 Arthrobacter sp. 100% | + | − | − | + | − | − |
| OR539574 Arthrobacter humicola 100% | + | + | − | − | − | − |
| KY753214 Arthrobacter crystallopoietes 100% | + | − | − | − | − | − |
| MN931288 Bacillus coreaensis 100% | + | − | − | − | − | − |
| MG705966 Bacillus flexus 100% | + | − | + | + | − | − |
| LN995471 Bacillus firmus 100% | − | − | − | − | + | + |
| MH819519 Bacillus sp. 100% | + | + | + | + | + | + |
| MH806388 Bacillus cereus 100% | − | + | + | + | − | − |
| MF620082 Bacillus paralicheniformis 100% | − | − | − | + | − | − |
| MT804101 Pontibacter sp. 98% | − | − | + | − | − | − |
| KC354466 Agrococcus citreus 100% | + | − | − | − | − | − |
| KU204869 Pseudarthrobacter siccitolerans 99% | + | − | − | − | − | − |
| NR_113620 Oxalicibacterium horti 100% | + | − | − | − | − | − |
| MG254794 Kocuria sp. 99% | + | − | − | − | − | − |
| KT922050 Rhodococcus erythropolis 100% | + | + | − | − | − | − |
| KF040418 Rhodococcus sp. 99% | + | + | − | − | − | − |
| KT003514 Pseudomonas sp. 100% | + | + | − | + | − | − |
| MW089200 Microbacterium sp. 100% | − | + | − | − | − | − |
| LC565814 Paraliobacillus sp. 100% | − | + | − | − | − | − |
| KF876899 Nocardiopsis flavescens 100% | − | − | + | + | − | − |
| KC493983 Nocardiopsis aegyptia 100% | − | − | − | + | − | − |
| JQ885684 Nocardiopsis sp. 99% | − | − | − | + | − | − |
| KC336307 Streptomyces sp. 100% | − | − | + | − | − | − |
| KY952739 Halomonas sp. 99% | − | − | + | − | − | − |
| MH392690 Sphingomonas sp. 99% | − | − | − | − | + | − |
| MH813363 Rathayibacter sp. 99% | − | − | − | − | + | − |
| FR727710 Paenisporosarcina sp. 99% | − | − | − | − | − | + |
| ∑ 18 genera/28 species | 15 | 8 | 7 | 9 | 4 | 3 |
| A1a | A1b | A2a | A2b | A3a | A3b | |
|---|---|---|---|---|---|---|
| A1a | - | S/J = 0.54 | S/J = 0.65 | S/J = 0.62 | S/J = 0.75 | S/J = 0.79 |
| A1b | S/J = 0.54 | - | S/J = 0.47 | S/J = 0.40 | S/J = 0.61 | S/J = 0.67 |
| A2a | S/J = 0.65 | S/J = 0.47 | - | S/J = 0.35 | S/J = 0.58 | S/J = 0.64 |
| A2b | S/J = 0.62 | S/J = 0.40 | S/J = 0.35 | - | - | S/J = 0.69 |
| A3a | S/J = 0.75 | S/J = 0.61 | S/J = 0.58 | S/J = 0.58 | - | S/J = 0.44 |
| A3b | S/J = 0.79 | S/J = 0.67 | S/J = 0.64 | S/J = 0.69 | S/J = 0.44 | - |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Šimonovičová, A.; Pauditšová, E.; Nosalj, S.; Oteuliev, M.; Klištincová, N.; Maisto, F.; Kraková, L.; Pavlović, J.; Šoltys, K.; Pangallo, D. Fungal and Prokaryotic Communities in Soil Samples of the Aral Sea Dry Bottom in Uzbekistan. Soil Syst. 2024, 8, 58. https://doi.org/10.3390/soilsystems8020058
Šimonovičová A, Pauditšová E, Nosalj S, Oteuliev M, Klištincová N, Maisto F, Kraková L, Pavlović J, Šoltys K, Pangallo D. Fungal and Prokaryotic Communities in Soil Samples of the Aral Sea Dry Bottom in Uzbekistan. Soil Systems. 2024; 8(2):58. https://doi.org/10.3390/soilsystems8020058
Chicago/Turabian StyleŠimonovičová, Alexandra, Eva Pauditšová, Sanja Nosalj, Medetbay Oteuliev, Nikola Klištincová, Francesca Maisto, Lucia Kraková, Jelena Pavlović, Katarína Šoltys, and Domenico Pangallo. 2024. "Fungal and Prokaryotic Communities in Soil Samples of the Aral Sea Dry Bottom in Uzbekistan" Soil Systems 8, no. 2: 58. https://doi.org/10.3390/soilsystems8020058
APA StyleŠimonovičová, A., Pauditšová, E., Nosalj, S., Oteuliev, M., Klištincová, N., Maisto, F., Kraková, L., Pavlović, J., Šoltys, K., & Pangallo, D. (2024). Fungal and Prokaryotic Communities in Soil Samples of the Aral Sea Dry Bottom in Uzbekistan. Soil Systems, 8(2), 58. https://doi.org/10.3390/soilsystems8020058







