Structural Shifts in the Soil Prokaryotic Communities Marking the Podzol-Forming Process on Sand Dumps
Abstract
1. Introduction
2. Materials and Methods
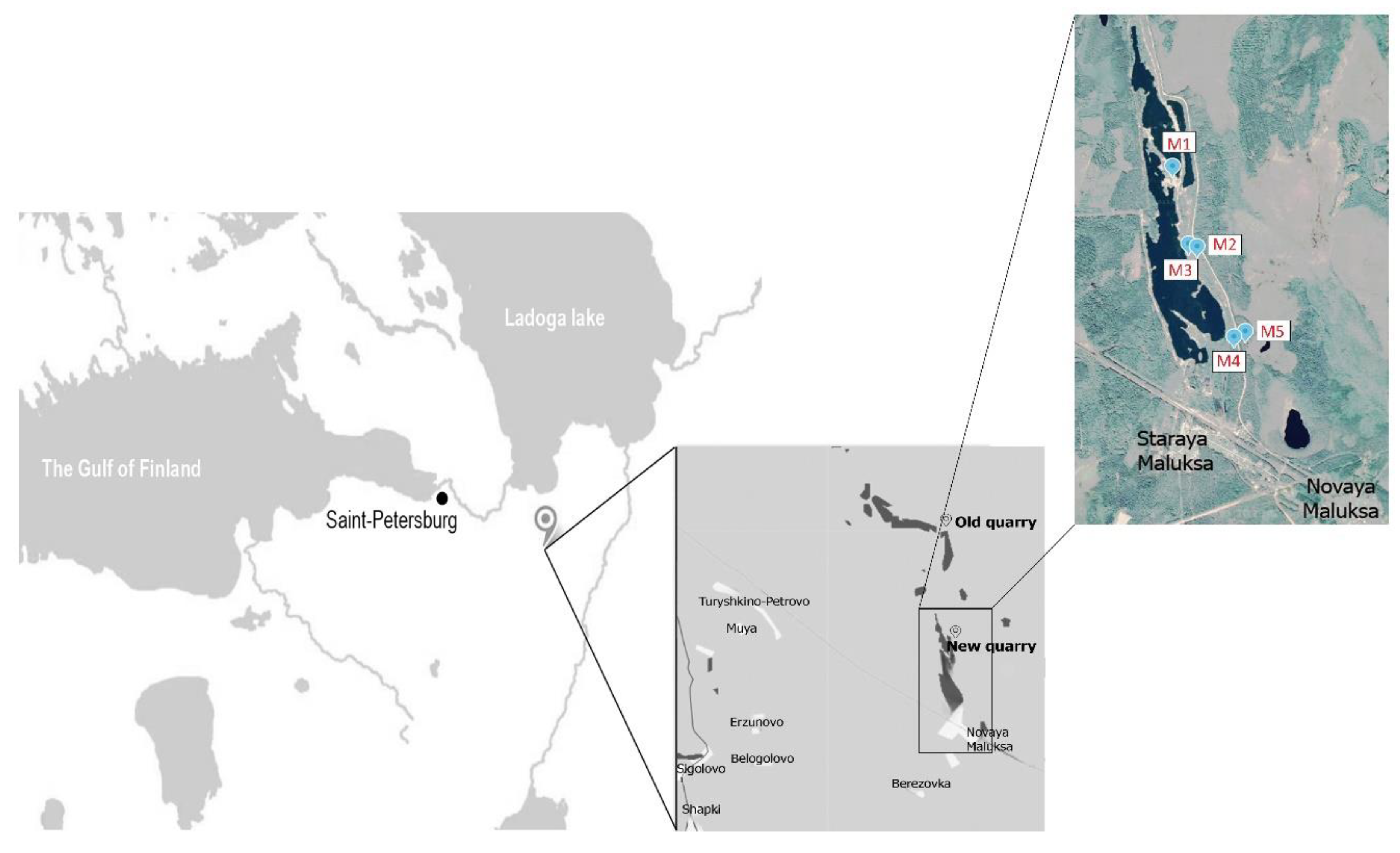
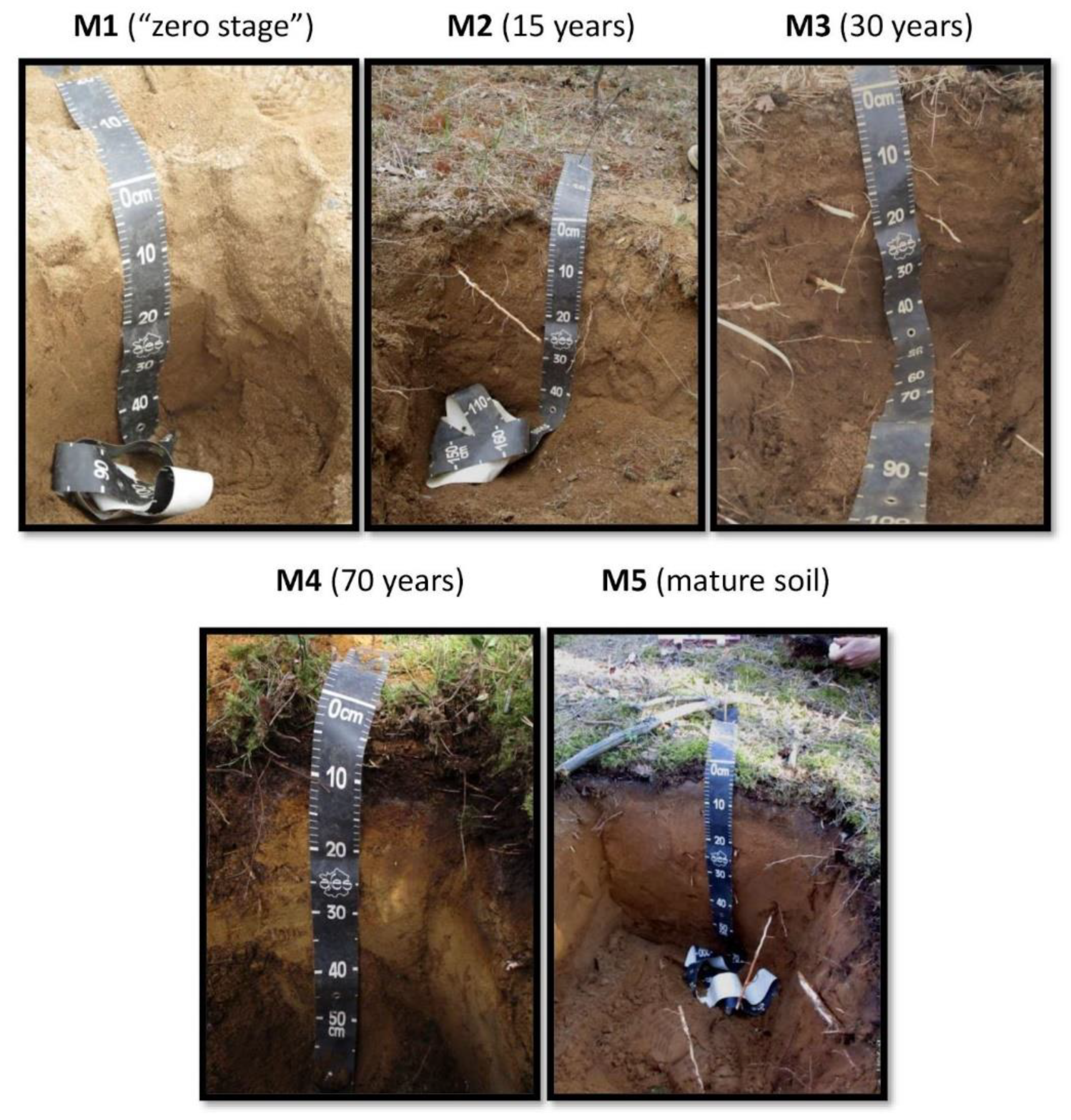
2.1. Study Sites
2.2. Soil Analyses
2.3. Soil DNA Extraction
2.4. Next-Generation Sequencing (NGS) of Amplicon Libraries of the 16S rRNA Gene
2.5. Bioinformatics and Statistical Analysis
3. Results
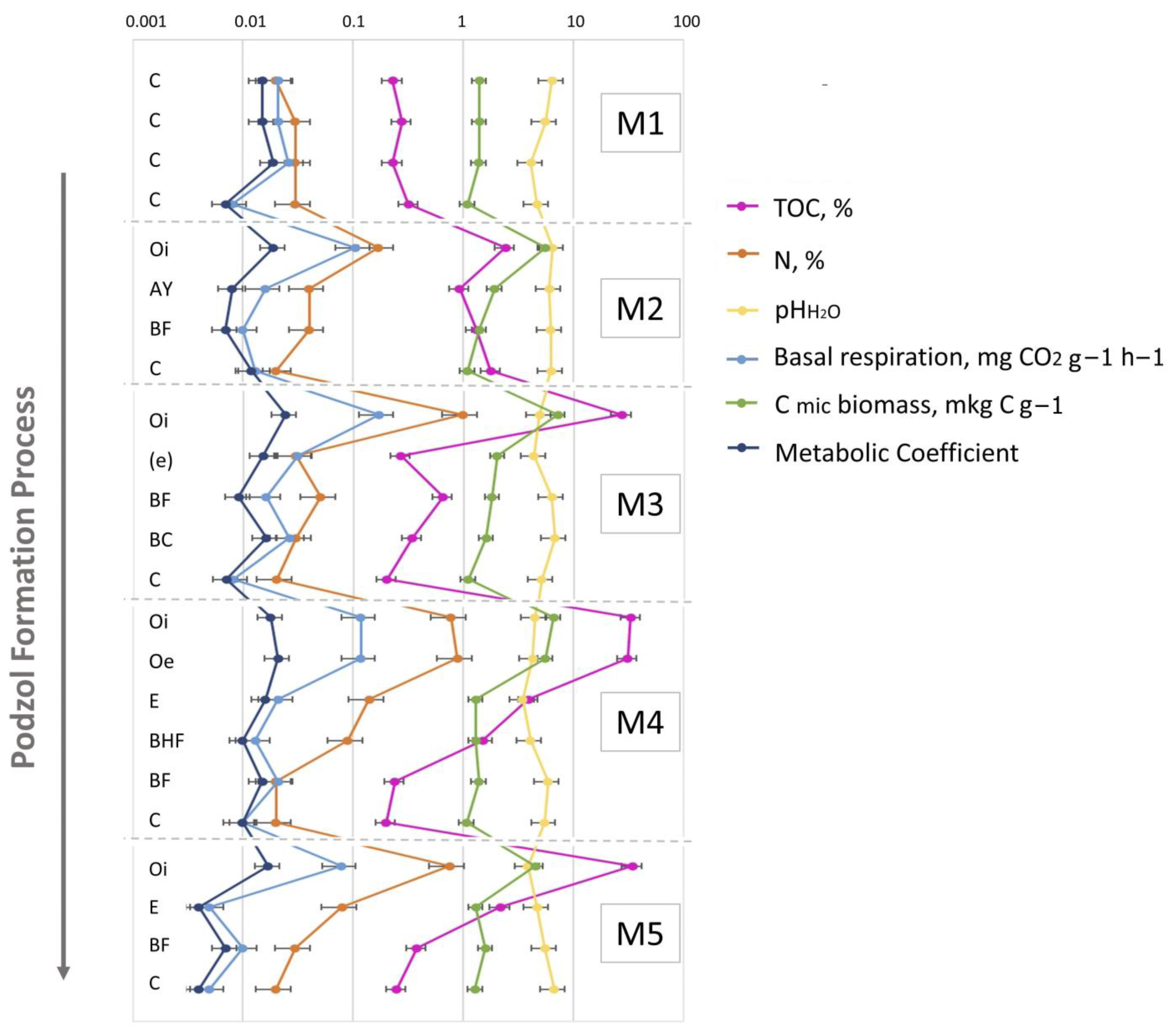
3.1. General Soil Characteristics
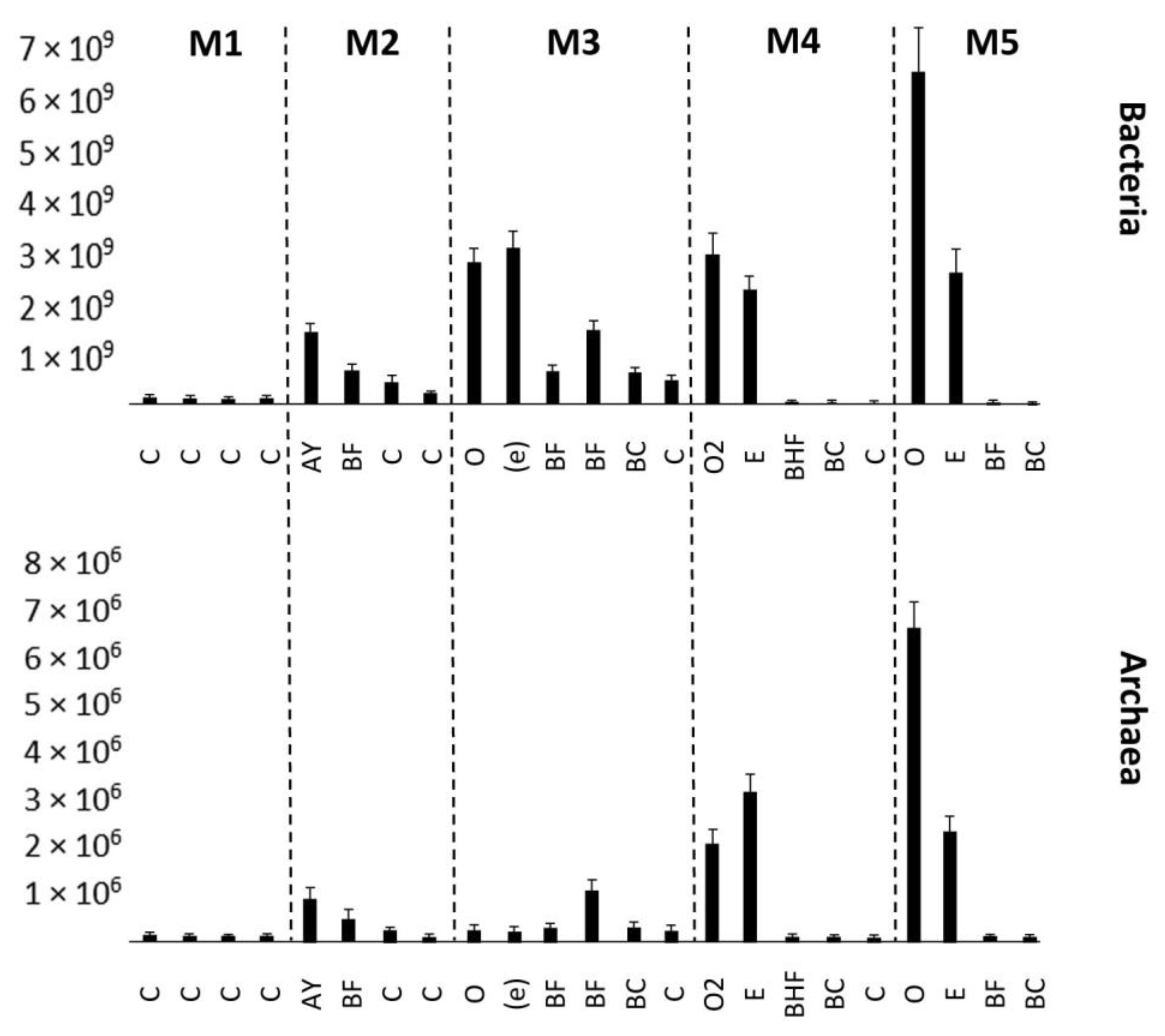
3.2. Real-Time PCR
3.3. Alpha Diversity of Soil Microbiomes
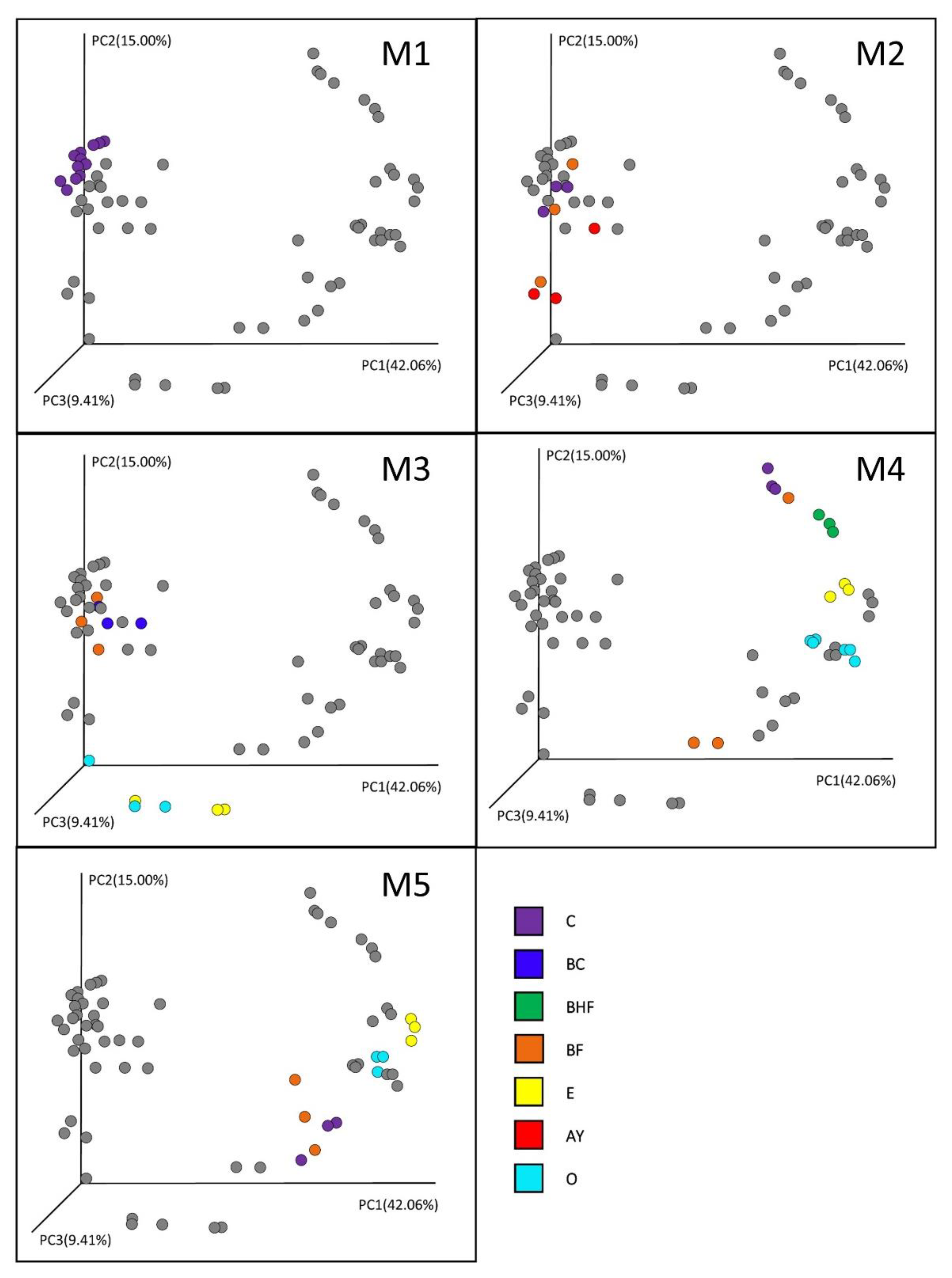
3.4. Beta Diversity of Soil Prokaryotic Communities
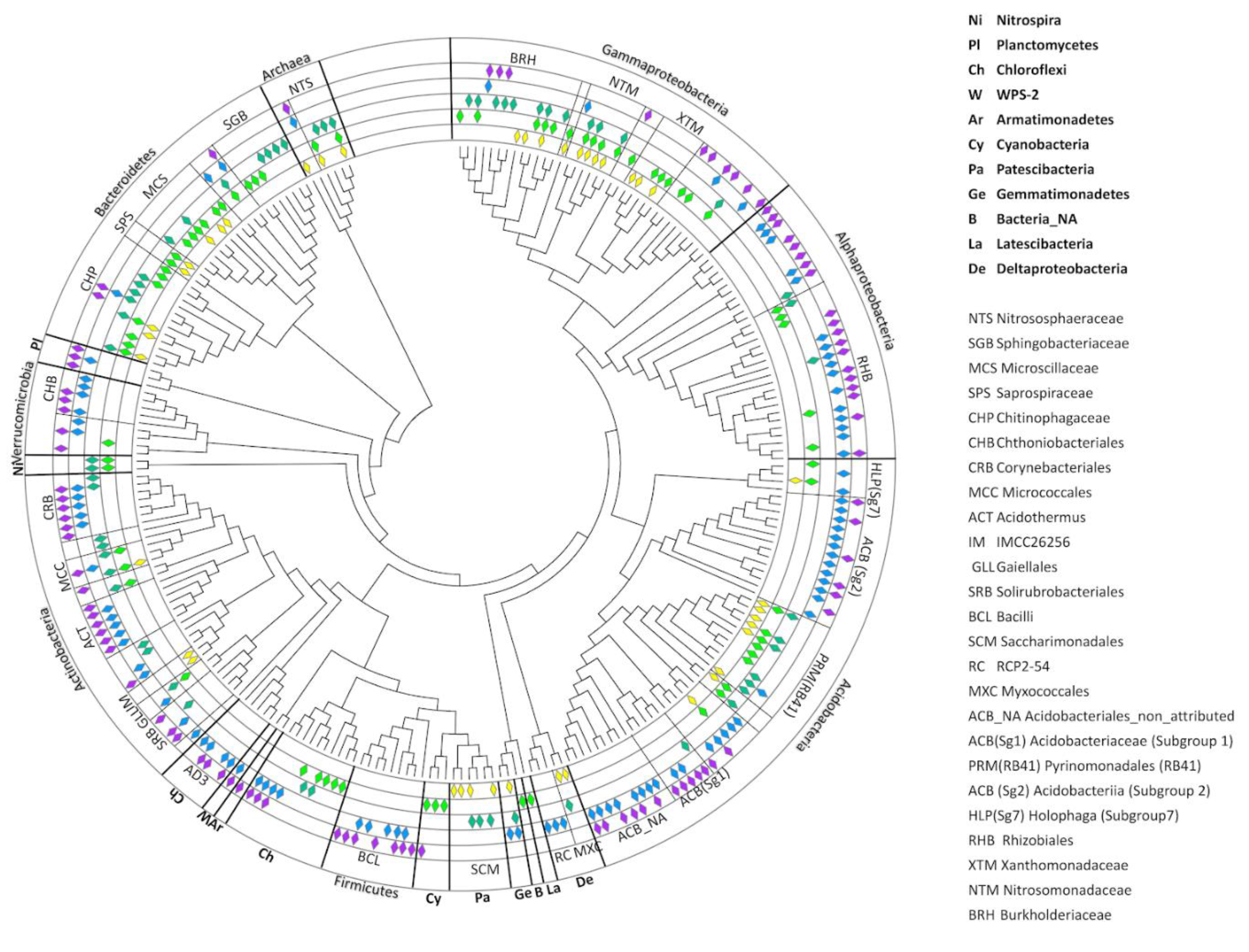
3.5. Taxonomic Characteristics of the Entire Dataset
3.6. Taxonomic Composition of the Core Prokaryotic Communities in the Different Stages of Soil Succession
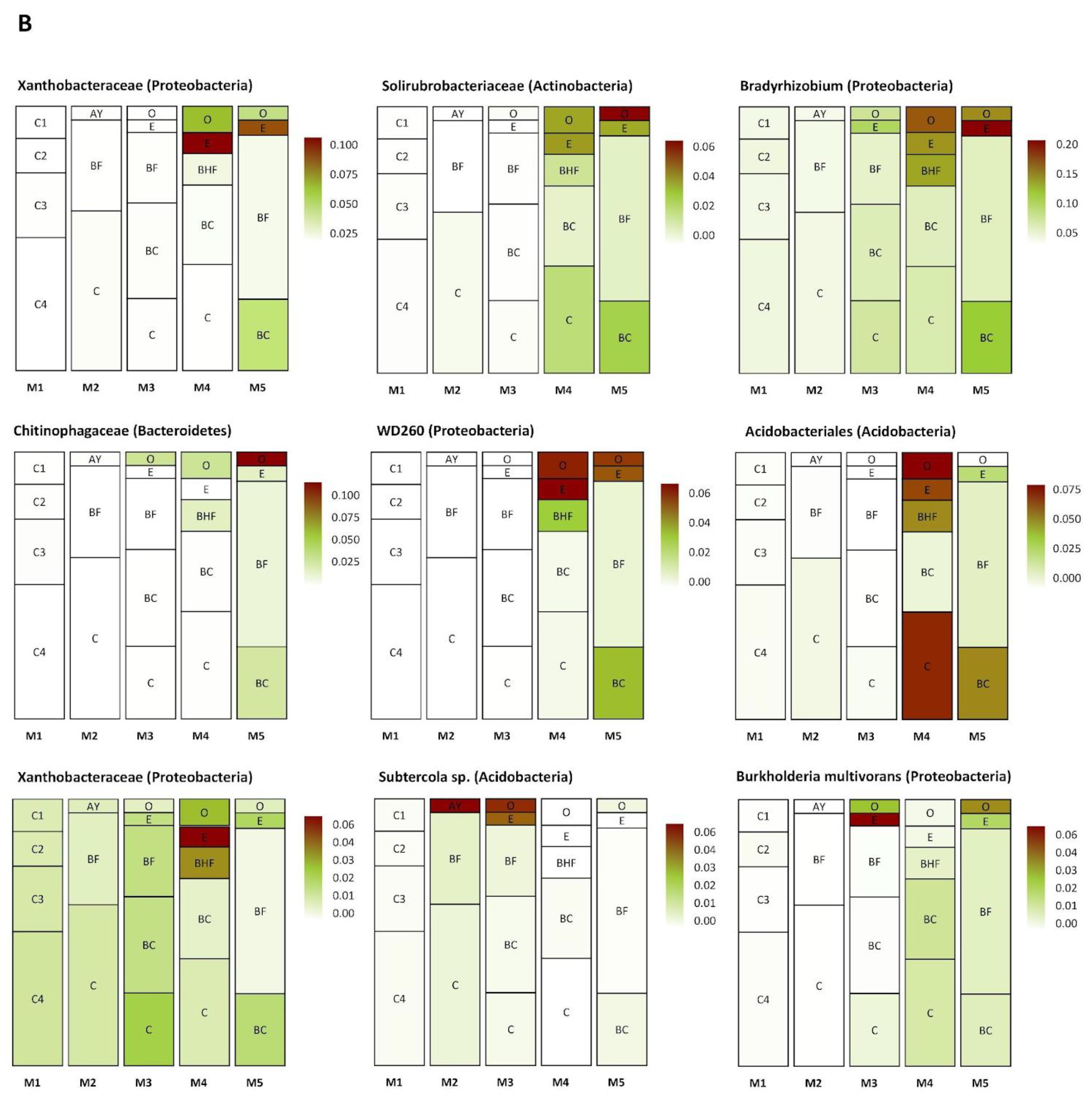
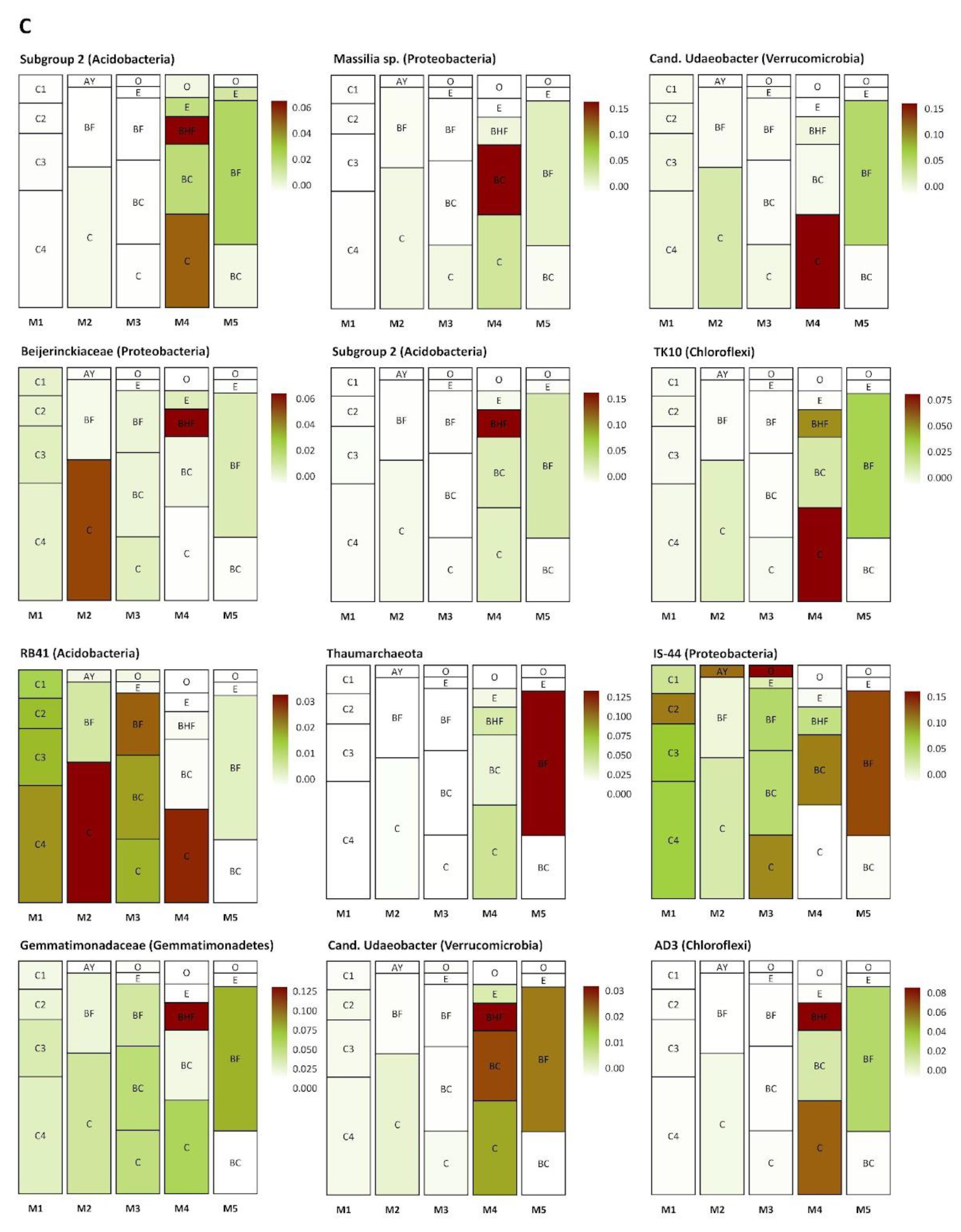
3.7. Dynamic Changes in the Compositions of the Prokaryotic Communities Inhabiting Different Soil Horizons through Time
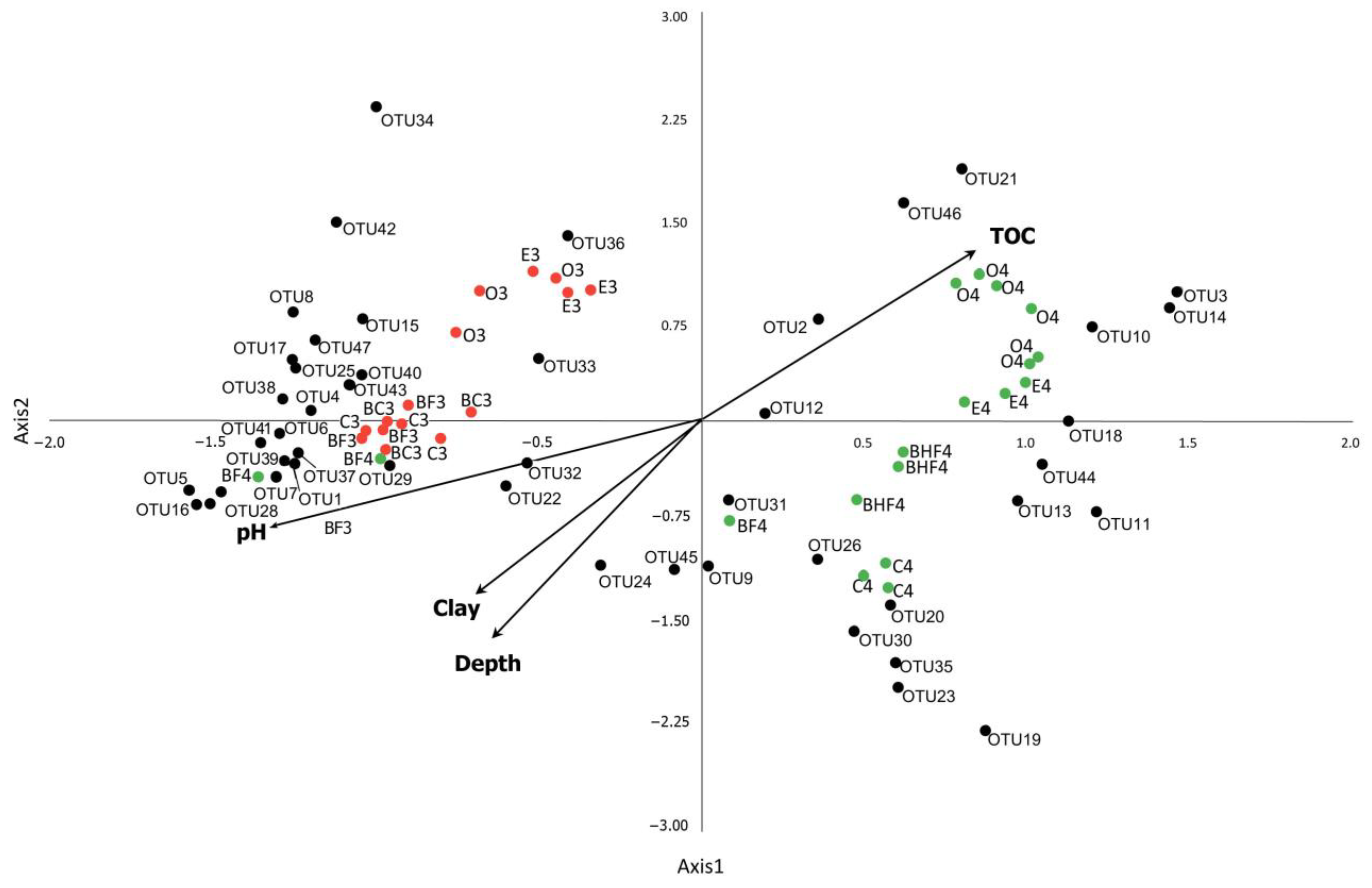
3.8. Influence of Physicochemical Factors in Shaping Prokaryotic Communities’ Structure in the Course of Podzol Profile Evolution
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Targulian, V.O.; Krasilnikov, P.V. Soil System and Pedogenic Processes: Self-Organization, Time Scales, and Environmental Significance. Catena 2007, 71, 373–381. [Google Scholar] [CrossRef]
- Abakumov, E.; Parnikoza, I.Y.; Vlasov, D.Y.; Lupachev, A. Biogenic–Abiogenic Interaction in Antarctic Ornithogenic Soils. In Biogenic—Abiogenic Interactions in Natural and Anthropogenic Systems; Mergelov, N.S., Shorkunov, I.G., Targulian, V.O., Dolgikh, A.V., Abrosimov, K.N., Zazovskaya, E.P., Goryachkin, S.V., Eds.; Springer International Publishing: Cham, Switzerland, 2016; pp. 205–222. ISBN 978-3-319-24985-8. [Google Scholar]
- Abakumov, E.V. Accumulation and Transformation of Organic Matter in Different-Aged Dumps from Sand Quarries. Eurasian Soil Sci. 2008, 41, 844–851. [Google Scholar] [CrossRef]
- Mokma, D.L.; Yli-halla, M.; Lindqvist, K. Podzol Formation in Sandy Soils of Finland. Geoderma 2004, 120, 259–272. [Google Scholar] [CrossRef]
- Bowman, G.M. Podzol Development in a Holocene Moruya Heads, New South Wales. Aust. J. Soil Res. 1989, 27, 607–628. [Google Scholar] [CrossRef]
- Abakumov, E.V.; Frouz, J. Evolution of the Soil Humus Status on the Calcareous Neogene Clay Dumps of the Sokolov Quarry Complex in the Czech Republic. Eurasian Soil Sci. 2009, 42, 718–724. [Google Scholar] [CrossRef]
- Huggett, R.J. Soil Chronosequences, Soil Development, and Soil Evolution: A Critical Review. Catena 1998, 32, 155–172. [Google Scholar] [CrossRef]
- Rosling, A.; Landeweert, R.; Lindah, B.D.; Larsson, K.H.; Kuyper, T.W.; Taylor, A.F.S.; Finlay, R.D. Vertical distribution of ectomycorrhizal fungal taxa in a podzol soil profile. New Phytol. 2003, 159, 775–783. [Google Scholar] [CrossRef]
- Naylor, D.; McClure, R.; Jansson, J. Trends in Microbial Community Composition and Function by Soil Depth. Microorganisms 2022, 10, 540. [Google Scholar] [CrossRef]
- Gagelidze, N.A.; Amiranashvili, L.L.; Sadunishvili, T.A.; Kvesitadze, G.I.; Urushadze, T.F.; Kvrivishvili, T.O. Bacterial Composition of Different Types of Soils of Georgia. Ann. Agrar. Sci. 2018, 16, 17–21. [Google Scholar] [CrossRef]
- Geisen, S. The Future of Soil Microbiome Studies: Current Limitations, Integration, and Perspectives. mSystems 2021, 6, e00613-21. [Google Scholar] [CrossRef]
- Fierer, N.; Schimel, J.P.; Holden, P.A. Variations in microbial community composition through two soil depth profiles. Soil Biol. Biochem. 2003, 35, 167–176. [Google Scholar] [CrossRef]
- Hansel, C.M.; Fendorf, S.; Jardine, P.M.; Francis, C.A. Changes in Bacterial and Archaeal Community Structure and Functional Diversity along a Geochemically Variable Soil Profile. Appl. Environ. Microbiol. 2008, 74, 1620–1633. [Google Scholar] [CrossRef] [PubMed]
- Will, C.; Thürmer, A.; Wollherr, A.; Nacke, H.; Herold, N.; Schrumpf, M.; Gutknecht, J.; Wubet, T.; Buscot, F.; Daniell, R. Horizon-Specific Bacterial Community Composition of German Grassland Soils, as Revealed by Pyrosequencing-Based Analysis of 16S RRNA Genes. Appl. Environ. Microbiol. 2010, 76, 6751–6759. [Google Scholar] [CrossRef] [PubMed]
- Richter, D.D.; Markewitz, D. How Deep Is Soil? BioScience 1995, 45, 600–609. [Google Scholar] [CrossRef]
- Eilers, K.G.; Debenport, S.; Anderson, S.; Fierer, N. Digging Deeper to Find Unique Microbial Communities: The Strong Effect of Depth on the Structure of Bacterial and Archaeal Communities in Soil. Soil Biol. Biochem. 2012, 50, 58–65. [Google Scholar] [CrossRef]
- Hartmann, A.; Schmid, M.; van Tuinen, D.; Berg, G. Plant-Driven Selection of Microbes. Plant Soil 2009, 321, 235–257. [Google Scholar] [CrossRef]
- Hartmann, M.; Howes, C.G.; Vaninsberghe, D.; Yu, H.; Bachar, D.; Christen, R.; Henrik Nilsson, R.; Hallam, S.J.; Mohn, W.W. Significant and Persistent Impact of Timber Harvesting on Soil Microbial Communities in Northern Coniferous Forests. ISME J. 2012, 6, 2199–2218. [Google Scholar] [CrossRef] [PubMed]
- Agnelli, A.; Ascher, J.; Corti, G.; Ceccherini, M.T.; Nannipieri, P.; Pietramellara, G. Distribution of Microbial Communities in a Forest Soil Profile Investigated by Microbial Biomass, Soil Respiration and DGGE of Total and Extracellular DNA. Soil Biol. Biochem. 2004, 36, 859–868. [Google Scholar] [CrossRef]
- Sánchez-Marañón, M.; Miralles, I.; Aguirre-Garrido, J.F.; Anguita-Maeso, M.; Millán, V.; Ortega, R.; García-Salcedo, J.A.; Martínez-Abarca, F.; Soriano, M. Changes in the Soil Bacterial Community along a Pedogenic Gradient. Sci. Rep. 2017, 7, 14593. [Google Scholar] [CrossRef]
- Turner, S.; Meyer-Stüve, S.; Schippers, A.; Guggenberger, G.; Schaarschmidt, F.; Wild, B.; Richter, A.; Dohrmann, R.; Mikutta, R. Microbial Utilization of Mineral-Associated Nitrogen in Soils. Soil Biol. Biochem. 2017, 104, 185–196. [Google Scholar] [CrossRef]
- Baldrian, P.; Kolaiřík, M.; Štursová, M.; Kopecký, J.; Valášková, V.; Větrovský, T.; Žifčáková, L.; Šnajdr, J.; Rídl, J.; Vlček, Č.; et al. Active and Total Microbial Communities in Forest Soil Are Largely Different and Highly Stratified during Decomposition. ISME J. 2012, 6, 248–258. [Google Scholar] [CrossRef] [PubMed]
- Dangi, S.R.; Stahl, P.D.; Wick, A.F.; Ingram, L.J.; Buyer, J.S. Soil Microbial Community Recovery in Reclaimed Soils on a Surface Coal Mine Site. Soil Sci. Soc. Am. J. 2012, 76, 915–924. [Google Scholar] [CrossRef]
- Li, Y.; Wen, H.; Chen, L.; Yin, T. Succession of Bacterial Community Structure and Diversity in Soil along a Chronosequence of Reclamation and Re-Vegetation on Coal Mine Spoils in China. PLoS ONE 2014, 9, e115024. [Google Scholar] [CrossRef] [PubMed]
- Sprocati, A.R.; Alisi, C.; Tasso, F.; Fiore, A.; Marconi, P.; Langella, F.; Haferburg, G.; Nicoara, A.; Neagoe, A.; Kothe, E. Bioprospecting at Former Mining Sites across Europe: Microbial and Functional Diversity in Soils. Environ. Sci. Pollut. Res. 2014, 21, 6824–6835. [Google Scholar] [CrossRef] [PubMed]
- Escobar, I.E.C.; Santos, V.M.; da Silva, D.K.A.; Fernandes, M.F.; Cavalcante, U.M.T.; Maia, L.C. Changes in Microbial Community Structure and Soil Biological Properties in Mined Dune Areas During Re-Vegetation. Environ. Manag. 2015, 55, 1433–1445. [Google Scholar] [CrossRef] [PubMed]
- Nemergut, D.R.; Anderson, S.P.; Cleveland, C.C.; Martin, A.P.; Miller, A.E.; Seimon, A.; Schmidt, S.K. Microbial Community Succession in an Unvegetated, Recently Deglaciated Soil. Microb. Ecol. 2007, 53, 110–122. [Google Scholar] [CrossRef]
- Schütte, U.M.E.; Abdo, Z.; Foster, J.; Ravel, J.; Bunge, J.; Solheim, B.; Forney, L.J. Bacterial Diversity in a Glacier Foreland of the High Arctic. Mol. Ecol. 2010, 19, 54–66. [Google Scholar] [CrossRef]
- Bradley, J.A.; Singarayer, J.S.; Anesio, A.M. Microbial Community Dynamics in the Forefield of Glaciers. Proc. Biol. Sci./R. Soc. 2014, 281, 20140882. [Google Scholar] [CrossRef]
- Goberna, M.; García, C.; Insam, H.; Hernández, M.T.; Verdú, M. Burning Fire-Prone Mediterranean Shrublands: Immediate Changes in Soil Microbial Community Structure and Ecosystem Functions. Microb. Ecol. 2012, 64, 242–255. [Google Scholar] [CrossRef]
- Sun, H.; Santalahti, M.; Pumpanen, J.; Köster, K.; Berninger, F.; Raffaello, T.; Asiegbu, F.O.; Heinonsalo, J. Bacterial Community Structure and Function Shift across a Northern Boreal Forest Fire Chronosequence. Sci. Rep. 2016, 6, 32411. [Google Scholar] [CrossRef]
- Gomez-Alvarez, V.; King, G.M.; Nüsslein, K. Comparative Bacterial Diversity in Recent Hawaiian Volcanic Deposits of Different Ages. FEMS Microbiol. Ecol. 2007, 60, 60–73. [Google Scholar] [CrossRef] [PubMed]
- Stevens, P.R.; Walker, T.W. The Chronosequence Concept and Soil Formation. Q. Rev. Biol. 1970, 45, 333–350. [Google Scholar] [CrossRef]
- Lewis, D.E.; White, J.R.; Wafula, D.; Athar, R.; Dickerson, T.; Williams, H.N.; Chauhan, A. Soil Functional Diversity Analysis of a Bauxite-Mined Restoration Chronosequence. Microb. Ecol. 2010, 59, 710–723. [Google Scholar] [CrossRef] [PubMed]
- Urbanová, M.; Kopecký, J.; Valášková, V.; Ságová-Marečková, M.; Elhottová, D.; Kyselková, M.; Moёnne-Loccoz, Y.; Baldrian, P. Development of Bacterial Community during Spontaneous Succession on Spoil Heaps after Brown Coal Mining. FEMS Microbiol. Ecol. 2011, 78, 59–69. [Google Scholar] [CrossRef] [PubMed]
- Selenska-Pobell, S.; Kampf, G.; Flemming, K.; Radeva, G.; Satchanska, G. Bacterial Diversity in Soil Samples from Two Uranium Waste Piles as Determined by Rep-APD, RISA and 16S RDNA Retrieval. Antonie Van Leeuwenhoek Int. J. Gen. Mol. Microbiol. 2001, 79, 149–161. [Google Scholar] [CrossRef] [PubMed]
- Kaplan, C.W.; Kitts, C.L. Bacterial Succession in a Petroleum Land Treatment Unit. Appl. Environ. Microbiol. 2004, 70, 1777–1786. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Lewis, D.E.; Chauhan, A.; White, J.R.; Overholt, W.; Green, S.J.; Jasrotia, P.; Wafula, D.; Jagoe, C. Microbial and Geochemical Assessment of Bauxitic Un-Mined and Post-Mined Chronosequence Soils from Mocho Mountains, Jamaica. Microb. Ecol. 2012, 64, 738–749. [Google Scholar] [CrossRef] [PubMed]
- Frouz, J.; Nováková, A. Development of Soil Microbial Properties in Topsoil Layer during Spontaneous Succession in Heaps after Brown Coal Mining in Relation to Humus Microstructure Development. Geoderma 2005, 129, 54–64. [Google Scholar] [CrossRef]
- Schulz, S.; Brankatschk, R.; Dümig, A.; Kögel-Knabner, I.; Schloter, M.; Zeyer, J. The Role of Microorganisms at Different Stages of Ecosystem Development for Soil Formation. Biogeosciences 2013, 10, 3983–3996. [Google Scholar] [CrossRef]
- Brankatschk, R.; Töwe, S.; Kleineidam, K.; Schloter, M.; Zeyer, J. Abundances and Potential Activities of Nitrogen Cycling Microbial Communities along a Chronosequence of a Glacier Forefield. ISME J. 2011, 5, 1025–1037. [Google Scholar] [CrossRef]
- Turner, B.L.; Wells, A.; Condron, L.M. Soil Organic Phosphorus Transformations along a Coastal Dune Chronosequence under New Zealand Temperate Rain Forest. Biogeochemistry 2014, 121, 595–611. [Google Scholar] [CrossRef]
- Lynch, J.M.; Benedetti, A.; Insam, H.; Nuti, M.P.; Smalla, K.; Torsvik, V.; Nannipieri, P. Microbial Diversity in Soil: Ecological Theories, the Contribution of Molecular Techniques and the Impact of Transgenic Plants and Transgenic Microorganisms. Biol. Fertil. Soils 2004, 40, 363–385. [Google Scholar] [CrossRef]
- Tscherko, D.; Rustemeier, J.; Richter, A.; Wanek, W.; Kandeler, E. Functional Diversity of the Soil Microflora in Primary Succession across Two Glacier Forelands in the Central Alps. Eur. J. Soil Sci. 2003, 54, 685–696. [Google Scholar] [CrossRef]
- Zumsteg, A.; Luster, J.; Göransson, H.; Smittenberg, R.H.; Brunner, I.; Bernasconi, S.M.; Zeyer, J.; Frey, B. Bacterial, Archaeal and Fungal Succession in the Forefield of a Receding Glacier. Microb. Ecol. 2012, 63, 552–564. [Google Scholar] [CrossRef] [PubMed]
- Uroz, S.; Tech, J.J.; Sawaya, N.A.; Frey-Klett, P.; Leveau, J.H.J. Structure and Function of Bacterial Communities in Ageing Soils: Insights from the Mendocino Ecological Staircase. Soil Biol. Biochem. 2014, 69, 265–274. [Google Scholar] [CrossRef]
- Tarlera, S.; Jangid, K.; Ivester, A.H.; Whitman, W.B.; Williams, M.A. Microbial Community Succession and Bacterial Diversity in Soils during 77 000 Years of Ecosystem Development. FEMS Microbiol. Ecol. 2008, 64, 129–140. [Google Scholar] [CrossRef]
- Fraterrigo, J.M.; Balser, T.C.; Turner, M.G. Microbial Community Variation and Its Relationship with Nitrogen Mineralization in Historically Altered Forests. Ecology 2014, 87, 570–579. [Google Scholar] [CrossRef]
- Turner, S.; Mikutta, R.; Meyer-Stüve, S.; Guggenberger, G.; Schaarschmidt, F.; Lazar, C.S.; Dohrmann, R.; Schippers, A. Microbial Community Dynamics in Soil Depth Profiles over 120,000 Years of Ecosystem Development. Front. Microbiol. 2017, 8, 874. [Google Scholar] [CrossRef]
- Mikutta, R.; Turner, S.; Schippers, A.; Gentsch, N.; Meyer-Stüve, S.; Condron, L.M.; Peltzer, D.A.; Richardson, S.J.; Eger, A.; Hempel, G.; et al. Microbial and Abiotic Controls on Mineral-Associated Organic Matter in Soil Profiles along an Ecosystem Gradient. Sci. Rep. 2019, 9, 10294. [Google Scholar] [CrossRef]
- Kleber, M.; Eusterhues, K.; Keiluweit, M.; Mikutta, C.; Mikutta, R.; Nico, P.S. Mineral-Organic Associations: Formation, Properties, and Relevance in Soil Environments; Elsevier Ltd.: Amsterdam, The Netherlands, 2015; Volume 130. [Google Scholar]
- Ortiz-Álvarez, R.; Fierer, N.; De Los Ríos, A.; Casamayor, E.O.; Barberán, A. Consistent Changes in the Taxonomic Structure and Functional Attributes of Bacterial Communities during Primary Succession. ISME J. 2018, 12, 1658–1667. [Google Scholar] [CrossRef]
- Pennanen, T. Microbial Communities in Boreal Coniferous Forest Humus Exposed to Heavy Metals and Changes in Soil PH—A Summary of the Use of Phospholipid Fatty Acids, Biolog® and 3H-Thymidine Incorporation Methods in Field Studies. Geoderma 2001, 100, 91–126. [Google Scholar] [CrossRef]
- Chodak, M.; Gołębiewski, M.; Morawska-Płoskonka, J.; Kuduk, K.; Niklińska, M. Soil Chemical Properties Affect the Reaction of Forest Soil Bacteria to Drought and Rewetting Stress. Ann. Microbiol. 2015, 65, 1627–1637. [Google Scholar] [CrossRef] [PubMed]
- Jangid, K.; Whitman, W.B.; Condron, L.M.; Turner, B.L.; Williams, M.A. Soil Bacterial Community Succession during Long-Term Ecosystem Development. Mol. Ecol. 2013, 22, 3415–3424. [Google Scholar] [CrossRef] [PubMed]
- Abakumov, E.; Trubetskoj, O.; Demin, D.; Celi, L.; Cerli, C.; Trubetskaya, O. Humic Acid Characteristics in Podzol Soil Chronosequence. Chem. Ecol. 2010, 26, 59–66. [Google Scholar] [CrossRef]
- Rastvorova, O.T.; Andreev, D.P. Preparation of soil samples for the analysis and methods of expressing the results of analysis. Theory Pract. Chem. Anal. Soils 2006, 1, 103–111. (In Russian) [Google Scholar]
- Vance, E.D.; Brookes, P.C.; Jenkinson, D.S. An extraction method for measuring soil microbial biomass C. Soil Biol. Biochem. 1987, 19, 703–707. [Google Scholar] [CrossRef]
- Lane, D.J. 16S/23S rRNA sequencing. In Nucleic acid Techniques in Bacterial Systematics; Stackebrandt, E., Goodfellow, M., Eds.; Wiley: New York, NY, USA, 1991; pp. 115–175. [Google Scholar]
- Yu, Y.; Lee, C.; Hwang, S. Analysis of community structures in anaerobic processes using a quantitative real-time PCR method. Water Sci. Technol. 2005, 52, 85–91. [Google Scholar] [CrossRef]
- Andronov, E.E.; Petrova, S.N.; Pinaev, A.G.; Pershina, E.V.; Rakhimgalieva, S.Z. Analysis of the structure of microbial community in soils with different degrees of salinization using T-RFLP and real-time PCR techniques. Eur. Soil Sci. 2012, 45, 147–156. [Google Scholar] [CrossRef]
- Bates, S.T.; Berg-Lyons, D.; Caporaso, J.G.; Walters, W.A.; Knight, R.; Fierer, N. Examining the Global Distribution of Dominant Archaeal Populations in Soil. ISME J. 2011, 5, 908–917. [Google Scholar] [CrossRef]
- Caporaso, J.G.; Kuczynski, J.; Stombaugh, J.; Bittinger, K.; Bushman, F.D.; Costello, E.K.; Fierer, N.; Peña, A.G.; Goodrich, J.K.; Gordon, J.I.; et al. QIIME allows analysis of high-throughput community sequencing data. Nat. Methods. 2010, 7, 335–336. [Google Scholar] [CrossRef]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A Flexible Trimmer for Illumina Sequence Data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef]
- Davis, N.M.; Proctor, D.M.; Holmes, S.P.; Relman, D.A.; Callahan, B.J. Simple Statistical Identification and Removal of Contaminant Sequences in Marker-Gene and Metagenomics Data. Microbiome 2018, 6, 226. [Google Scholar] [CrossRef] [PubMed]
- Lozupone, C.; Hamady, M.; Knight, R. UniFrac—An Online Tool for Comparing Microbial Community Diversity in a Phylogenetic Context. BMC Bioinform. 2006, 7, 371. [Google Scholar] [CrossRef]
- Pershina, E.V.; Ivanova, E.A.; Korvigo, I.O.; Chirak, E.L.; Sergaliev, N.H.; Abakumov, E.V.; Provorov, N.A.; Andronov, E.E. Investigation of the Core Microbiome in Main Soil Types from the East European Plain. Sci. Total Environ. 2018, 631–632, 1421–1430. [Google Scholar] [CrossRef]
- McMurdie, P.J.; Holmes, S. Phyloseq: An R Package for Reproducible Interactive Analysis and Graphics of Microbiome Census Data. PLoS ONE 2013, 8, e61217. [Google Scholar] [CrossRef] [PubMed]
- Clements, F.E. Nature and Structure of the Climax. J. Ecol. 1936, 24, 252. [Google Scholar] [CrossRef]
- Aristovskaya, T.V. The Microbiology of Podzolic Soils; Nauka: Moscow, Russia, 1965; 188p. [Google Scholar]
- Ponomareva, V.V. Theory of Podzolization; Israel Progr. Sci. Transr. Jerusalem; Nauka: Leningrad, Russia, 1969; 309p. [Google Scholar]
- Abakumov, E.; Polyakov, V.; Orlova, K. Podzol development on different aged coastal bars of Lake Ladoga. Tomsk State Univ. J. Biol. 2019, 48, 6–31. [Google Scholar] [CrossRef]
- Hillel, D.; Hatfield, J.H.; Powlson, D.S.; Rosenzweig, C.; Scow, K.M.; Singer, M.J.; Sparks, D.L. (Eds.) Encyclopedia of Soils in the Environment; Elsevier: Amsterdam, The Netherlands; Academic Press: Cambridge, MA, USA, 2005. [Google Scholar]
- Zhao, S.; Banerjee, S.; White, J.F.; Liu, J.J.; Zhou, N.; Tian, C.Y. High salt stress increases archaeal abundance and network connectivity in saline agricultural soils. Catena 2022, 217, 106520. [Google Scholar] [CrossRef]
- Cai, P.; Huang, Q.Y.; Zhang, X.W. Interactions of DNA with Clay Minerals and Soil Colloidal Particles and Protection against Degradation by DNase. Environ. Sci. Technol. 2006, 40, 2971–2976. [Google Scholar] [CrossRef]
- Ivanova, E.A.; Pershina, E.V.; Kutovaya, O.V.; Sergalieva, N.K.; Nagieva, A.G.; Zhiengaliev, A.T.; Provorov, N.A.; Andronov, E.E. Comparative analysis of microbial communities of contrasting soil types in different plant communities. Russ. J. Ecol. 2018, 49, 30–39. [Google Scholar] [CrossRef]
- Ivanova, E.A.; Kutovaya, O.V.; Tkhakakhova, A.K.; Chernov, T.I.; Pershina, E.V.; Markina, L.G.; Andronov, E.E.; Kogut, B.M. The Structure of Microbial Community in Aggregates of a Typical Chernozem Aggregates under Contrasting Variants of Its Agricultural Use. Eur. Soil Sci. 2015, 48, 1242–1256. [Google Scholar] [CrossRef]
- Zhou, J.; Xia, B.; Huang, H.; Treves, D.S.; Hauser, L.J.; Mural, R.J.; Palumbo, A.V.; Tiedje, J.M. Bacterial Phylogenetic Diversity and a Novel Candidate Division of Two Humid Region, Sandy Surface Soils. Soil Biol. Biochem. 2003, 35, 915–924. [Google Scholar] [CrossRef]
- Moreno-Espíndola, I.P.; Ferrara-Guerrero, M.J.; Luna-Guido, M.L.; Ramírez-Villanueva, D.A.; De León-Lorenzana, A.S.; Gómez-Acata, S.; González-Terreros, E.; Ramírez-Barajas, B.; Navarro-Noya, Y.E.; Sánchez-Rodríguez, L.M.; et al. The Bacterial Community Structure and Microbial Activity in a Traditional Organic Milpa Farming System under Different Soil Moisture Conditions. Front. Microbiol. 2018, 9, 2737. [Google Scholar] [CrossRef] [PubMed]
- Silveira Sartori Silva, M.R.; Pereira de Castro, A.; Krüger, R.H.; Bustamante, M. Soil Bacterial Communities in the Brazilian Cerrado: Response to Vegetation Type and Management. Acta Oecologica 2019, 100, 103463. [Google Scholar] [CrossRef]
- Lauber, C.L.; Hamady, M.; Knight, R.; Fierer, N. Pyrosequencing-Based Assessment of Soil PH as a Predictor of Soil Bacterial Community Structure at the Continental Scale. Appl. Environ. Microbiol. 2009, 75, 5111–5120. [Google Scholar] [CrossRef]
- Chan, O.C.; Yang, X.; Fu, Y.; Feng, Z.; Sha, L.; Casper, P.; Zou, X. 16S RRNA Gene Analyses of Bacterial Community Structures in the Soils of Evergreen Broad-Leaved Forests in South-West China. FEMS Microbiol. Ecol. 2006, 58, 247–259. [Google Scholar] [CrossRef] [PubMed]
- Yeoh, Y.K.; Dennis, P.G.; Paungfoo-Lonhienne, C.; Weber, L.; Brackin, R.; Ragan, M.A.; Schmidt, S.; Hugenholtz, P. Evolutionary Conservation of a Core Root Microbiome across Plant Phyla along a Tropical Soil Chronosequence. Nat. Commun. 2017, 8, 215. [Google Scholar] [CrossRef] [PubMed]
- Naz, I.; Mirza, M.; Bano, A. Molecular Characterization of Rhizosphere Bacterial Communities Associated With Wheat (Triticum aestivum L.) Cultivars at flowering stage. JAPS J. Anim. Plant 2014, 24, 1123–1134. [Google Scholar]
- Chen, J.; Xu, D.; Chao, L.; Liu, H.; Bao, Y. Microbial Assemblages Associated with the Rhizosphere and Endosphere of an Herbage, Leymus Chinensis. Microb. Biotechnol. 2020, 13, 1390–1402. [Google Scholar] [CrossRef]
- Pankratova, E.M. Functioning of cyanobacteria in soil ecosystems. Eur. Soil Sci. 2006, 39 (Suppl. 1), S118–S127. [Google Scholar] [CrossRef]
- Harantová, L.; Mudrák, O.; Kohout, P.; Elhottová, D.; Frouz, J.; Baldrian, P. Development of Microbial Community during Primary Succession in Areas Degraded by Mining Activities. Land Degrad. Dev. 2017, 28, 2574–2584. [Google Scholar] [CrossRef]
- Mehmood, M.A.; Fu, Y.; Zhao, H.; Cheng, J.; Xie, J.; Jiang, D. Enrichment of bacteria involved in the nitrogen cycle and plant growth promotion in soil by sclerotia of rice sheath blight fungus. Stress Biol. 2022, 2, 32. [Google Scholar] [CrossRef] [PubMed]
- Stursová, M.; Zifčáková, L.; Leigh, M.B.; Burgess, R.; Baldrian, P. Cellulose utilization in forest litter and soil: Identification of bacterial and fungal decomposers. FEMS Microbiol. Ecol. 2012, 80, 735–746. [Google Scholar] [CrossRef]
- Waksman, S.A. Decomposition of the various chemical constituents etc of complex plant materials by pure cultures of fungi and bacteria. Arch. Mikrobiol. 1931, 2, 136–154. [Google Scholar] [CrossRef]
- Yin, C.; Mueth, N.; Hulbert, S.; Schlatter, D.; Paulitz, T.C.; Schroeder, K.; Prescott, A.; Dhingra, A. Bacterial Communities on Wheat Grown under Long-Term Conventional Tillage and No-till in the Pacific Northwest of the United States. Phytobiomes J. 2017, 1, 83–90. [Google Scholar] [CrossRef]
- Bárta, J.; Tahovská, K.; Šantrůčková, H.; Oulehle, F. Microbial Communities with Distinct Denitrification Potential in Spruce and Beech Soils Differing in Nitrate Leaching. Sci. Rep. 2017, 7, 9738. [Google Scholar] [CrossRef] [PubMed]
- Bedogni, G.L.; Massello, F.L.; Giaveno, A.; Donati, E.R.; Urbieta, M.S. A Deeper Look into the Biodiversity of the Extremely Acidic Copahue Volcano-Río Agrio System in Neuquén, Argentina. Microorganisms 2020, 8, 58. [Google Scholar] [CrossRef]
- Opelt, K.; Berg, G. Biodiversity and Antagonistic Potential of Bacteria Associated with Bryophytes from Nutrient-Poor Habitats of the Baltic Sea Coast. Appl. Environ. Microbiol. 2004, 70, 6569–6579. [Google Scholar] [CrossRef]


| Succession Stage | Horizon | Chao1 | Good’s_Coverage | Observed_OTUs | Shannon Index | Faith’s Diversity Index |
|---|---|---|---|---|---|---|
| M1 | C1 | 2592.5 ± 4.4 a | 0.905 ± 0.002 a | 1639.6 ± 32.1 a | 8.7 ± 0.1 a | 77.6 ± 0.4 a |
| C2 | 2717.2 ± 114.4 a | 0.901 ± 0.005 a | 1705.2 ± 66.6 a | 9.1 ± 0.2 b | 81.6 ± 3.3 a | |
| C3 | 2555.9 ± 108.8 a | 0.909 ± 0.003 a | 1637.4 ± 80.7 a | 9.2 ± 0.3 b | 77.9 ± 3.9 a | |
| C4 | 2623.3 ± 57.9 a | 0.905 ± 0.002 a | 1681.6 ± 29.3 a | 9.2 ± 0.1 b | 79.9 ± 0.3 a | |
| M2 | AY | 2228.9 ± 143.9 b | 0.92 ± 0.005 b | 1330.6 ± 79.4 b | 7.9 ± 0.3 c | 61.8 ± 1.9 b |
| BF | 2144.5 ± 110.8 b | 0.924 ± 0.004 b | 1345.9 ± 59.5 b | 8.5 ± 0.2 a | 67.8 ± 3.0 b | |
| C | 2071.5 ± 89.8 b | 0.927 ± 0.003 b | 1328.4 ± 49.8 b | 8.5 ± 0.1 a | 68.8 ± 2.7 b | |
| M3 | Oi | 2814.8 ± 59.5 a | 0.899 ± 0.001 a | 1731.5 ± 41.8 a | 9.2 ± 0.2 b | 67.7 ± 2.7 b |
| (e)* | 2138.3 ± 87.9 b | 0.925 ± 0.004 b | 1338.2 ± 101.0 b | 8.5 ± 0.3 a | 52.9 ± 4.2 c | |
| BF | 2507.0 ± 194.9 a | 0.91 ± 0.008 a | 1547.7 ± 139.8 a | 8.9 ± 0.2 b | 72.1 ± 4.9 a | |
| BC | 2626.5 ± 181.2 a | 0.905 ± 0.006 a | 1599.3 ± 84.7 a | 9.0 ± 0.2 b | 75.4 ± 2.8 a | |
| C | 2292.4 ± 226.6 b | 0.919 ± 0.009 b | 1431.1 ± 74.3 ab | 8.7 ± 0.1 a | 67.7 ± 3.6 b | |
| M4 | Oi | 1086.4 ± 55.5 c | 0.966 ± 0.002 c | 792.1 ± 44.8 c | 7.7 ± 0.2 c | 35.9 ± 1.5 d |
| Oe | 739.2 ± 48.7 d | 0.978 ± 0.002 d | 542.2 ± 26.5 d | 6.9 ± 0.1 d | 27.6 ± 1.5 e | |
| E | 1078.5 ± 149.6 c | 0.966 ± 0.006 c | 735.4 ± 111.6 c | 7.6 ± 0.2 c | 35.9 ± 4.4 d | |
| BHF | 1199.7 ± 20.2 c | 0.959 ± 0.001 e | 792.8 ± 31.1 c | 7.3 ± 0.3 d | 40.8 ± 2.0 d | |
| BF | 1197.5 ± 161.8 c | 0.962 ± 0.007 e | 810.5 ± 134.2 c | 7.6 ± 0.4 c | 38.7 ± 8.9 d | |
| C | 1038.3 ± 59.2 c | 0.968 ± 0.002 ec | 691.8 ± 21.6 cd | 7.6 ± 0.1 c | 38.2 ± 1.3 d | |
| M5 | Oi | 908.1 ± 67.8 c | 0.972 ± 0.002 cd | 629.6 ± 44.8 d | 7.2 ± 0.1 d | 28.6 ± 0.9 e |
| E | 845.4 ± 18.8 c | 0.975 ± 0.001 cd | 610.3 ± 18.1 d | 7.3 ± 0.1 d | 29.8 ± 1.2 e | |
| BF | 1097.3 ± 14.3 c | 0.969 ± 0.001 d | 791.9 ± 16.5 c | 8.0 ± 0.1 c | 38.6 ± 0.9 d | |
| C | 984.8 ± 77.8 c | 0.974 ± 0.003 cd | 734.5 ± 69.7 c | 8.0 ± 0.2 c | 37.6 ± 3.1 d |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Evdokimova, E.; Ivanova, E.; Gladkov, G.; Zverev, A.; Kimeklis, A.; Serikova, E.; Pinaev, A.; Kichko, A.; Aksenova, T.; Andronov, E.; et al. Structural Shifts in the Soil Prokaryotic Communities Marking the Podzol-Forming Process on Sand Dumps. Soil Syst. 2024, 8, 9. https://doi.org/10.3390/soilsystems8010009
Evdokimova E, Ivanova E, Gladkov G, Zverev A, Kimeklis A, Serikova E, Pinaev A, Kichko A, Aksenova T, Andronov E, et al. Structural Shifts in the Soil Prokaryotic Communities Marking the Podzol-Forming Process on Sand Dumps. Soil Systems. 2024; 8(1):9. https://doi.org/10.3390/soilsystems8010009
Chicago/Turabian StyleEvdokimova, Elizaveta, Ekaterina Ivanova, Grigory Gladkov, Aleksei Zverev, Anastasiia Kimeklis, Elena Serikova, Alexandr Pinaev, Arina Kichko, Tatiana Aksenova, Evgeny Andronov, and et al. 2024. "Structural Shifts in the Soil Prokaryotic Communities Marking the Podzol-Forming Process on Sand Dumps" Soil Systems 8, no. 1: 9. https://doi.org/10.3390/soilsystems8010009
APA StyleEvdokimova, E., Ivanova, E., Gladkov, G., Zverev, A., Kimeklis, A., Serikova, E., Pinaev, A., Kichko, A., Aksenova, T., Andronov, E., & Abakumov, E. (2024). Structural Shifts in the Soil Prokaryotic Communities Marking the Podzol-Forming Process on Sand Dumps. Soil Systems, 8(1), 9. https://doi.org/10.3390/soilsystems8010009








