Abstract
Microbial communities throughout the 6.5 m depth profile of a boreal ombrotrophic bog were characterized using amplicon sequencing of archaeal, fungal, and bacterial marker genes. Microbial populations and their relationship to oxic and anoxic batch sorption of radionuclides (using radioactive tracers of I, Se, Cs, Ni, and Ag) and the prevailing metal concentrations in the natural bog was investigated. The majority of the detected archaea belonged to the Crenarchaeota, Halobacterota, and Thermoplasmatota, whereas the fungal communities consisted of Ascomycota, Basidiomycota, and unclassified fungi. The bacterial communities consisted mostly of Acidobacteriota, Proteobacteria, and Chloroflexi. The occurrence of several microbial genera were found to statistically significantly correlate with metal concentrations as well as with Se, Cs, I, and Ag batch sorption data. We suggest that the metal concentrations of peat, gyttja, and clay layers affect the composition of the microbial populations in these nutrient-low conditions and that particularly parts of the bacterial and archaeal communities tolerate high concentrations of potentially toxic metals and may concurrently contribute to the total retention of metals and radionuclides in this ombrotrophic environment. In addition, the varying metal concentrations together with chemical, mineralogical, and physical factors may contribute to the shape of the total archaeal and bacterial populations and most probably shifts the populations for more metal resistant genera.
1. Introduction
Heavy metals and radionuclides polluting the environment originate mainly from anthropogenic sources, including uranium mining, coal combustion, and accidental or purposeful release of radionuclides. Heavy metals and radioactive isotopes may present variable threats to biota, due to, e.g., atmospheric deposition and enrichment in soils and sediments, build-up into water bodies, and their high toxicity [1,2,3,4,5,6]. Radioactive isotopes and heavy metals can have toxic effects on soil microorganism activity if bioavailable and present in sufficient concentrations [7]. Heavy metals may also inhibit the growth and vitality of microorganisms and cause changes in the community structure [6,8].
In the Finnish concept for the disposal of spent nuclear fuel, the fuel will be placed in copper-sheeted canisters and disposed of in a deep bedrock repository constructed on Olkiluoto Island. During the last glacial period, this area was covered by the Scandinavian Ice Sheet and after the ice retreated, gradual post-glacial land uplift still continues in the area. The land uplift will result in a formation of new bogs in the area on which the first possible releases from the deep nuclear repository to the upper biosphere (through the clay, gyttja and peat layers in this order) would, based on biosphere safety assessment calculations, be possible if some of the copper canisters would leak. The Lastensuo Bog, sampled in our study, approximates the bog type likely to form in this area and has therefore been used as an analogue biotope in the biosphere safety assessments of the disposal of spent nuclear fuel in Finland [9]. 59Ni, 108mAg, 135Cs, 79Se, and 129I, with calculated potential geosphere release rates between 101 and 103 Bq/a starting 103 years after the start of disposal, are among the most important radionuclides, as the potential radiation doses for humans in the biosphere safety assessment calculations of spent nuclear fuel are considered [10].
Non-radioactive Ni may end up in the environment from antropogenic sources, see, e.g., in [11,12,13,14]. Ni occurs in several forms in soils, e.g., as inorganic crystalline minerals or precipitates, adsorbed or complexed on various organic or inorganic cation exchange surfaces, chelated to metal complexes in soil solution where a decrease in soil pH increases its mobility, or as a water-soluble free ion [15]. From the radioecological point of view, radioactive isotope of Ni, 59Ni, with long half-life of 76,000 years has classified as one of the high priority radionuclides in the biosphere safety assessments of the disposal of SNF in Finland due to the high calculated potential biosphere release rates of this nuclide (maximum of ~103 Bq a−1 at t = 105 years after the beginning of the disposal) [10]. Ni has high metabolic impact on living organisms because it is an important component in many enzymes [16,17]. In addition, many microorganisms can bind Ni by using various uptake mechanisms, including intracellular accumulation, straight biosorption on cell surfaces (through physical adsorption, complexation, or ion-exchange), and extracellular precipitation [18,19,20,21,22]. Retardation may occur through cell surface metal-binding functional groups, like carboxyl, hydroxyl, phosphate, and sulfate groups [18,19,20]. In addition, many microbes are able to accumulate Ni also inside the cells (see, e.g., in [22]) and until this date at least two different uptake mechanisms have been reported: ATP-binding cassette (ABC-type) transporters as well as Ni-specific permeases [23]. The possible toxic effects of Ni may be suppressed during active accumulation inside bacterial cells through detoxification reactions and previously we observed that Ni(II) concentrations between 0.01 and 1 mM had no toxic effects on Pseudomonas sp. strains isolated from the same Lastensuo Bog [22] studied in present study.
Silver (Ag) has been released to the biosphere especially through the photographic and imaging industry [24]. Ag is a non-essential metal with a high toxicity to many organisms already at very low concentrations [25] and is used as an effective bactericide [26,27]. Ag accumulates effectively in food chains and can cause various diseases and disorders, and even death [28,29,30]. It has been estimated that 5% of the total soil Ag is bioavailable [25]. In heavily contaminated soils, this proportion may be enough to harmfully affect the soil micro- and macrobiological populations. The radioisotope of Ag, 108mAg (T½ 418 years), is found in SNF and is assumed to be one of the instant release fraction (IRF) radionuclides if the copper canisters of the SNF are compromised and thus 108mAg can cause a substantial part of the radiation dose from potentially released SNF to the biosphere [31].
Radioisotopes of cesium, including 137Cs and 135Cs, are typically considered a greater health risk than stable cesium [32]. In soils, cesium is relatively rare and the stable cesium (133Cs) concentrations are roughly 5 mg/kg [33]. In addition to the above-mentioned isotopes of Ni and Ag, 135Cs, with its long half-life of 2.3 My and large inventory in SNF, is also among the most important radionuclides in the SNF repository long-term biosphere safety assessments [34]. Typical for alkali metals, cesium is highly soluble and therefore in SNF it is expected to occur in the instant release fraction [33]. Cesium occurs at oxidation state I (Cs+), and in soils it is typically sorbed by outer-sphere complexation, although in mica and clay minerals also inner-sphere complexes are formed [34,35,36]. In addition, cesium retains, e.g., on organic matter an on iron, manganese and aluminum oxides. Bacteria, including boreal Pseudomonas sp. Paenibacillus sp., Burkholderia sp., Rhodococcus sp. [37] and anaerobic iron- and sulfate-reducing bacterial mixtures [38] have been reported to accumulate low (Kd values below 100 L/kg) concentrations of cesium. Arbusculat mycorrhizal fungi have been shown to facilitate the uptake of the radioactive 137Cs and translocate it to a plant host [39] or concentrate in the fungal fruiting bodies [40]. Mycorrhizal fungi have also been shown to transport radio-cesium between plant individuals connected via the same hyphal network [41].
79Se is a high-priority radionuclide in the long-term safety assessments of the disposal of SNF into deep bedrock repositories [33]. Therefore, the possible radiation doses caused by radioactive selenium for humans are taken into consideration in the long-term safety calculations of SNF disposal [33]. The radioactive selenium isotope 79Se (T½ 65,000 years) is a fission product of uranium used in nuclear reactors and is also formed by neutron activation from stable selenium. Moreover, substantial amounts of stable selenium enter the biosphere through anthropogenic activities including coal combustion, mining, refining of sour crude oils, and agricultural irrigation of seleniferous soils [42,43,44,45,46]. Selenium is a vital micronutrient for humans and other mammals, but it is toxic at higher concentrations and the range between deficient and toxic dosed is characteristically narrow [47,48]. For humans, the effective daily dietary doses vary between 40 and 400 µg per day [49] and the mean abundance of selenium in the soil crust is 0.083 mg/kg [50]. In most soils, its concentrations are between 0.1 and 2 mg/kg [51]. However, exceptionally high Se concentrations up to 1200 mg/kg have been reported from different areas of the world (like organic-rich soils in Ireland) [52]. In soils, selenium occurs in different oxidation states, and in alkaline agricultural soils, selenium mainly occurs as selenate (Se(VI), SeO42−) as for acidic forest soils its most profound form is selenite (Se(IV), SeO32−) [52]. The retention and mobility of selenium is dependent, in addition to other factors like pH and organic matter content of the soil, on its speciation [53]. Especially, selenite is highly reactive and may react with glutathione thiol groups (GSH), explaining, at least in partly, its toxicisity [54]. Selenite spontaneously reacts with glutathione producing, e.g., glutathioselenol and selenodigluthatione [55]. It has been also reported that different species of Se-reducing bacteria produce elemental Se0 biominerals, which can have different atomic structures [56,57]. In soils, selenite and selenate are quickly reduced (also under oxic conditions), and this occurs mostly through microbial reduction [58,59,60]. Certain bacteria may use selenate and selenite as terminal electron acceptor [61] and may facilitate selenium uptake in plants [62,63]. Fungi have also been found to reduce selenite and selenate in oxic conditions [64]. In addition, under highly reducing conditions slow abiotic reduction is mediated by inorganic Fe(II,III) [65]. Previously, we showed that Pseudomonas sp. strains isolated from the same Lastensuo bog as was studied in the present study, were able to reduce selenite at 6 mM concentrations into reduced elemental selenium inside the bacterial cells even though the bacterial growth rate (as increased lag phase duration) was affected by the Se(IV) amendment [63].
129I, with its very long half-life of 15.7 My, large inventory in SNF, and high mobility, is one of the most significant radionuclides in the long-term safety assessments of SNF [33]. Iodine is also vastly biophilic, and via the food chain or by inhalation can bioaccumulate, particularly in the thyroid glands of both humans and other mammalians [48]. The migration and sorption behavior of iodine in the bio- and geosphere is affected by several factors, such as chemical speciation, organic matter content, mineral properties, redox potential, pH, and microorganisms [66,67,68,69,70,71]. Although iodine is primarily retained in soil organic matter (SOM) [72,73,74,75,76], microorganisms may affect its sorption, see, e.g., in [77,78,79].
Copper may also leach into the biosphere from the spent nuclear fuel canisters (the copper canisters in the KBS-3 model) through both abiotic and microbially induced corrosion mechanisms [80,81], locally increasing the environmental Cu load. Cu is a cofactor in many enzymes and is an essential micronutrient but is generally toxic to microorganisms, see, e.g., in [82,83]. However, many microorganisms have developed resistance mechanisms for coping with elevated Cu concentrations [83,84]. Elevated concentration is a relative term, but for, e.g., E. coli, the minimal inhibitory concentration (MIC) of Cu2+ has been determined to be 1 mM [85].
The resident microbial community has different mechanisms to interact with radionuclides and heavy metals, including biosorption, accumulation, precipitation, enzymatic transformations (redox reactions), and detoxification reactions [84,85,86]. Biosorption may occur both in living cells as well as in dead cells and cell fragments, but other mechanisms in which uptake occurs through active biological processes are present only in living cells [84,86,87]. For example, in prokaryotic cells, Ni and Se can be transported inside the cells using ABC transporters [84,87]. In the active uptake mechanisms, Ni is incorporated into Ni-dependent enzymes like urease, carbon monoxide dehydrogenase or certain superoxide dismutases and methylenediureases [84]. In the case of Se, the use of various enzymes like sulfite reductases and OYE enzymes is also possible [87], and often uptake is followed by detoxification reactions utilizing reduction (see, e.g., in [58,64]). For iodine, bacterial oxidation processes are well documented (see, e.g., in [88,89,90,91]) and H2O2-dependent iodide oxidation mechanism has been reported, e.g., in bacteria from the Savannah River Site [72]. In soil, organic matter iodine sorption has been suggested to occur through catalytic oxidation of I− into reactive iodine species, such as I2 and HIO, by environmental (microbial) peroxides, which after reactive iodine is bound into organic matter [80]. However, uptake (probably through I2 and HIO oxidation) of iodine has been described in fewer studies (see, e.g., in [88,92,93,94]). However, filamentous fungi have also been shown to bioaccumulate both iodide and iodate, but the mobilization of iodate is more efficient [95]. Typical for the metabolism-dependent mechanisms is that they are slower than biosorption on cell walls [22]. Bacterial and fungal surfaces contain multiple functional groups that may serve as sites for adsorption (and possibly subsequent desorption) (see, e.g., in [96,97]). Such functional groups, found both in Gram-negative and in Gram-positive bacteria, may be carboxylic, phosphate, and hydroxyl sites that deprotonate with increasing pH and therefore straight sorption on to the cellular structures is considered pH dependent [98]. On these sites the uptake follows the displacement of protons and therefore depends on the protonation [99,100]. However, typical bacterial cells have high buffering capacity, which can range, e.g., through the whole environmentally relevant pH range from pH 3 to pH 9 [100]. Vast prokaryotic buffering capacity results from distinct acidic sites that are located in the cell walls [100] and is of great importance in chancing environmental conditions. Microorganisms also have mechanisms for removing heavy metals, metalloids and transition metals from their cells, by using e.g., ATP-coupled pumps more or less specific for different ions, such as copper or silver ATPase transporters in bacteria, archaea and fungi [101,102,103].
In the present study, the objective was to study the effects of the total microbiome on the sorption behavior of the radioactive isotopes of Ni, Ag, Se, I, and Cs in an ombrotrophic boreal bog, and vice versa. In addition, the relationship between the microbiome and measured soil metal concentrations and correlations between soil metal concentrations, pH, water content and organic matter content and bacterial, archaeal, and fungal populations was studied.
2. Materials and Methods
2.1. Sampling Site and Sample Pretreatment
The sampling site, Lastensuo Bog, is an ombrotrophic, nutrient-poor bog with a maximum peat thickness of 5–6 m with a bottom soil consisting of gyttja and clay [9,104]. The bog is located on the western coast of Finland and our sampling point (Figure 1, WGS84 coordinates 61°17′31″ N, 21°50′22″ E) is located in the center parts of the bog, which consist of treeless or near treeless Sphagnum fuscum bog. The surrounding bog area also contain ridge hollow pine bog and hollow bog, with low sedge bog, cotton grass pine bog, tall sedge pine fen, and forested peatland at the edges [105]. The main peat types include Sphagnum peat (58%), sedge peat (19%), and few-flowered sedge (15%).
Figure 1.
(A) Map of Lastensuo bog, (B) sampling point in the center of the bog, and (C) sampling with a peat corer.
Discontinuous depth-wise samples of peat, gyttja and clay for oxic and anoxic studies were obtained in 2013 [80]. In addition, surface Sphagnum moss was collected. Values for pH, humification degree, sulfur content (g/kg DW), organic matter, and water content of the studied moss, peat, gyttja, and clay layers and bog water samples were determined previously [80,104,106] (Table 1). X-ray diffraction analysis for the mineral gyttja and clay layer samples was performed to examine the presence of clay minerals as described previously in [80] (Table 1).

Table 1.
pH of bog water and bog layer, humification degree (%), organic matter content (%), water content (%), and number of bacterial 16S rRNA genes g−1 DW of the bog samples from Lastensuo bog [80,106]. S concentrations (g/kg DW) were calculated as average concentrations obtained from the data of Aro et al. [104]. Data for mineral fractions and exchangeable cations (meq/100 g) from 1 M ammonium acetate extractions as well as the sum of extractable cations (meq/100 g) were obtained from in [38,80]. ND = no determination.
2.2. Nucleic Acid Isolation
Microbial community DNA was isolated from two parallel samples, A and B, of approximately 0.5 g subsamples of thawed surface moss, peat, gyttja, and clay samples with a NucleoSpin® Soil DNA extraction kit (MACHEREY-NAGEL GmbH & Co. KG, Dürren, Germany). Sample types with different characteristics, such as high content of organic material, high mineral content, varying pH, etc., may require dissimilar lysis conditions for optimal results. In order to increase the possibility to lyse as many different types of microbial cells as possible and detect the widest microbial diversity, the replicate samples were treated with different lysis buffers provided in the DNA extraction kit. Sample A was extracted with Lysis Buffer SL1 and sample B with Lysis Buffer 2. Enhancer solution SE was used with both samples and the extraction proceeded according to the manufacturer’s instructions. Negative DNA isolation controls were included in the isolation protocol. The DNA was eluted in 100 μL elution buffer and the DNA concentration of each extract was measured using the NanoDrop-1000 spectrophotometer (Thermo Fisher Scientific, Waltham, MA, USA).
2.3. Amplicon Library Preparation
The bacterial community was studied by high-throughput amplicon sequencing using the 454 Titanium FLX technology (Roche) and was previously reported [106]. The data were re-analyzed here and combined with the fungal and archaeal data. The archaeal and fungal communities were analyzed with high-throughput sequencing of the archaeal v4 region of the 16S rRNA gene and fungal internal transcribed spacer 1 (ITS1) region using the Ion Torrent PGM platform previously described [107]. The archaeal primers were the S-D-Arch-0349-a-S-17 (5′- GYGCASCAGKCGMGAAW-3′) and S-D-Arch-0787-a-A-20 (5′- GGACTACVSGGGTATCTAAT -3′) [108] and the fungal primers the ITS1 (5′- CTTGGTCATTTAGAGGAAGTA-3′) and ITS2 (5′- GCTGCGTTCTTCATCGATGC-3′) [109,110]. Primers 8F (5′- AGAGTTTGATCCTGGCTCAG-3′) and P2 (5′- CAG GCC TAA CAC ATG CAA GTC-3′) were used for amplifying the v1–v3 region of the bacterial 16S rRNA gene [111,112].
2.4. Sequence Processing and Analysis
The sequence reads were analyzed using the Mothur software (v.1.43.0) [113] as earlier described [107]. The archaeal and bacterial 16S rRNA sequences were aligned to the Silva version 138 reference alignment [114,115] using Mothur. The fungal ITS1 sequences were identified using the dynamic version of the UNITE ITS data base version 6 [116,117,118], as previously described [107]. Sequence reads were clustered into operational taxonomic units (I) sharing 97% identity. For comparing alphadiversity metrices the fungal ITS and for bacterial 16S rRNA gene sequence data sets were normalized to a subset of 2000 random sequences for sequences per sample and 1000 sequences per sample for the archaea. The alphadiversity analyses included the Shannon diversity indices and the estimated ChaoI OTU richness.
The bacterial sequences have previously been deposited to the European Nucleotide Archive (ENA, http://www.ebi.ac.uk/ena) under Study accession number PRJEB6875. The archaeal and fungal sequences have been submitted to the ENA under Study accession number PRJEB41440.
2.5. Sorption Batch Experiments Data and Metal Concentrations
Ni, Ag, Se, I, and Cs sorption data were obtained as previously described [119]. Briefly, batch sorption experiments were used to determine the distribution coefficients (Kd) of subsurface peat, gyttja, and clay. The experiments were set up aseptically by mixing 0.5 g of fresh sample and 25 mL of sterilized artificial bog water (see in [119]) and incubating for one week in order for the cation and anion concentrations to stabilize. The stabilization was confirmed by inductively coupled plasma mass spectrometry (ICP-MS) (Agilent 7500ce, Agilent Technologies, Inc., Santa Clara, CA, USA) measurement of the cations in the model bog water solution. After this, the tracers of 63Ni,110mAg, 134Cs, 125I, and 75Se were added with activities of 200 Bq per sample for 63Ni, 110mAg, 125I, and 75Se. For 134Cs 1000 Bq per sample was used. Incubation of the samples was continued at 20 °C for 7 days under constant mixing. Thereafter, the samples were submitted to a 20 min centrifugation at 20,000 rpm (Beckman Coulter J-26 XPI, rotor JA-25.50), filtered through a 0.2 µm syringe filter, the filtrate pH was measured whereafter the filtered solution was used for gamma spectrometric determination of 63Ni, 110mAg, 75Se, 125I and 134Cs activity as previously described [119]. The batch Kd values of the experiment were calculated as previously described [119]. Three parallel samples were used for all measurements. Batch sorption data for Se, I, and anoxic Se and I were obtained from our previous studies [80,119] (Table 2). For the anoxic determinations, only the innermost parts of the peat corer nest samples were used and transferred tightly sealed in plastic from the sampling site into a nitrogen cabinet [119]. The samples were further handled as described above, except under anoxic conditions. The redox potential (Eh (V)) was recorded for the redox sensitive species at the end of the incubation period.

Table 2.
Geometrical mean of sorption data (Kd values L/kg DW, three replicate samples) for Ni, Ag, and Cs (this paper), and Se, I, anoxic Se, and anoxic I (data from in [80,119]) after 7 days of incubation at 20 °C.
Microwave-assisted HNO3 extraction was used to determine the overall sodium (Na), magnesium (Mg), aluminium (Al), potassium (K), calsium (Ca), iron (Fe), cobolt (Co), Ni, copper (Cu), zinc (Zn), Se, molybdenum (Mo), silver (Ag), cesium (Cs), thorium (Th), and uranium (U) concentrations in the surface moss, peat, gyttja, and bottom clay samples. A 0.5 g subsample of fresh soil was weighed into a Teflon tube to which 10 mL HNO3 (concentrated) was added, and the sample was wet digested in a microwave oven using the EPA3051 method (Milestone Inc., Shelton, CT, USA). The extract solutions were filtered through a 0.2 mm Supor membrane filter (Pall Corp., Port Washington, NY, USA), the samples were diluted to 1:33, and the metal concentrations were determined using ICP-MS (Agilent 7500ce, Agilent Technologies, Inc., Santa Clara, CA, USA). No-sample negative control samples, containing only extraction solutions, were used in the extractions to detect possible reagent contamination. All ICP-MS determinations were performed using two parallel samples, and control samples with known concentrations were included in all ICP-MS runs. The microwave-assisted conc. HNO3 extraction was able to totally dissolve the organic peat layer fractions, but for the lowest mineral clay layer a small proportion of siliceous residue remained after digestion. This was removed in the following filtration step. Unlike sequential extractions, HNO3 extraction does not distinguish between exchangeable fraction, metals bound on oxide minerals (Fe(III)), organics, or primary sulfides.
2.6. Statistical Analyses
The relationships between the measured metal concentrations and Ni, Ag, Cs, Se, and I sorption data and microbial communities were examined with univariate analysis (Pearson’s r). The relationships between pH, water content, and humification degree versus bacterial, archaeal, and fungal communities; community richness (Chao 1); and diversity (Shannon index H′) of the bacterial, archaeal, and fungal communities were also examined using univariate analysis (Pearson’s r) with all samples and separately with only peat samples. In addition, the correlation between number of bacteria (as number of bacterial 16S rRNA gene copies g−1 dry weight sample) reported by Tsitko et al. [106] and sorption of radionuclides and metal concentrations was tested. All comparisons were Bonferroni corrected in order to minimize false positive correlations. The normal distribution of the data was evaluated by the Shapiro–Wilk test (p < 0.05) before the analysis. The normality hypothesis was rejected for all used data. All statistical analyses were performed using PAST v. 4 [120]. The statistical analyses were performed on the genus rank. Only groups with relative abundances of > 0.1% in at least one of the studied bog layers were included in the analyses. The differences in the microbial communities of the different bog layers were analyzed with Principal Coordinates Analysis (PCoA) using Phyloseq in R [121,122]. The PCoA were performed on the relative abundance of OTUs, including only samples with at least 1000 sequence reads, using the Bray–Curtis distance model. The statistical significance of environmental parameters on the distribution of samples on the PCoA chart was tested with 999 permutations and parameters with p < 0.05 were plotted as vectors on the plots.
3. Results
3.1. Sorption Experiments
The sorption (reported as Kd values liters (L) per dry weight kg (DW)) of Ni in all studied bog samples was found somewhat lower than the corresponding Kd values for Ag after one week stabilization period (Table 2, Figure 2A). Ni Kd values varied from 190 L/kg DW detected in the clay layer to 19,900 L/kg DW in the gyttja layer after one week equilibrium time. Ag Kd values ranged from 10,000 L/kg DW to 33,400 L/kg DW, with the highest values recorded for the peat layer from 2.5 m depth. Cs Kd values were found several orders of magnitude lower than those observed for Ag and Ni. Highest values (1600 L/kg DW) were observed for the clay layer (Figure 2B).
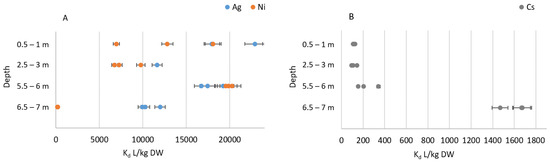
Figure 2.
Ag, Ni (A) and Cs (B) Kd values (L/kg DW) in fresh Lastensuo bog samples from 0.5–1 to 6.5–7 m. Layers from 0.5 m to 3.0 m consisted of peat, layer 5.5 m of gyttja, and layer 6.5 of clay. Error bars indicate 5% measurement uncertainty. The figure shows all three parallel samples.
In comparison, the sorption values for Se and I (reported in [80,119]) were 696–4388 L/kg and 24–1797 L/kg, respectively, in the aerobic incubations and 0–18 L/kg and 194–1629 L/kg, respectively, in the anaerobic incubations (Table 2). The recorded average Eh values for 0.5 m peat, 2.5 m peat, 5.5 m gyttja, and 6.5 m clay layers were 0.40 ± 0.05 V, 0.41 ± 0.06 V, 0.29 ± 0.08 V, and 0.12 ± 0.06 V, respectively, indicating more reducing conditions at the lower layers, compared to the upper layers.
3.2. Metal Concentrations
The metal concentrations varied between different bog layers and for all samples, and the highest concentration of metals was found in the lowest clay layer (Table 3). In addition, high concentrations of Na, Mg, Al, K, and Ca were found in the surface moss, 433 mg/kg DW, 634 mg/kg DW, 273 mg/kg DW, 4673 mg/kg DW, and 1619 mg/kd DW, respectively. Naturally occurring Ni, Ag, Cs, Se, and Th were generally detected at low concentrations, or not at all. With the exception of Ag, the highest concentrations were found in the clay layer (Table 3). Ni was also detected in the peat layers at 0.5 m and 1.5 m. Th was also detected in the gyttja layer.

Table 3.
Metal concentrations from the microwave assistant extraction of Lastensuo bog surface moss, peat, gyttja, and clay layers (mg/kg DW).
3.3. Archaeal, Fungal, and Bacterial Diversity
A total of 1617 archaeal 16S rRNA genes and 1670 fungal ITS1 Operational Taxonomic Units (OTUs) were identified from the amplicon sequence data, when clustered at 97% sequence homology. The bacterial 16S rRNA gene sequence data from [106] were reanalyzed and contained a total of 5029 bacterial OTUs. The highest number of 293 identified and 513 Chao1 estimated average number archaeal OTUs in the rarefied data was obtained from the surface moss sample (Table 4). The highest archaeal diversity index (H′ (average) = 4.8) was also seen in the surface moss. The lowest average number of identified (129) and estimated archaeal OTUs (294) was observed in the peat sample from 3.7 m. The lowest Shannon diversity index (H′ (average) = 1.6) was detected in the peat from 0.5–1.0 m depth. For all peat layer samples from 0.5 m to 3.7 m the number of observed and Chao1 estimated OTUs was below 150 and 180, respectively (Table 2). In the lowest clay layer (6.5 m), the number of archaeal OTUs was higher compared to the peat layers, with on average 198 observed and up to 314 Chao1 estimated archaeal OTUs.

Table 4.
The total number of archaeal 16S rRNA gene sequence reads (Sequences), and the number of observed (OTUs) and Chao1 estimated archaeal OTUs (Chao1), Good’s coverage (%) of detected OTUs estimated from the Chao1 (Coverage), and Shannon’s diversity indices (Shannon H′) calculated from the rarefied (1000 sequence reads) data. Samples A and B are the duplicate samples from the same layer and the DNA was extracted using Extraction Buffer SL1 and SL2 for sample A and B, respectively.
The highest average number of fungal ITS OTUs in the rarefied data was observed in the peat from 0.5 m (Table 5), with an average of 297 identified and 323 Chao1 estimated fungal OTUs. The highest fungal diversity index (H′(average) = 3.8) was found in the surface moss and the lowest (H′(average = 2.8) in the bog middle layers from 2.5 m to 3.5 m, where also the lowest average number of identified (160) and estimated fungal OTUs (256) were detected. The mean number of fungal OTUs and Chao1 estimated OTUs in the clay layer was 276 and 303, respectively, with a mean H′ of 3.2.

Table 5.
Obtained number of fungal ITS1 region sequence reads (Sequences), observed (OTUs) and estimated (Chao1) number of fungal OTUs, Good’s OTU coverage (%) estimated from the Chao1 (Coverage), and diversity index (Shannon H′) calculated from the sequence data. The data were rarefied to 2000 sequences for calculating the number of OTUs, Shannon index, Chao1 estimate, and Good’s coverage. The coverage describes the percentage of OTUs detected from the rarefied data set compared to the Chao1 estimated number of OTUs determined from the rarefied data. Samples A and B are the duplicate samples from the same layer and the DNA was extracted using Extraction Buffer SL1 and SL2 for sample A and B, respectively.
The highest number of rarefied bacterial OTUs and Chao1 estimated OTUs was observed in the peat layer from 1.5 m (Table 6). The surface moss has the highest Shannon’s H′ of 5.5. The lowest number of identified (191) and Chao1 estimated OTUs (385) were observed in the peat sample from 2.5 m depth.

Table 6.
Obtained number of bacterial 16S rRNA gene sequence reads (Sequences), observed (OTUs) and estimated (Chao1) number of bacterial OTUs, Good’s OTU coverage (%) estimated from the Chao1 (Coverage), and diversity index (Shannon) of the communities. The data were rarefied to 2000 sequences for calculating the number of OTUs, Shannon index, Chao1 estimate, and Good’s coverage. The coverage describes the percentage of OTUs detected from the rarefied data set compared to the Chao1 estimated number of OTUs determined from the rarefied data. Samples A and B are the duplicate samples from the same layer and the DNA was extracted using Extraction Buffer SL1 and SL2 for sample A and B, respectively.
3.4. Archaeal, Fungal, and Bacterial Community Compositions
The archaeal community in the Lastensuo bog depth profile, contained a total of 37 different genera belonging to 10 archaeal phyla. The major part of the community affiliated with the Crenarchaeota, Halobacterota, and Thermoplasmatota (Figure 3). Crenarchaeota dominated the archaeal community especially in the lower peat, gyttja, and clay layers from 2.5 m to 6.5 m (45–83% of sequence reads). Archaea remaining without classification (unclassified archeota) were clearly most common in the surface moss layer (on average 85% of sequence reads). The relative abundance of Halobacterota and Thermoplasmatota decreased with depth, the highest relative abundance observed in the 0.5 m peat layer (43% and 41% of sequence reads, respectively).
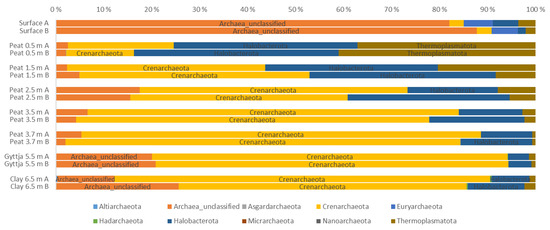
Figure 3.
Relative distribution of archaeal phyla in surface moss, peat, gyttja and clay samples collected from Lastensuo bog. Each depth is represented by duplicate samples A and B.
Candidatus Altiarchaeum, Nitrososphaeria (Marine Benthic Group A), Methanomicrobiaceae (uncultured), and unclassified Methanomicrobiales were only found in the lowest clay sample (6.5 m) (Figure A1), whereas Bathyarchaeia contributed with on average 68% of sequence reads. The clay layer contained the highest number (19) of archaeal genera (Figure A1), whereas the lowest number of eight archaeal genera was found in the surface layer (unclassified archaea, Bathyarchaeia, Nitrososphaeria Group 1.1c, unclassified Methanobacteriaceae, Methanobacterium, Methanomicrobiales Rice Cluster II, Methanomassiliicoccus, unclassified Thermoplasmata). In the surface moss layer, 85% of archaeal sequence reads consisted of unclassified archeota. Unclassified Nitrososphaeria was the most common archaeal lineage in the 5.5 m gyttja layer with 35% of sequences belonging to this genus.
Five fungal phyla (Figure 4), representing 145 different genera (Figure A2), were found throughout the Lastensuo Bog profile. Ascomycota generally dominated the fungal communities, with the exception of the surface moss, where unclassified fungi dominated with relative abundaces >50%. Basidiomycota were most commonly found in the surface moss sample, representing on average 13% of the fungal community, and were less common in the deeper layers. In the peat, gyttja, and clay layers, Ascomycota formed 77–78% of the fungal population.
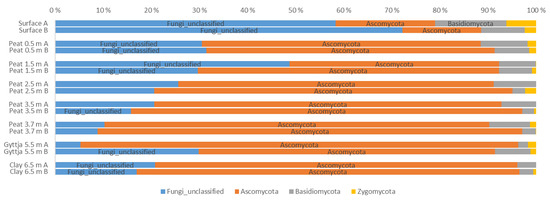
Figure 4.
Relative distribution of fungal phyla in surface moss, peat, gyttja, and clay samples collected from Lastensuo bog. Each depth is represented by duplicate samples A and B.
Of the detected genera (Figure A2), Phaeoacremonium formed 37% of the fungal community in the Lastensuo Bog, whereas 26% of the sequence reads on genus level belonged to unclassified fungi. The highest mean relative abundance of Phaeoacremonium (53%) was observed in the clay layer.
Acidobacteriota, Proteobacteria, and Chloroflexi were the most common bacterial taxa in the bog samples (Figure 5, Figure A3). Spirochaetota were found especially in the lowest gyttja (5.5 m) and clay (6.5 m) layers. Acidobacteriota covered 21–87% of the sequence reads of each sample, with the lowest numbers in the clay. Proteobacteria were common in the surface moss samples, contributing with 41% of the bacterial 16S rRNA gene sequence reads. Chloroflexi covered 35% of bacterial community in the 3.7 m peat layer. Spirochaetota contributed with 17% of the bacterial community in the clay layer (6.5 m). Other bacterial phyla detected in the bog samples included, e.g., Planctomycetota (11% in the 6.5 m clay layer) and Actinobacteria (6.3%, 2.7% an d2.2% in the 3.5 m and 3.7 m peat and surface moss layer, respectively). In the clay layer, 11% of the bacterial sequence reads remained without further classification.
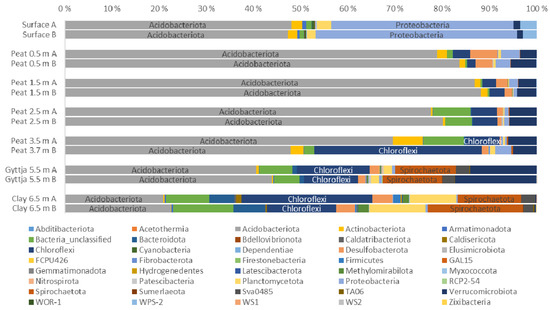
Figure 5.
Relative distribution of bacterial phyla in surface moss, peat, gyttja, and clay samples collected from Lastensuo Bog. Each depth is represented by duplicate samples A and B.
A total of 333 bacterial genera were identified from the whole bog profile. Acidobacteriota were represented by 33 genera. The most prominent genus belonged to Acidobacteriae Subgroup 13 (Figure A4), which was most common in the 1.5 m peat sample covering a mean 64% of the bacterial community. Granulicella (14%) and Candidatus Solibacter (12%) were observed especially in the 2.5 m peat sample. The clay layer bacterial community contined 6.3% Aminicenantales, whereas the surface layer was colonized by Occallatibacter (9.0%) and uncultured Acidobacteriales (8.4%).
Proteobacteria were represented mainly by Alphaproteobacteria (75% of the Proteobacteria) (Figure A5) and Gammaproteobacteria (25% of the proteobacteria, Figure A6). In the surface moss, alphaproteobacteial contributed with 19% of the bacterial community, of which Roseiarcus and a genus belonging to uncultured Acetobacteraceae contributed with 4.9% and 3.5% of the bacterial sequence reads, respectively. In all peat layers from 0.5 m to 3.5 m, uncultured Acetobacteraceae dominated the alphaproteobacterial community. In the gyttja and clay layers alphaproteobacterial represented less than 0.1% of the bacterial communities.
Gammaproteobacteria were most commonly found in the surface moss (~6.5% of all sequence reads). The gammaproteobacterial sequence reads were mostly assigned to the WD260 group (6.6% in total in all samples), unclassified Burkholderiales (in total 2.1%), and Acidibacter (in total 1.6%). The relative abundance of all other observed gammaproteobacterial genera was <1%.
Chloroflexi were most common in the peat layer from 3.7 m, where they contributed with 35% of the sequence reads. The most prominent genus, covering 99% of the Chloroflexi, was the Dehalococcoidia GIF9 (Figure A7). In all other peat layers, Chloroflexi contributed with <8% of the bacterial sequence reads and were not found in the surface layer. In contrast, the gyttja and clay layers contained several Chloroflexi genera, with Dehalococcoidia GIF9 contributing with 31% of the bacterial community of the gyttja and Dehalococcoidia SCGC-AB-539-J10 and unclassified Dehalococcoidia, together contributing with 43% the bacterial community in the clay layer.
Spirochaetota were represented by four groups, i.e., the Brevinema, Spirochaeta, unclassified Spirochaetaceae, and uncultured Spirochaetaceae, the most prominent group being Spirochaeta which covered 99.9% of all Spirochaetota reads. Spirochaetota were mainly found in the lowest gyttja and clay layers, with low abundances, covering on average 3.9 % of all bacterial sequence reads.
3.5. Correlation between Microbial Communities, Alphadiversity, and Physicochemical Parameters
No correlations between archaeal genera or archaeal out numbers, Chao1 richness, or Shannon’s H′ and sorption data or between fungal OTU numbers, Chao1 richness, or H′ index and metal concentrations or bog characteristics data (pH, etc.) were found. However, several archaeal genera showed positive correlation (based on Bonferroni-corrected p-values) with pH, Eh, and different metal concentrations (Table A1). Lokiarchaeia, Hadarchaeales, uncultured Methanomicrobiaceae, Methanosaeta, and uncultured Methanomassiliicoccaceae showed strong positive correlation to pH (p < 0.05–0.00001), whereas Lokiarchaeia correlated negatively (p < 0.05) to Eh). An unclassified crenarchaeotal genus correlated positively to the humification degree (p < 0.05). Interestingly, Hadarchaeales, uncultured Methanomicrobiaceae, Methanosaeta, and uncultured Methanomassiliicoccaceae correlated negatively with the amount of organic matter in the samples (p < 0.05–0.00001). Candidatus Altiarchaeum, Lokiarchaeia, Hadarchaeales, uncultured Methanomicrobiaceae, Methanosaeta, and uncultured Methanomassiliicoccaceae correlated positively with concentrations of Fe, Co, Se, Cs, Th, and U. Lokiarchaeia, Hadarchaeales, uncultured Methanomicrobiaceae, Methanosaeta, and uncultured Methanomassiliicoccaceae correlated positively with Mg. In addition, Lokiarchaeia and Methanosaeta correlated positively with Cu and Hadarchaeales and uncultured Methanomicrobiaceae with Zn. The presence of Lokiarchaeia and Methanosaeta correlated negatively with Ag sorption (Kd values) in the Lastensuo bog samples, whereas Hadarchaeales correlated positively with Cs sorption (Table 7).

Table 7.
Pearson correlation coefficients of the Bonferroni corrected correlations between relative abundance of bacterial genera or bacterial numbers and sorption data. Correlation coefficients for pairs with p < 0.05 are shown.
No statistically significant correlations were observed between fungal genera and pH, water content, humification degree, depth, organic matter content, nor metal concentrations. Neither were correlations between fungal genera or fungal OTU numbers, Chao1 richness, or Shannon’s H′ and sorption data found.
The metal concentrations found in the bog samples did not correlate significantly with the number of bacterial 16S rRNA genes, OTUs, Shannon’s diversity index (H′), or Chao1 richness. However, several strong positive correlations between bacterial genera (Table A2) and metal concentrations were observed. The strongest positive correlations were observed between Pelolinea and Phyciphaerae DG-20 and Fe, Se, and Cs (r > 0.999, p < 0.05). Strong positive correlations (r ≥ 0.997) were in addition observed between several other bacterial genera (e.g., Acetothermiia, Latereibacteraceae, Polyangia, and Phycisphaerae) and Fe, Co, Se, and Cs (Table A2).
Strong positive correlations (r ≥ 0.995, p < 0.01) between bacterial genera and Cs sorption (Kd values) were observed (Table 7). The bacteria with strong positive correlations with Cs sorption data included Aceothermiia, Acidobacteriae GOUTB8, Thermoanaerobaculum, Pelolinea, Latescibacteraceae, Polyangia MidBa8, and Phycisphaerae. In addition, anoxic sorption data for Se correlated positively (r = 0.993, p = 0.04) with uncultured Anaerolineaceae. No correlations between any of the other sorption data (I, Se, Ni, and Ag) and bacterial genera were found.
Several bacterial genera correlated with organic matter content (%) (Table A2). All of the statistically significant correlations were negative and included, e.g., Aceothermiia, Acidobacteriae GOUTB8, Thermoanaerobaculum, Pelolinea, Latescibacteraceae, Polyangia MidBa8, and Phycisphaerae (Table A2).
The similarity between the archaeal, fungal, and bacterial communities of the different bog layers was tested using Principal Coordinates analysis (PCoA) and the Bray–Curtis dissimilarity model (Figure 6). For all microbial groups, the surface samples separated from the rest of the samples. The archaeal community in the surface samples (the highest relative abundance of unclassified archaea) appeared to be affected by the concentration of Ag, whereas the fungal and bacterial communities of the same sample were not (Figure 6A). The archaeal communities of the peat samples also clearly clustered according to depth, with samples from similar depths falling closer together. The gyttja and clay communities were clearly different from the surface and peat communities, and although not clustering together, both sample types fell in the upper left quadrant of the plot where the metal concentrations had the highest influence. In the fungal PCoA analysis, the surfaces samples separated to the opposite side of the plot from the rest of the samples (Figure 6B). All peat, gyttja, and clay samples fell to the right side of the plot and did not clearly separate from each other. No environmental parameters appeared to affect the fungal communities with any statistical significance. The bacterial communities separated into three distinct groups: the surface communities, the peat and gyttja communities, and the clay communities. The clay community was most strongly affected by the measured metals.
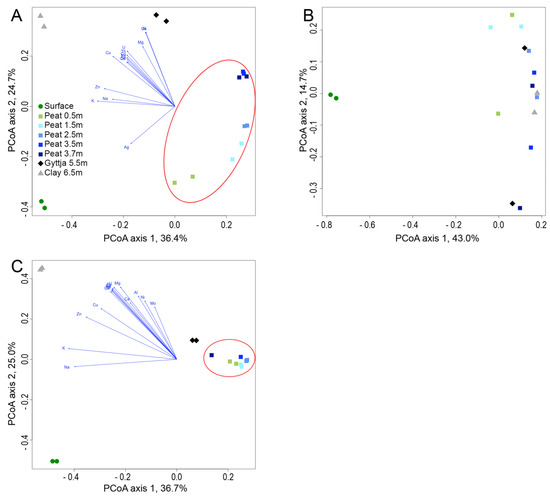
Figure 6.
Principal Coordinates Analysis (PCoA) of the (A) archaeal, (B) fungal, and (C) bacterial communities in relation to environmental parameters. Vectors for metal concentrations having statistically significant (p < 0.05) effect on the microbial communities are shown. For fungi, no significant metal concentrations were identified. In A and C, the peat samples are indicated with a red oval.
No statistically significant correlation between bacterial numbers or microbial taxa and the concentration of Ni or Ag, or sorption of Ni and Ag in the tested samples, was seen. In the regression analysis, the sorption of Ni did not appear to be affected by bacterial numbers or by sample type. However, the higher sorption of Ag was seen in the samples with the highest number of bacteria, and lowest in the clay sample, with the lowest number of bacteria and generally different conditions compared to peat (Figure 7A,B).
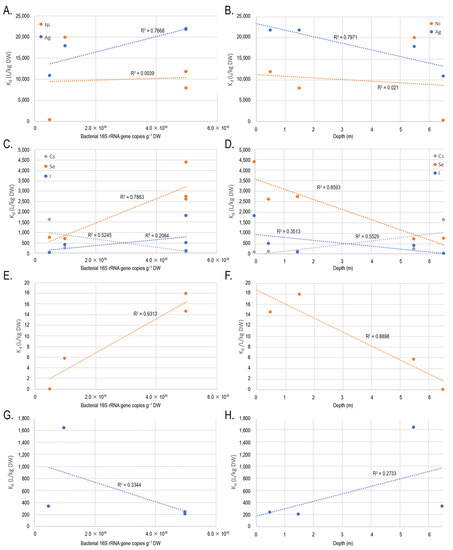
Figure 7.
The sorption (Kd) of (A,B) Ni and Ag; (C,D) Cs, Se, and Ag; (E,F) anoxic Se; and (G,H) anoxic I plotted against (A,C,E,G) bacterial numbers (16S rRNA gene copies g−1 DW) and (B,D,F,H) sampling depth, i.e., sample type.
The concentration and sorption of Cs correlated strongly with several microbial groups (Table 7, Table A1 and Table A2), but not with the number of bacterial 16S rRNA gene copies. The regression analysis is in agreement with these results, showing highest sorption of Cs in the clay layer (Figure 7D). In contrast, the sorption of Se correlated strongly and significantly with the number of bacterial 16S rRNA genes (Table 7) under oxic (p < 0.00001) and anoxic (p < 0.05) conditions, which is also shown by the regression analysis, where the highest Kd values for Se coincides with the highest bacterial numbers (Figure 7). The sorption of I did not show statistically significant correlation with any microbial groups or bacterial numbers under aerobic conditions, but the regression analysis indicated slightly higher Kd values in samples with high bacterial counts, and lower Kd values in the clay layer. The sorption of I in anoxic conditions was low and now correlation with the bacterial numbers was seen (Table 7). However, uncultured Anaerolineaceae correlated positively with anaerobic iodine sorption (Table 7).
4. Discussion
Microbiological processes may affect the solubility and bioavailability of heavy metals and radionuclides, and consequently affect the overall behavior of these elements in the biosphere. Simultaneously, heavy metals and radionuclides and low pH, typical for ombrotrophic boreal bog environments, can select for microbial communities of more resilient types able to survive in these acidic and nutrient-poor environments [38,58,80]. In the present study, several factors, including variable metal concentrations, pH, organic matter content, humification degree, and bacterial numbers, were shown to influence the microbial community composition and/or vice versa in the acidic bog environment with high diversity of bacteria, archaea, and fungi (Table 1 and Table 4, Table 5, Table 6). In addition, specific parts of the microbial communities were observed to correlate with radioactive Se, Cs, and Ag sorption behavior, as well as retention of I under anoxic conditions, which has also been reported earlier [40,46,65].
The alkali and heavy metal concentrations varied depending on the bog layer. However, the concentrations of, e.g., Cu, Pb, and Ni were not different from concentrations measured from four other Finnish oligotrophic bogs to a depth of 80 cm, but the concentration of Zn was elevated [2]. For all alkali and heavy metals, the highest concentrations were found in the bottom clay layer (Table 3), which was formed during the post-glacial land uplift in this area and once was part of the upper seabed [9]. High concentrations of alkali metals were also found especially in the surface moss sample and at the depth of 3.5 m in the peat. This ombrotrophic region is assumed to obtain all its nutrients through rainfall [9], which may explain the high alkali metal concentrations in the upper layers, from where they gradually diffuse through the peat layers eventually ending up to the lowest gyttja and clay layers. Interestingly, the highest H′ values of the archaeal, fungal, and bacterial communities were observed in the surface moss layer with high alkali metal concentrations, which all are important nutrients. Heavy metals from the acid digestion including Fe, Co, and Se were concentrated in the bottom clay layer and several statistically significant positive correlations between archaeal and bacterial genera and these metals were found. Especially high positive correlations were also found between radionuclides U and Th, which are part of the natural decay series of uranium and were clearly concentrated to the bottom clay layer.
Ni was found at 0.5–1.5 m depth in the peat samples and in the clay layer at concentrations of 15–34 mg kg−1 DW (Table Metals), and below the detection limit of the assay in the other samples. Ni is a key component in a functioning methyl-coenzyme M reductase (MCR) of methanogens [123], but no methanogenic archaea correlated with the Ni concentrations in Lastensuo. Many plant growth-promoting bacteria have the capacity to reduce the toxicity of Ni and other heavy metals by accumulating the metals in the cell mass, thus allowing for better plant growth at mM concentrations of heavy metals [124]. The amount of Ni in Lastensuo did not appear to have any effect on bacterial numbers, indicating that the concentrations found were not high enough to be toxic to the microorganisms (Figure 7). Commercially available peat has successfully been used as sorbent for the removal of Ni from aqueous solutions [125], indicating that the peat itself may sorb the Ni and perhaps that is why there is a specific layer where the Ni appears to be enriched in the peat profile. Nevertheless, bacterial strains originating from Lastensuo have been shown to accumulate Ni intracellularly as well as by sorption to cell surfaces [23].
Silver appeared to be much more influenced by the number of bacteria in the samples, compared to Ni (Figure 7). Although the number of bacteria did not correlate positively or negatively with the concentration of Ag, nor did any microbial taxa, the highest Ag sorption values coincided with the highest bacterial numbers. Only Lokiarchaeia and Methanosaeta correlated negatively with Ag+ sorption data, and they were present only in the lowest gyttja and clay layers, where Ag+ sorption was on average only 65% of the sorption observed in the upper peat layers. Ag was present in all bog layers evenly distributed in low concentration (0.01–0.02 mg/kg DW), which was possibly too low to have toxic effects on the microbial communities [126]. The Ag+ ion is a known bactericide but reports about its efficiency against archaea is scarce. Silver nanoparticles have however shown potent antimicrobial activity against, e.g., Haloarchaea [80] at the concentrations exceeding the extracted Ag concentrations observed in our study (300–400 µg/mL vs. ~0.01–0.02 µg/mL). Nevertheless, bacterial strains isolated from the Lastensuo Bog have been shown to remove Ag from solution by yet unknown mechanisms [127]. The same study estimated that less than 0.2% of the Ag sorption in the peat, gyttja, and clay layers is due to bacteria, whereas in the surface moss, this proportion was 2.3%
In addition to positive correlations between Cs concentration and archaeal and bacterial taxa, specific parts of the archaeal and bacterial communities also correlated positively and significantly with Cs (Cs+) batch sorption data. However, the microorganisms correlating with Cs sorption were almost exclusively detected in the gyttja and clay layers (Figure A3 and Figure A4), where also the highest Cs+ retention (Kd value) and Cs concentrations were recorded. However, the bacterial numbers in the different bog layers did not correlate with the concentration or retention of Cs. The accumulation of Cs on the lowest clay layer is expected, as clays have high cation exchange capacity and the pH of the layer is higher. This is because the surface charge is dependent on the pH and cation sorption typically decreases as the pH decreases. Previously, we observed the deepest clay layer behaving somewhat differently from the upper layers as the effect of pH change on Cs sorption was less prominent than in the upper organic layers [38]. However, in the upper organic layers, the sorption Kd values reached those of the lowest clay layer, but not until alkaline pH was recorded [38]. In the clay layer, Cs retention maximum was found at pH 8.9 as in the upper layers the corresponding maximum was between pH 6.7 and 9.6 depending on the layer [38]. Peat typically has a broad spectrum of pKa values, due to various protonating groups like carboxylic groups, alcoholic, and phenolic OH groups as well as amino groups (see, e.g., in [128,129,130,131]). For clay minerals, like illite present in the lowest layer, pKa values from 4.2 to 9.0 have been reported (see, e.g., in [132]). On mineral surfaces sorption takes place via two separate mechanisms namely outer-sphere and inner-sphere complexation on which sorption occurs on hydroxyl groups or through sorption on interlayers and frayed edge sites (FES sites) of clay minerals [133]. Sorption takes place between charged surfaces and partially or fully hydrated ions. Clay minerals are characterized by permanent negative charge [35], which is stabilized by exchangeable cations adsorbed on the basal planes or interlayer spaces of these minerals. In the case of partly or fully dehydrated Cs+ ions, inner-sphere complexes are formed directly to the siloxane groups of clay minerals (especially illite) within the interlayer or at the FES site [35,133]. Microorganisms are not expected to affect the formation of these inner-sphere complexes, and typically the higher clay content increases the straight sorption of various metals on clay mineral ion-exchange sites. Therefore, it seems most probable, that the change in the pH and mineral content controls the sorption of Cs in this layer and at the same time the change in the microbial community is seen because of the changes in the layer chemical, physical and mineralogical properties, resulting in positive correlation between bacterial and archaeal taxa and Cs concentrations.
Previously, microbiota have been shown to affect the retention of SeO32− in this acidic nutrient-poor bog environment under oxic conditions and the retention mechanisms most likely include the formation of reduced Se0 [58,64]. In present study, strong correlation between bacterial numbers and selenium retention both under oxic and anoxic conditions was seen. The mechanism here is not known, but under aerobic or microaerophilic conditions, SeO32− can be reduced by several bacterial strains using detoxification or redox homoeostasis, possible reactions including, e.g., Painter-type reactions, thioredoxin reductase system, and sulfide-mediated reduction [62] facilitating Se retention. In the present study, the speciation of selenium was not determined, but, e.g., Pseudomonas spp. And Burkholderia spp. Strains isolated from Lastensuo peat grown with SeO32− under oxic conditions effectively remove SeO32− from the nutrient solutions, presumably by a detoxification reaction and Pseudomonas sp. Has been shown to accumulate reduced Se0 inside the cell [58,64,134]. However, these bacteria were present at only very low relative abundances of <3%, and no correlation with SeO32− retention under aerobic conditions and these bacterial groups was observed. In addition, under anoxic conditions the number of bacteria correlated with the retention of Se (Table 7, Figure 7). Under anoxic (reducing) conditions, slow abiotic reduction (>1month) in the presence of Fe(II) is possible [66], and in addition several bacteria may use SeO32− or SeO42− as terminal electron acceptors also under anoxic conditions and reduce soluble SeO32−/SeO42− to Se0 (see, e.g., in [134,135]). This occurs predominantly via microbial dissimilatory reduction with organic substrates, such as acetate, lactate and ethanol, or hydrogen as electron donors [61]. Dissimilatory Se reduction to Se0 one of the most important sinks for Se oxyanions under anoxic environments [136] and SeO42− reductases have been identified in the genomes of, e.g., Anaerolineaceae [137], which in present study were present mainly in the lowest gyttja layer, with 2% abundance. However, correlation between these bacteria and selenium retention was not seen in present study. Microbial reduction of oxyanionic selenium species (SeO32− and SeO42−) into insoluble elemental Se0 is important especially in the organic wetland environments affecting selenium mobility [138]. However, in addition to the microbial-mediated processes, in suboxic and anoxic Fe(II,III) oxide- and sulfide (S2−)-containing environments, gradual abiotic reduction of SeO32− to Se0 is also important [66,139].
No statistically significant correlation between microbial groups or bacterial numbers and sorption of I in aerobic conditions was identified. However, the regression analysis showed slightly higher I sorption with higher bacterial numbers and low sorption in the clay layer (Figure 7C,D), indicating that bacterial biomass may somewhat affect the I retention in aerobic conditions. In soil organic matter, iodine sorption has been suggested to occur through catalytic oxidation of I− into reactive iodine species, such as I2 and HIO, by environmental (microbial) peroxides, which after reactive iodine is bound into organic matter (forming org-I), see, e.g., in [77]. Peatlands are considered the most important sinks of terrestrial iodine and peatland micro- and macroalgae, fungi, and bacteria facilitate the iodine redox cycle by I− oxidation and formation of org-I, but also by dehalogenation of org-I, and reduction of oxidizes iodine species to I− [140], e.g., Anaerolineae (a genus of Anaerolinaceae) has been previously shown to reductively deiodinize benzene derivates [140]. However, in present study, we observed a strong positive correlation (Table 7) between Anaerolinaceae and anaerobic iodine retention. These bacteria were present mainly in the gyttja layer, where interactions between organic matter and increasing proportion of clay may occur, resulting in complex iodine geochemistry as well as changes in the microbial populations. The lack of correlation between bacterial numbers and iodine sorption data may result from the more important role of fungi in the iodine cycle, which has also been suggested in previous studies, see, e.g., in [140].
Cu is known to be toxic for bacteria and fungi and the toxicity increases with decreasing pH [141]. In our study, only Lokiarchaeia and the MSBL5 of the Chloroflexi showed any effects to Cu and both correlated positively with the highest concentration of Cu in the clay layer. However, the concentration of Cu in Lastensuo was low or similar compared to boreal peatlands and podzols and agricultural soils [2,142]. However, Cu concentrations may increase in the vicinity of SNF repositories if Cu is leached from exposed waste canisters. Zn is another putatively toxic metal for microorganisms. Zn may reduce the microbial diversity in agricultural soils at levels of 400 mg kg−1 [143]. One study showed that culturable soil bacteria may be susceptible to zinc at concentrations of 2.5 mM (approximately 180 mg L−1) in their growth medium [144]. However, this is still a significantly higher concentrations than what we found in the bog samples, which are in line with Zn concentrations found in other boreal peatlands [2]. In Lastensuo, only Hadarchaeales and uncultured Methanomicrobiaceae correlated with the concentration of Zn, but these archaea were mostly found in the clay layer, where the Zn concentration also was the highest, indicating that the clay layer itself may be a stronger driver for these archaea than the concentration of Zn.
Our present study supports our previous assumptions [38,64] that microbial populations can contribute partly to Se and I retention in this ombrotrophic boreal environment. The role of microbial populations in the retention behavior of Cs, Ag, and Ni appears to be less pronounced although certain archaea may affect to Ag environmental behavior.
Finally, fungi did not correlate strongly with any physicochemical parameter or with sorption data, although fungi were detected throughout the bog profile. Our work is one of the few that has investigated the relative abundances of fungi present throughout the depth profile of an oligotrophic boreal bog. Nevertheless, the importance of fungi as decomposers increases in nutrient-poor conditions [145]. Phaeoacremonium was the most prominent fungal genus in all other samples but the surface moss with an increasing relative abundance in the deeper peat layers (Figure A2). This fungus has been shown to produce high amounts of manganese peroxidases in anoxic lake sediment, thus promoting anoxic degradation of lignin [146]. Although anaerobic degradation of recalcitrant carbon compounds in deep peat environments has been shown to be low [147], anaerobic degradation of recalcitrant organic matter to more easily utilized sugars by fungi could support a diverse prokaryotic community in oligotrophic and anaerobic bogs [148].
5. Conclusions
The Lastensuo Bog area has high bacterial, archaeal, and fungal diversity, and especially several bacterial and archaeal groups seemed adapted to the variable concentrations of heavy metals, alkali metals, as well as radionuclides from natural decay chains of uranium (U and Th). The present data indicate that the archaeal and bacterial communities in the deep bog layers (namely gyttja and clay) tolerate high concentrations of metals and may concurrently contribute to the total retention of metals and radionuclides in these layers. At the same time, it is probable that a change in chemical, mineralogical, and physical factors (pH, redox potential, and mineral content) present in this ombrotrophic boreal environment is affecting both the microbial population and the retention of metals. However, more research on the toxicity of the metals on various parts of the microbial community present in this area is still needed.
Author Contributions
Conceptualization, M.L. and M.B.; methodology, M.L. and M.B.; software, M.L. and M.B.; formal analysis, M.L. and M.B.; investigation, M.L.; writing—original draft preparation, M.L.; writing—review and editing, M.L. and M.B. All authors have read and agreed to the published version of the manuscript.
Funding
This research received no external funding.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
The data sequence data sets presented in this study are available from European Nucleotide Archive (ENA, http://www.ebi.ac.uk/ena) under Study accession numbers PRJEB6875 (bacteria) and PRJEB41440 (archaea, fungi). All other used data is presented in the tables and references of the paper.
Conflicts of Interest
The authors declare no conflict of interest.
Appendix A
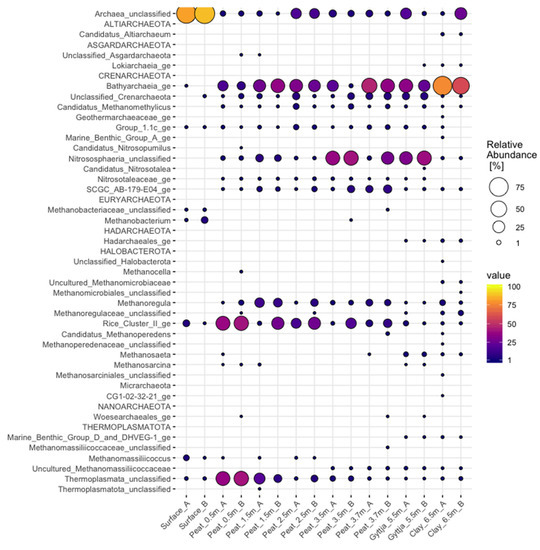
Figure A1.
Relative abundance of archaeal genera in surface moss, peat, gyttja, and clay samples collected from Lastensuo bog.
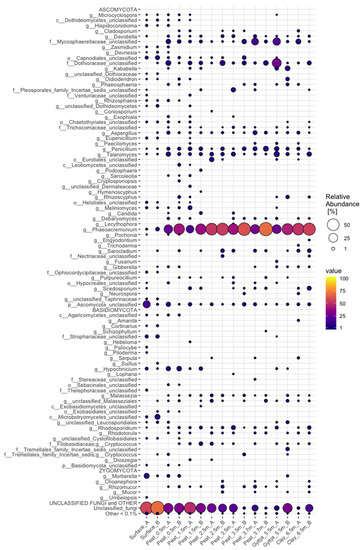
Figure A2.
Relative abundance of fungal genera in surface moss, peat, gyttja, and clay samples collected from Lastensuo bog.
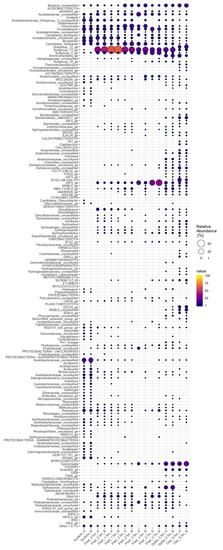
Figure A3.
The relative abundance of the bacterial genera (% of sequence reads) in the Lastensuo bog samples.
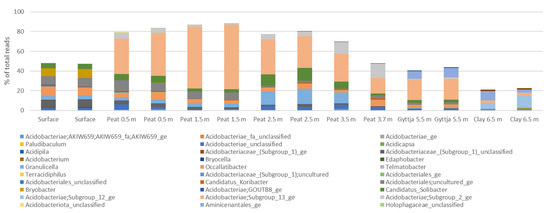
Figure A4.
The relative abundance of Acidobacteriotal genera in Lastensuo Bog samples.
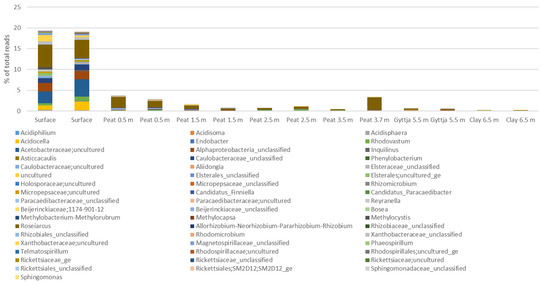
Figure A5.
The relative abundance of Alphaproteobacterial genera in Lastensuo Bog samples.
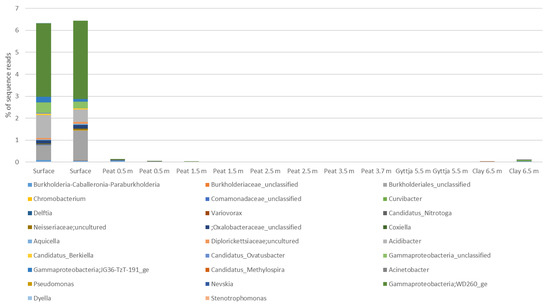
Figure A6.
The relative abundance of Gammaproteobacterial genera in Lastensuo Bog samples.
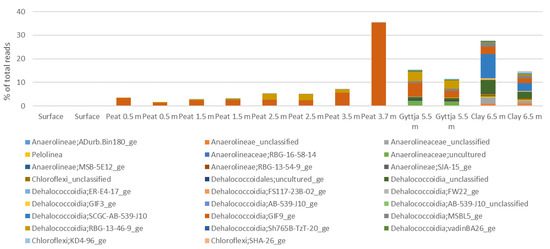
Figure A7.
The relative abundance of Chloroflexi genera in Lastensuo Bog samples.

Table A1.
Pearson correlation coefficients of the Bonferroni corrected correlations between relative abundance of archaeal genera and physicochemical parameters and metal concentrations. Correlation coefficients for pairs with p < 0.05 are shown.
Table A1.
Pearson correlation coefficients of the Bonferroni corrected correlations between relative abundance of archaeal genera and physicochemical parameters and metal concentrations. Correlation coefficients for pairs with p < 0.05 are shown.
| Candidatus Altiarchaeum | Lokiarchaeia Genus | Crenarchaeota Unclassified | HADARCHAEALES GENUS | Methanomicrobiaceae Uncultured | Methanosaeta | Methanomassiliicoccaceae Uncultured | |
|---|---|---|---|---|---|---|---|
| pH | 0.98 *** | 0.92 ** | 0.85 | 0.98 *** | 0.86 | ||
| Eh | −0.98 | ||||||
| Humification degree | 0.88 | ||||||
| Organic matter | −0.98 *** | −0.96 *** | −0.92 ** | −0.90 | |||
| Mg | 0.88 | 0.91 * | 0.89* | 0.85 | 0.93 ** | ||
| Fe | 0.88 | 0.94 ** | 0.98 *** | 0.96 *** | 0.92 * | 0.90 * | |
| Co | 0.88 | 0.96 ** | 0.99 *** | 0.96 *** | 0.93 ** | 0.90 * | |
| Cu | 0.90 * | 0.91 * | |||||
| Zn | 0.88 | 0.88 | |||||
| Se | 0.88 | 0.93 ** | 0.98 *** | 0.96 *** | 0.90 * | 0.89 * | |
| Cs | 0.88 | 0.93 ** | 0.98 *** | 0.96 *** | 0.90 * | 0.89 * | |
| Th | 0.87 | 0.97 *** | 0.99 *** | 0.95 *** | 0.95 *** | 0.90 * | |
| U | 0.86 | 0.98 *** | 0.98 *** | 0.93 ** | 0.97 *** | 0.90 * |
* p < 0.001, ** p < 0.0001, *** p < 0.00001.

Table A2.
Pearson correlation coefficients of the Bonferroni-corrected correlations between relative abundance of bacterial genera and physicochemical parameters and metal concentrations. Correlation coefficients for pairs with p < 0.05 are shown.
Table A2.
Pearson correlation coefficients of the Bonferroni-corrected correlations between relative abundance of bacterial genera and physicochemical parameters and metal concentrations. Correlation coefficients for pairs with p < 0.05 are shown.
| pH | Organic Matter | Mg | Al | Fe | Co | Cu | Se | Cs | Th | U | |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Acetothermiia | 0.95 | −1.0 *** | 0.92 | 1.0 *** | 0.99 *** | 1.0 *** | 1.0 *** | 0.99 *** | 0.97 *** | ||
| GOUTB8 | −1.0 *** | 0.92 | 1.0 *** | 1.0 *** | 0.99 *** | 0.99 *** | 1.0 *** | 0.98 *** | |||
| Acidobacteriae Subgroup 12 | 0.94 | 0.93 | 0.94 | 0.94 | 0.93 | ||||||
| Thermoanaerobaculum | −1.0 *** | 0.92 | 1.0 *** | 0.99 *** | 1.0 *** | 1.0 *** | 0.99 *** | 0.97 *** | |||
| Vicinamibacterales uncultured | 0.92 | 0.92 | 0.93 | 0.92 | 0.92 | ||||||
| Bacteroidales unclassified | 0.94 | 0.93 | 0.94 | 0.94 | 0.93 | ||||||
| Bacteroidetes vadinHA17 | −0.98 *** | 0.98 *** | 0.98 *** | 0.98 *** | 0.98 *** | 0.98 *** | 0.97 *** | ||||
| Prolixibacteraceae BSV13 | −0.98 *** | 0.98 *** | 0.98 *** | 0.98 *** | 0.98 *** | 0.97 *** | 0.95 *** | ||||
| BSV26 | 0.92 | ||||||||||
| Anaerolineae unclassified | −0.98 *** | 0.98 *** | 0.98 *** | 0.98 *** | 0.98 *** | 0.97 *** | 0.95 * | ||||
| Pelolinea | −1.0 *** | 0.93 | 1.0 *** | 1.0 *** | 1.0 *** | 1.0 *** | 0.99 *** | 0.97 *** | |||
| SJA-15 | −0.96 | 0.96 * | 0.95 * | 0.98 * | 0.96 * | 0.95 | 0.93 | ||||
| Chloroflexi unclassified | 0.91 | 0.92 | 0.91 | ||||||||
| Dehalococcoidia unclassified | 0.92 | 0.93 | 0.94 | 0.94 | |||||||
| GIF3 | 0.98 *** | 0.94 | 0.94 | 0.93 | 0.93 | 0.95 | 0.95 * | ||||
| MSBL5 | 0.94 | 0.92 | 0.96 * | ||||||||
| vadinBA26 | 0.94 | ||||||||||
| Desulfatiglans | 0.93 | 0.92 | 0.93 | 0.93 | 0.92 | ||||||
| Smithella | 0.93 | 0.92 | 0.93 | 0.93 | 0.92 | ||||||
| Syntrophus | −0.99 *** | 0.92 | 1.0 *** | 0.99 *** | 0.99 *** | 0.99 *** | 0.98 *** | 0.96 * | |||
| Firmicutes unclassified | −0.96 | 0.96 * | 0.95 * | 0.96 * | 0.96 * | 0.96 | 0.93 | ||||
| TSAC18 | −0.96 | 0.96 * | 0.95 * | 0.96 * | 0.96 * | 0.95 | 0.93 | ||||
| Hydrogenedensaceae | −0.99 *** | 0.91 | 0.99 *** | 0.98 *** | 0.99 *** | 0.99 *** | 0.98 *** | 0.96 * | |||
| Candidatus Latescibacter | −0.98 ** | 0.98 *** | 0.98 *** | 0.98 *** | 0.98 *** | 0.97 *** | 0.95 | ||||
| Latescibacteraceae unclassified | 0.94 | −1.0 *** | 0.92 | 1.0 *** | 0.99 *** | 1.0 *** | 1.0 *** | 0.99 *** | 0.97 *** | ||
| Methylomirabilaceae Sh765B-TzT-35 | −0.97 * | 0.92 | 0.97 *** | 0.97 *** | 0.96 *** | 0.96 * | 0.97 *** | 0.97 *** | |||
| MidBa8 | −1.0 *** | 0.92 | 1.0 *** | 0.99 *** | 1.0 *** | 1.0 *** | 0.99 *** | 0.97 *** | |||
| DG-20 | −1.0 *** | 0.92 | 1.0 *** | 0.99 *** | 1.0 *** | 1.0 *** | 0.99 *** | 0.97 *** | |||
| MSBL9 unclassified | 0.93 | 0.92 | 0.93 | 0.93 | 0.92 | ||||||
| AKAU3564 sediment group | −0.96 | 0.95 * | 0.95 * | 0.95 * | 0.95 * | 0.95 | 0.93 | ||||
| Planctomycetota unclassified | −0.94 | ||||||||||
| Spirochaeta | 0.93 | ||||||||||
| Sva0485 | 0.93 | ||||||||||
| WCHB1-41 | −0.96 | 0.96 * | 0.95 * | 0.96 * | 0.96 * | 0.95 | 0.93 |
* p < 0.001, ** p < 0.0001, *** p < 0.00001.
References
- Hovmand, M.F.; Kemp, K.; Kystol, J.; Johnsen, I.; Riis-Nielsen, T.; Pacyna, J.M. Atmospheric heavy metal deposition accumulated in rural forest soils of southern Scandinavia. Environ. Pollut. 2008, 155, 537–541. [Google Scholar] [CrossRef] [PubMed]
- Ukonmaanaho, L.; Nieminen, T.M.; Rausch, N.; Shotyk, W. Heavy metal and arsenic profiles in ombrogenous peat cores from four differently loaded areas in Finland. Water Air Soil Pollut. 2004, 158, 277–294. [Google Scholar] [CrossRef]
- Laird, K.R.; Das, B.; Cumming, B.F. Enrichment of uranium, arsenic, molybdenum, and selenium in sediment cores from boreal lakes adjacent to northern Saskatchewan uranium mines. Lake Reserv. Manag. 2014, 30, 344–357. [Google Scholar] [CrossRef]
- Roberts, S.; Kirk, J.L.; Wiklund, J.A.; Muir, D.C.; Keating, J.; Yang, F.; Gleason, A.; Lawson, G.; Wang, X.; Evans, M. Sources of atmospheric metal (loid) pollution recorded in Thompson Manitoba lake sediment cores within the Canadian boreal biome. Sci. Total Environ. 2020, 732, 139043. [Google Scholar] [CrossRef] [PubMed]
- Tchounwou, P.B.; Yedjou, C.G.; Patlolla, A.K.; Sutton, D.J. Heavy metal toxicity and the environment. Mol. Clin. Environ. Toxicol. 2012, 101, 133–164. [Google Scholar]
- Sigee, D.C.; Al-Rabaee, R.H. Nickel toxicity inPseudomonas tabaci: Single cell and bulk sample analysis of bacteria cultured at high cation levels. Protoplasma 1986, 130, 171–185. [Google Scholar] [CrossRef]
- Rajapaksha, R.M.C.P.; Tobor-Kapłon, M.A.; Bååth, E. Metal toxicity affects fungal and bacterial activities in soil differently. Appl. Environ. Microbiol. 2004, 70, 2966–2973. [Google Scholar] [CrossRef]
- Pennanen, T.; Frostegard, A.S.A.; Fritze, H.; Baath, E. Phospholipid fatty acid composition and heavy metal tolerance of soil microbial communities along two heavy metal-polluted gradients in coniferous forests. Appl. Environ. Microbiol. 1996, 62, 420–428. [Google Scholar] [CrossRef]
- Haapanen, R.; Aro, L.; Lahdenperae, A.M.; Helin, J.; Ikonen, A.T.K. Studies on Reference Mires: 1. Lastensuo and Pesaensuo in 2010–2011; No. POSIVA-WR--12-102; Posiva Oy: Eurajoki, Finland, 2013. [Google Scholar]
- Hjerpe, T.; Broed, R.; Ikonen, A.T.K. Biosphere Assessment Report 2009; No. POSIVA--10-03; Posiva Oy: Eurajoki, Finland, 2010. [Google Scholar]
- Dönmez, G.Ç.; Aksu, Z.; Öztürk, A.; Kutsal, T. A comparative study on heavy metal biosorption characteristics of some algae. Process Biochem. 1999, 34, 885–892. [Google Scholar] [CrossRef]
- Wong, J.P.K.; Wong, Y.S.; Tam, N.F.Y. Nickel biosorption by two chlorella species, C. Vulgaris (a commercial species) and C. Miniata (a local isolate). Bioresour. Technol. 2000, 73, 133–137. [Google Scholar] [CrossRef]
- Aksu Ksu, Z. Determination of the equilibrium, kinetic and thermodynamic parameters of the batch biosorption of nickel (II) ions onto Chlorella vulgaris. Process Biochem. 2002, 38, 89–99. [Google Scholar] [CrossRef]
- Grandjean, P. Human Exposure to Nickel; IARC Scientific Publications: Lyon, France, 1984; pp. 469–485. [Google Scholar]
- Scott-Fordsmand, J.J. Toxicity of nickel to soil organisms in Denmark. Rev. Environ. Contam. Toxicol. 1997, 148, 1–34. [Google Scholar]
- Gupta, V.K.; Rastogi, A.; Nayak, A. Biosorption of nickel onto treated alga (Oedogonium hatei): Application of isotherm and kinetic models. J. Colloid Interface Sci. 2010, 342, 533–539. [Google Scholar] [CrossRef] [PubMed]
- Akhtar, N.; Iqbal, J.; Iqbal, M. Removal and recovery of nickel (II) from aqueous solution by loofa sponge-immobilized biomass of Chlorella sorokiniana: Characterization studies. J. Hazard. Mater. 2004, 108, 85–94. [Google Scholar] [CrossRef] [PubMed]
- White, C.E.; Wilkinson, S.C.; Gadd, G.M. The role of microorganisms in biosorption of toxic metals and radionuclides. Intern. Biodeterior. Biodegrad. 1995, 35, 17–40. [Google Scholar] [CrossRef]
- Ledin, M.; Pedersen, K. The environmental impact of mine wastes—Roles of microorganisms and their significance in treatment of mine wastes. Earth-Sci. Rev. 1996, 41, 67–108. [Google Scholar] [CrossRef]
- López, A.; Lázaro, N.; Priego, J.M.; Marqués, A.M. Effect of pH on the biosorption of nickel and other heavy metals by Pseudomonas fluorescens 4F39. J. Ind. Microbiol. Biotechnol. 2000, 24, 146–151. [Google Scholar] [CrossRef]
- Sar, P.; Kazy, S.K.; Singh, S.P. Intracellular nickel accumulation by Pseudomonas aeruginosa and its chemical nature. Lett. App. Microbiol. 2001, 32, 257–261. [Google Scholar] [CrossRef]
- Mulrooney, S.B.; Hausinger, R.P. Nickel uptake and utilization by microorganisms. FEMS Microbiol. Rev. 2003, 27, 239–261. [Google Scholar] [CrossRef]
- Knuutinen, J.; Bomberg, M.; Kemell, M.; Lusa, M. Ni(II) interactions in boreal Peanibacillus sp., Methylobacterium sp., Paraburkholderia sp. and Pseudomonas sp. strains isolated from an acidic, ombrotrophic bog. Front. Microbiol. 2019. [Google Scholar] [CrossRef]
- Eitinger, T.; Mandrand-Berthelot, M.-A. Nickel transport systems in microorganisms. Arch. Microbiol. 2000, 173, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Pedroso, M.S.; Pinho, G.L.; Rodrigues, S.C.; Bianchini, A. Mechanism of acute silver toxicity in the euryhaline copepod Acartia tonsa. Aquat. Toxicol. 2007, 82, 173–180. [Google Scholar] [CrossRef] [PubMed]
- Jacobson, A.R.; McBride, M.B.; Baveye, P.; Steenhuis, T.S. Environmental factors determining the trace-level sorption of silver and thallium to soils. Sci. Total Environ. 2005, 345, 191–205. [Google Scholar] [CrossRef] [PubMed]
- Smith, I.; Carson, B. Silver; MI7 Ann Arbor Science Publishers: Ann Arbor, MI, USA, 1977. [Google Scholar]
- Hadrup, N.; Sharma, A.K.; Loeschner, K. Toxicity of silver ions, metallic silver, and silver nanoparticle materials after in vivo dermal and mucosal surface exposure: A review. Regul. Toxicol. Pharmacol. 2018, 98, 257–267. [Google Scholar] [CrossRef]
- Rosenman, K.D.; Seixas, N.; Jacobs, I. Potential nephrotoxic effects of exposure to silver. Occup. Environ. Med. 1987, 44, 267–272. [Google Scholar] [CrossRef]
- Das, D.; Das, N.; Mathew, L. Kinetics, equilibrium and thermodynamic studies on biosorption of Ag (I) from aqueous solution by macrofungus Pleurotus platypus. J. Hazard. Mater. 2010, 184, 765–774. [Google Scholar] [CrossRef]
- SKB. Long-Term Safety for the Final Repository for Spent Nuclear Fuel at Forsmark Main Report of the SR-Site Project Volume III; Errata 2011-10, SKB TR-11-01; Svensk Kärnbränslehantering AB: Stockholm, Sweden, 2011. [Google Scholar]
- Williams, M.; Wohlers, D.W.; Citra, M.; Diamond, G.L.; Swarts, S.G. Toxicological profile of Cesium; U.S. Department of Health and Human Services, Public Health Service Agency for Toxic Substances and Disease Registry: Atlanta, GA, USA, 2004.
- Sparks, D.L. Environmental Soil Chemistry; Elsevier: Amsterdam, The Netherlands, 2003. [Google Scholar]
- Helin, J.; Ikonen, A.T.K.; Hjerpe, T. Review of Element-Specific Data for Biosphere Assessment BSA-2009; Working Report 2010-37; Posiva Oy: Eurajoki, Finland, 2010. [Google Scholar]
- Bostick, B.C.; Vairavamurthy, M.A.; Karthikeyan, K.G.; Chorover, J. Cesium adsorption on clay minerals: An EXAFS spectroscopic investigation. Environ. Sci. Technol. 2002, 36, 2670–2676. [Google Scholar] [CrossRef]
- Chang, K.P.; Hsu, C.N.; Tamaki, H. Basic study of 137Cs sorption on soil. J. Nucl. Sci. Technol. 1993, 30, 1243–1247. [Google Scholar] [CrossRef]
- Zhuang, J.; Flury, M.; Jin, Y. Colloid-facilitated Cs transport through water-saturated Hanford sediment and Ottawa sand. Environ. Sci. Technol. 2003, 37, 4905–4911. [Google Scholar] [CrossRef]
- Lusa, M.; Bomberg, M.; Virtanen, S.; Lempinen, J.; Aromaa, H.; Knuutinen, J.; Lehto, J. Factors affecting the sorption of cesium in a nutrient-poor boreal bog. J. Environ. Radioact. 2015, 147, 22–32. [Google Scholar] [CrossRef]
- Sasaki, T.; Kubota, T.; Mito, S.; Kauri, T.; Kudo, A. Radionuclide sorption to a mixture of anaerobic bacteria in the repository environment. J. Nucl. Sci. Technol. 2002, 39 (Suppl. 3), 954–957. [Google Scholar] [CrossRef]
- Higo, M.; Kang, D.J.; Isobe, K. First report of community dynamics of arbuscular mycorrhizal fungi in radiocesium degradation lands after the Fukushima-Daiichi Nuclear disaster in Japan. Sci. Rep. 2019, 9, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Ohnuki, T.; Sakamoto, F.; Kozai, N.; Nanba, K.; Neda, H.; Sasaki, Y.; Niizato, T.; Watanabe, N.; Kozaki, T. Role of filamentous fungi in migration of radioactive cesium in the Fukushima forest soil environment. Environ. Sci. Process. Impacts 2019, 21, 1164–1173. [Google Scholar] [CrossRef] [PubMed]
- Gyuricza, V.; Thiry, Y.; Wannijn, J.; Declerck, S.; Dupré de Boulois, H. Radiocesium transfer between Medicago truncatula plants via a common mycorrhizal network. Environ. Microbiol. 2010, 12, 2180–2189. [Google Scholar] [PubMed]
- Coppin, F.; Chabroullet, C.; Martin-Garin, A. Selenite interactions with some particulate organic and mineral fractions isolated from a natural grassland soil. Eur. J. Soil Sci. 2009, 60, 369–376. [Google Scholar] [CrossRef]
- Sharmasarkar, S.; Vance, G.F. Soil and plant selenium at a reclaimed uranium mine. J. Environ. Qual. 2002, 31, 1516–1521. [Google Scholar] [CrossRef]
- Manceau, A.; Gallup, D.L. Removal of Selenocyanate in Water by Precipitation: Characterization of Copper− Selenium Precipitate by X-ray Diffraction, Infrared, and X-ray Absorption Spectroscopy. Environ. Sci. Technol. 1997, 31, 968–976. [Google Scholar] [CrossRef]
- Yasin, M.; El Mehdawi, A.F.; Jahn, C.E.; Anwar, A.; Turner, M.F.; Faisal, M.; Pilon-Smits, E.A. Seleniferous soils as a source for production of selenium-enriched foods and potential of bacteria to enhance plant selenium uptake. Plant Soil 2015, 386, 385–394. [Google Scholar] [CrossRef]
- De Souza, M.P.; Chu, D.; Zhao, M.; Zayed, A.M.; Ruzin, S.E.; Schichnes, D.; Terry, N. Rhizosphere bacteria enhance selenium accumulation and volatilization by Indian mustard. Plant Physiol. 1999, 119, 565–574. [Google Scholar] [CrossRef]
- Barceloux, D.G. Selenium. J. Toxicol. Clin. Toxicol. 1999, 37, 145–172. [Google Scholar] [CrossRef]
- Xu, C.; Zhang, S.; Ho, Y.-F.; Miller, E.J.; Roberts, K.A.; Li, H.-P.; Schwehr, K.A.; Otosaka, S.; Kaplan, D.I.; Brinkmeyer, R.; et al. Is soil natural organic matter a sink or source for mobile radioiodine (129I) at the Savannah River Site? Geochim. Cosmochim. Acta 2011, 75, 5716–5735. [Google Scholar] [CrossRef]
- World Health Organization. Trace Elements in Human Nutrition and Health; World Health Organization: Geneva, Switzerland, 1996. [Google Scholar]
- Antonyak, H.; Iskra, R.; Panas, N.; Lysiuk, R. Selenium. In Trace Elements and Minerals in Health and Longevity; Springer International Publishing: Berlin/Heidelberg, Germany, 2018. [Google Scholar]
- Oldfield, J.E. Selenium World Atlas; Selenium-Tellurium Development Association (STDA): Grimbergen, Belgium, 2002. [Google Scholar]
- Fleming, G.A.; Walsh, T. Selenium occurrence in certain Irish soils and its toxic effects on animals. In Proceedings of the Royal Irish Academy. Section B: Biological, Geological, and Chemical Science; Hodges, Figgis, Co.: Dublin, Ireland, 1956; pp. 151–166. [Google Scholar]
- Li, H.F.; McGrath, S.P.; Zhao, F.J. Selenium uptake, translocation and speciation in wheat supplied with selenate or selenite. New Phytol. 2008, 178, 92–102. [Google Scholar] [CrossRef]
- Bebien, M.; Chauvin, J.P.; Adriano, J.M.; Grosse, S.; Verméglio, A. Effect of selenite on growth and protein synthesis in the phototrophic bacterium Rhodobacter sphaeroides. Appl. Environ. Microbiol. 2001, 67, 4440–4447. [Google Scholar] [CrossRef]
- Tarze, A.; Dauplais, M.; Grigoras, I.; Lazard, M.; Ha-Duong, N.-T.; Barbier, F.; Blanquet, S.; Plateau, P. Extracellular production of hydrogen selenide accounts for thiol-assisted toxicity of selenite against Saccharomyces cerevisiae. J. Biol. Chem. 2007, 282, 8759–8767. [Google Scholar] [CrossRef]
- Oremland, R.S.; Herbel, M.J.; Blum, J.S.; Langley, S.; Beveridge, T.J.; Ajayan, P.M.; Sutto, T.; Ellis, A.V.; Curran, S. Structural and spectral features of selenium nanospheres produced by Se-respiring bacteria. Appl. Environ. Microbiol. 2004, 70, 52–60. [Google Scholar] [CrossRef]
- Lusa, M.; Knuutinen, J.; Bomberg, M. Uptake and reduction of Se (IV) in two heterotrophic aerobic Pseudomonads strains isolated from boreal bog environment. AIMS Microbiol. 2017, 3, 798. [Google Scholar] [CrossRef]
- Oremland, R.S.; Steinberg, N.A.; Maest, A.S.; Miller, L.G.; Hollibaugh, J.T. Measurement of in situ rates of selenate removal by dissimilatory bacterial reduction in sediments. Environ. Sci. Technol. 1990, 24, 1157–1164. [Google Scholar] [CrossRef]
- Zhang, Y.; Moore, J.N. Selenium fractionation and speciation in a wetland system. Environ. Sci. Technol. 1996, 30, 2613–2619. [Google Scholar] [CrossRef]
- Stolz, J.F.; Oremland, R.S. Bacterial respiration of arsenic and selenium. FEMS Microbiol. Rev. 1999, 23, 615–627. [Google Scholar] [CrossRef]
- Nancharaiah, Y.V.; Lens, P.N.L. Ecology and biotechnology of selenium-respiring bacteria. Microbiol. Mol. Biol. Rev. 2015, 79, 61–80. [Google Scholar] [CrossRef] [PubMed]
- De Souza, M.P.; Huang, C.P.A.; Chee, N.; Terry, N. Rhizosphere bacteria enhance the accumulation of selenium and mercury in wetland plants. Planta 1999, 209, 259–263. [Google Scholar] [CrossRef]
- Lusa, M.; Help, H.; Honkanen, A.P.; Knuutinen, J.; Parkkonen, J.; Kalasová, D.; Bomberg, M. The reduction of selenium (IV) by boreal Pseudomonas sp. strain T5-6-I—Effects on selenium (IV) uptake in Brassica oleracea. Environ. Res. 2019, 177, 108642. [Google Scholar] [CrossRef]
- Rosenfeld, C.E.; Kenyon, J.A.; James, B.R.; Santelli, C.M. Selenium (IV, VI) reduction and tolerance by fungi in an oxic environment. Geobiology 2017, 15, 441–452. [Google Scholar] [CrossRef] [PubMed]
- Charlet, L.; Scheinost, A.; Tournassat, C.; Greneche, J.-M.; Gehin, A.; Fernández-Martınez, A.; Coudert, S.; Tisserand, D.; Brendle, J. Electron transfer at the mineral/water interface: Selenium reduction by ferrous iron sorbed on clay. Geochim. Cosmochim. Acta 2007, 71, 5731–5749. [Google Scholar] [CrossRef]
- Assemi, S.; Erten, H. Sorption of radioiodine on organic rich soil, clay minerals and alumina. J. Radioanal. Nucl. Chem. 1994, 178, 193–204. [Google Scholar] [CrossRef]
- Evans, G.J.; Hammad, K.A. Radioanalytical studies of iodine behaviour in the environment. J. Radioanal. Nucl. Chem. 1995, 192, 239–247. [Google Scholar] [CrossRef]
- Sheppard, M.I.; Hawkins, J.L. Iodine and microbial interactions in an organic soil. J. Environ. Radioact. 1995, 29, 91–109. [Google Scholar] [CrossRef]
- Ashworth, D.J.; Shaw, G.; Butler, A.P.; Ciciani, L. Soil transport and plant uptake of radio-iodine from near-surface groundwater. J. Environ. Radioact. 2003, 70, 99–114. [Google Scholar] [CrossRef]
- Ashworth, D.J.; Shaw, G. A comparison of the soil migration and plant uptake of radioactive chlorine and iodine from contaminated groundwater. J. Environ. Radioact. 2006, 89, 61–80. [Google Scholar] [CrossRef]
- Li, H.-P.; Yeager, C.M.; Brinkmeyer, R.; Zhang, S.; Ho, Y.-F.; Xu, C.; Jones, W.L.; Schwehr, K.A.; Otosaka, S.; Roberts, K.A.; et al. Bacterial production of organic acids enhances H2O2-dependent iodide oxidation. Environ. Sci. Technol. 2012, 46, 4837–4844. [Google Scholar] [CrossRef]
- Bostock, A.C.; Shaw, G.; Bell, J.N.B. The volatilisation and sorption of 129I in coniferous forest, grassland and frozen soils. J. Environ. Radioact. 2003, 70, 29–42. [Google Scholar] [CrossRef]
- Yamaguchi, N.; Nakano, M.; Takamatsu, R.; Tanida, H. Inorganic iodine incorporation into soil organic matter: Evidence from iodine K-edge X-ray absorption near-edge structure. J. Environ. Radioact. 2010, 101, 451–457. [Google Scholar] [CrossRef] [PubMed]
- Xu, C.; Miller, E.J.; Zhang, S.; Li, H.-P.; Ho, Y.-F.; A Schwehr, K.; Kaplan, D.I.; Otosaka, S.; A Roberts, K.; Brinkmeyer, R.; et al. Sequestration and remobilization of radioiodine (129I) by soil organic matter and possible consequences of the remedial action at Savannah River Site. Environ. Sci. Technol. 2011, 45, 9975–9983. [Google Scholar] [CrossRef] [PubMed]
- Li, H.P.; Yeager, C.M.; Brinkmeyer, R.; Jones, W.L.; Zhang, S.J.; Xu, C.; Schwehr, K.A.; Santschi, P.H.; Kaplan, D.I.; Yeager, C.M. Iodide accumulation by aerobic bacteria isolated from subsurface sediments of an I-129 contaminated aquifer at the Savannah River Site, South Carolina. Appl. Environ. Microbiol. 2011, 77, 2153–2160. [Google Scholar] [CrossRef]
- Xu, C.; Chen, H.; Sugiyama, Y.; Zhang, S.; Li, H.P.; Ho, Y.F.; Chuang, C.Y.; Schwehr, K.A.; Kaplan, D.I.; Yeager, C.; et al. Novel molecular-level evidence of iodine binding to natural organic matter from Fourier transform ion cyclotron resonance mass spectrometry. Sci. Total Environ. 2013, 449, 244–252. [Google Scholar] [CrossRef]
- Amachi, S.; Mishima, Y.; Shinoyama, H.; Muramatsu, Y.; Fujii, T. Active transport and accumulation of iodide by newly isolated marine bacteria. Appl. Environ. Microbiol. 2005, 71, 741–745. [Google Scholar] [CrossRef] [PubMed]
- Ban-Nai, T.; Muramatsu, Y.; Amachi, S. Rate of iodine volatilization and accumulation by filamentous fungi through laboratory cultures. Chemosphere 2006, 65, 2216–2222. [Google Scholar] [CrossRef]
- Lusa, M.; Bomberg, M.; Aromaa, H.; Knuutinen, J.; Lehto, J. Sorption of radioiodide in an acidic, nutrient-poor boreal bog: Insights into the microbial impact. J. Environ. Radioact. 2015, 143, 110–122. [Google Scholar] [CrossRef]
- Dupont, C.L.; Grass, G.; Rensing, C. Copper toxicity and the origin of bacterial resistance—New insights and applications. Metallomics 2011, 3, 1109–1118. [Google Scholar] [CrossRef]
- Vincent, M.; Duval, R.E.; Hartemann, P.; Engels-Deutsch, M. Contact killing and antimicrobial properties of copper. J. Appl. Microbiol. 2018, 124, 1032–1046. [Google Scholar] [CrossRef]
- Antsotegi-Uskola, M.; Markina-Iñarrairaegui, A.; Ugalde, U. New insights into copper homeostasis in filamentous fungi. Int. Microbiol. 2020, 23, 65–73. [Google Scholar] [CrossRef]
- Richard, D.; Ravigné, V.; Rieux, A.; Facon, B.; Boyer, C.; Boyer, K.; Grygiel, P.; Javegny, S.; Terville, M.; Canteros, B.I.; et al. Adaptation of genetically monomorphic bacteria: Evolution of copper resistance through multiple horizontal gene transfers of complex and versatile mobile genetic elements. Mol. Ecol. 2017, 26, 2131–2149. [Google Scholar] [CrossRef]
- Nies, D.H. Bacterial transition metal homeostasis. In Molecular Microbiology of Heavy Metals; Springer: Berlin/Heidelberg, Germany, 2007; pp. 117–142. [Google Scholar]
- Rosen, B.P.; Liu, Z. Transport pathways for arsenic and selenium: A minireview. Environ. Int. 2009, 35, 512–515. [Google Scholar] [CrossRef]
- Turner, R.J.; Weiner, J.H.; Taylor, D.E. Selenium metabolism in Escherichia coli. Biometals 1998, 11, 223–227. [Google Scholar] [CrossRef] [PubMed]
- Amachi, S. Microbial contribution to global iodine cycling: Volatilization, accumulation, reduction, oxidation, and sorption of iodine. Microbes Environ. 2008, 23, 269–276. [Google Scholar] [CrossRef]
- Gozlan, R.S. Isolation of iodine-producing bacteria from aquaria. Antonie Leeuwenhoek 1968, 34, 226. [Google Scholar] [CrossRef] [PubMed]
- Amachi, S.; Muramatsu, Y.; Akiyama, Y.; Miyazaki, K.; Yoshiki, S.; Hanada, S.; Kamagata, Y.; Ban-Nai, T.; Shinoyama, H.; Fujii, T. Isolation of iodide-oxidizing bacteria from iodide-rich natural gas brines and seawaters. Microb. Ecol. 2005, 49, 547–557. [Google Scholar] [CrossRef] [PubMed]
- Li, H.-P.; Daniel, B.; Creeley, D.; Grandbois, R.; Zhang, S.; Xu, C.; Ho, Y.-F.; Schwehr, K.A.; Kaplan, D.I.; Santschi, P.H.; et al. Superoxide production by a manganese-oxidizing bacterium facilitates iodide oxidation. Appl. Environ. Microbiol. 2014, 80, 2693–2699. [Google Scholar] [CrossRef] [PubMed]
- Amachi, S.; Kimura, K.; Muramatsu, Y.; Shinoyama, H.; Fujii, T. Hydrogen peroxide-dependent uptake of iodine by marine Flavobacteriaceae bacterium strain C-21. Appl. Environ. Microbiol. 2007, 73, 7536–7541. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Amachi, S.; Minami, K.; Miyasaka, I.; Fukunaga, S. Ability of anaerobic microorganisms to associate with iodine: 125I tracer experiments using laboratory strains and enriched microbial communities from subsurface formation water. Chemosphere 2010, 79, 349–354. [Google Scholar] [CrossRef]
- Lusa, M.; Lehto, J.; Aromaa, H.; Knuutinen, J.; Bomberg, M. Uptake of radioiodide by Paenibacillus sp., Pseudomonas sp., Burkholderia sp. and Rhodococcus sp. isolated from a boreal nutrient-poor bog. J. Environ. Sci. 2016, 44, 26–37. [Google Scholar] [CrossRef]
- Duborská, E.; Urík, M.; Bujdoš, M. Comparison of iodide and iodate accumulation and volatilization by filamentous fungi during static cultivation. Water Air Soil Pollut. 2017, 228, 1–8. [Google Scholar] [CrossRef]
- Fang, L.; Cai, P.; Chen, W.; Liang, W.; Hong, Z.; Huang, Q. Impact of cell wall structure on the behavior of bacterial cells in the binding of copper and cadmium. Colloids Surf. A Physicochem. Eng. Asp. 2009, 347, 50–55. [Google Scholar] [CrossRef]
- Bhainsa, K.C.; D’Souza, S.F. Thorium biosorption by Aspergillus fumigatus, a filamentous fungal biomass. J. Hazard. Mater. 2009, 165, 670–676. [Google Scholar] [CrossRef]
- Fein, J.B.; Daughney, C.J.; Yee, N.; Davis, T.A. A chemical equilibrium model for metal adsorption onto bacterial surfaces. Geochim. Cosmochim. Acta 1997, 61, 3319–3328. [Google Scholar] [CrossRef]
- Gadd, G.M.; White, C. Microbial treatment of metal pollution—A working biotechnology? Trends Biotechnol. 1993, 11, 353–359. [Google Scholar] [CrossRef]
- Mishra, B.; Boyanov, M.; Bunker, B.A.; Kelly, S.D.; Kemner, K.M.; Fein, J.B. High-and low-affinity binding sites for Cd on the bacterial cell walls of Bacillus subtilis and Shewanella oneidensis. Geochim. Cosmochim. Acta 2010, 74, 4219–4233. [Google Scholar] [CrossRef]
- Antsotegi-Uskola, M.; Markina-Iñarrairaegui, A.; Ugalde, U. Copper resistance in Aspergillus nidulans relies on the PI-type ATPase CrpA, regulated by the transcription factor AceA. Front. Microbiol. 2017, 8, 912. [Google Scholar] [CrossRef] [PubMed]
- Orell, A.; Navarro, C.A.; Arancibia, R.; Mobarec, J.C.; Jerez, C.A. Life in blue: Copper resistance mechanisms of bacteria and archaea used in industrial biomining of minerals. Biotechnol. Adv. 2010, 28, 839–848. [Google Scholar] [CrossRef]
- Nies, D.H. Efflux-mediated heavy metal resistance in prokaryotes. FEMS Microbiol. Rev. 2003, 27, 313–339. [Google Scholar] [CrossRef]
- Aro, L.; Korpela, L.; Mäkinen, V.; Salemaa, M.; Saarinen, M.; Lahdenperä, A.M.; Parviainen, L.; Kuusisto, J. Studies on Reference Mires: 2. Lastensuo, Pesänsuo and Häädetkeidas in 2010–2015; Posiva WR 2017-15; Posiva Oy: Eurajoki, Finland, 2018. [Google Scholar]
- Mäkilä, M.; Grundström, A. Turpeen ikä ja Kerrostumisnopeus Lounais-Suomen Soilla; Working Report 2008–12; Posiva Oy: Eurajoki, Finland, 2008. (In Finnish) [Google Scholar]
- Tsitko, I.; Lusa, M.; Lehto, J.; Parviainen, L.; Ikonen, A.T.; Lahdenperä, A.M.; Bomberg, M. The variation of microbial communities in a depth profile of an acidic, nutrient-poor boreal bog in southwestern Finland. Open J. Ecol. 2014, 4, 832. [Google Scholar] [CrossRef]
- Lusa, M.; Knuutinen, J.; Lindgren, M.; Virkanen, J.; Bomberg, M. Microbial communities in a former pilot-scale uranium mine in Eastern Finland–Association with radium immobilization. Sci. Total Environ. 2019, 686, 619–640. [Google Scholar] [CrossRef]
- Klindworth, A.; Pruesse, E.; Schweer, T.; Peplies, J.; Quast, C.; Horn, M.; Glöckner, F.O. Evaluation of general 16S ribosomal RNA gene PCR primers for classical and next-generation sequencing-based diversity studies. Nucleic Acids Res. 2013, 41, e1. [Google Scholar] [CrossRef]
- Gardes, M.; Bruns, T.D. ITS primers with enhanced specificity for basidiomycetes-application to the identification of mycorrhizae and rusts. Mol. Ecol. 1993, 2, 113–118. [Google Scholar] [CrossRef]
- White, T.J.; Bruns, T.; Lee, S.J.W.T.; Taylor, J. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. PCR Protoc. Guide Methods Appl. 1990, 18, 315–322. [Google Scholar]
- Edwards, U.; Rogall, T.; Blöcker, H.; Emde, M.; Böttger, E.C. Isolation and direct complete nucleotide determination of entire genes. Characterization of a gene coding for 16S ribosomal RNA. Nucleic Acids Res. 1989, 17, 7843–7853. [Google Scholar] [CrossRef] [PubMed]
- Muyzer, G.; De Waal, E.C.; Uitterlinden, A.G. Profiling of complex microbial populations by denaturing gradient gel electrophoresis analysis of polymerase chain reaction-amplified genes coding for 16S rRNA. Appl. Environ. Microbiol. 1993, 59, 695–700. [Google Scholar] [CrossRef] [PubMed]
- Schloss, P.D.; Westcott, S.L.; Ryabin, T.; Hall, J.R.; Hartmann, M.; Hollister, E.B.; Lesniewski, R.A.; Oakley, B.B.; Parks, D.H.; Robinson, C.J.; et al. Introducing mothur: Open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl. Environ. Microbiol. 2009, 75, 7537–7541. [Google Scholar] [CrossRef]
- Pruesse, E.; Quast, C.; Knittel, K.; Fuchs, B.M.; Ludwig, W.; Peplies, J.; Glöckner, F.O. SILVA: A comprehensive online resource for quality checked and aligned ribosomal RNA sequence data compatible with ARB. Nucleic Acids Res. 2007, 35, 7188–7196. [Google Scholar] [CrossRef] [PubMed]
- Quast, C.; Pruesse, E.; Yilmaz, P.; Gerken, J.; Schweer, T.; Yarza, P.; Peplies, J.; Glöckner, F.O. The SILVA ribosomal RNA gene database project: Improved data processing and web-based tools. Nucleic Acids Res. 2012, 41, D590–D596. [Google Scholar] [CrossRef]
- Kõljalg, U.; Nilsson, R.H.; Abarenkov, K.; Tedersoo, L.; Taylor, A.F.S.; Bahram, M.; Bates, S.T.; Bruns, T.D.; Bengtsson-Palme, J.; Callaghan, T.M.; et al. Towards a unified paradigm for sequence-based identification of fungi. Mol. Ecol. 2013, 22, 5271–5277. [Google Scholar] [CrossRef]
- Nilsson, R.H.; Larsson, K.-H.; Taylor, A.F.S.; Bengtsson-Palme, J.; Jeppesen, T.S.; Schigel, D.; Kennedy, P.; Picard, K.; Glöckner, F.O.; Tedersoo, L.; et al. The UNITE database for molecular identification of fungi: Handling dark taxa and parallel taxonomic classifications. Nucleic Acids Res. 2019, 47, D259–D264. [Google Scholar] [CrossRef]
- Abarenkov, K.; Zirk, A.; Piirmann, T.; Pöhönen, R.; Ivanov, F.; Nilsson, R.H.; Kõljalg, U. UNITE Mothur Release for Fungi. Version 04.02.2020. UNITE Community. 2020. Available online: https://plutof.ut.ee/#/doi/10.15156/BIO/786381 (accessed on 20 April 2020).
- Lusa, M.; Bomberg, M.; Aromaa, H.; Knuutinen, J.; Lehto, J. The microbial impact on the sorption behaviour of selenite in an acidic, nutrient-poor boreal bog. J. Environ. Radioact. 2015, 147, 85–96. [Google Scholar] [CrossRef] [PubMed]
- Hammer, Ø.; Harper, D.A.; Ryan, P.D. PAST: Paleontological statistics software package for education and data analysis. Palaeontol. Electron. 2001, 4, 9. [Google Scholar]
- McMurdie, P.J.; Holmes, S. phyloseq: An R package for reproducible interactive analysis and graphics of microbiome census data. PLoS ONE 2013, 8, e61217. [Google Scholar] [CrossRef]
- R Core Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2014; Available online: http://www.R-project.org/ (accessed on 9 September 2020).
- Scheller, S.; Goenrich, M.; Boecher, R.; Thauer, R.K.; Jaun, B. The key nickel enzyme of methanogenesis catalyses the anaerobic oxidation of methane. Nature 2010, 465, 606–608. [Google Scholar] [CrossRef] [PubMed]
- Madhaiyan, M.; Poonguzhali, S.; Sa, T. Metal tolerating methylotrophic bacteria reduces nickel and cadmium toxicity and promotes plant growth of tomato (Lycopersicon esculentum L.). Chemosphere 2007, 69, 220–228. [Google Scholar] [CrossRef] [PubMed]
- Ho, Y.S.; Wase, D.J.; Forster, C.F. Batch nickel removal from aqueous solution by sphagnum moss peat. Water Res. 1995, 29, 1327–1332. [Google Scholar] [CrossRef]
- Vila Domínguez, A.; Ayerbe Algaba, R.; Miró Canturri, A.; Rodríguez Villodres, Á.; Smani, Y. Antibacterial activity of colloidal silver against gram-negative and gram-positive bacteria. Antibiotics 2020, 9, 36. [Google Scholar] [CrossRef]
- Lusa, M.; Lehto, J.; Bomberg, M. The uptake of Ni2+ and Ag+ by bacterial strains isolated from a boreal nutrient-poor bog. AIMS Microbiol. 2016, 2, 120–137. [Google Scholar] [CrossRef]
- Andreasson, A.; Jönsson, B.; Lindman, B. Surface and colloid chemistry of peat and peat dewatering. Electrostatic effects. Colloid Polym. Sci. 1988, 266, 164–172. [Google Scholar] [CrossRef]
- Strelko, V., Jr.; Malik, D.J. Characterization and metal sorptive properties of oxidized active carbon. J. Colloid Interface Sci. 2002, 250, 213–220. [Google Scholar] [CrossRef] [PubMed]
- Corapcioglu, M.O.; Huang, C.P. The adsorption of heavy metals onto hydrous activated carbon. Water Res. 1987, 21, 1031–1044. [Google Scholar] [CrossRef]
- Tan, K.H. Humic Matter in Soil and the Environment: Principles and Controversies, 2nd ed.; CRC Press, Taylor and Francis Group: Boca Raton, FL, USA, 2014; p. 324. [Google Scholar]
- Martinez, E.R.; Sharma, P.; Kappler, A. Surface binding site analysis of Ca2+-homoionized clay–humic acid complexes. J. Colloid Interface Sci. 2010, 352, 526–534. [Google Scholar] [CrossRef] [PubMed]
- Koch-Steindl, H.; Pröhl, G. Considerations on the behaviour of long-lived radionuclides in the soil. Radiat. Environ. Biophys. 2001, 40, 93–104. [Google Scholar] [CrossRef]
- Huber, R.; Sacher, M.; Vollmann, A.; Huber, H.; Rose, D. Respiration of arsenate and selenate by hyperthermophilic archaea. Syst. Appl. Microbiol. 2000, 23, 305–314. [Google Scholar] [CrossRef]
- Sarret, G.; Avoscan, L.; Carrière, M.; Collins, R.; Geoffroy, N.; Carrot, F.; Covès, J.; Gouget, B. Chemical forms of selenium in the metal-resistant bacterium Ralstonia metallidurans CH34 exposed to selenite and selenate. Appl. Environ. Microbiol. 2005, 71, 2331–2337. [Google Scholar] [CrossRef] [PubMed]
- Oremland, R.S.; Hollibaugh, J.T.; Maest, A.S.; Presser, T.S.; Miller, L.G.; Culbertson, C.W. Selenate reduction to elemental selenium by anaerobic bacteria in sediments and culture: Biogeochemical significance of a novel, sulfate-independent respiration. Appl. Environ. Microbiol. 1989, 55, 2333–2343. [Google Scholar] [CrossRef]
- Handley, K.M.; Verberkmoes, N.C.; I Steefel, C.; Williams, K.H.; Sharon, I.; Miller, C.S.; Frischkorn, K.R.; Chourey, K.; Thomas, B.C.; Shah, M.B.; et al. Biostimulation induces syntrophic interactions that impact C, S and N cycling in a sediment microbial community. ISME J. 2013, 7, 800–816. [Google Scholar] [CrossRef]
- Nakamaru, Y.M.; Altansuvd, J. Speciation and bioavailability of selenium and antimony in non-flooded and wetland soils: A review. Chemosphere 2014, 111, 366–371. [Google Scholar] [CrossRef]
- Pettine, M.; Gennari, F.; Campanella, L.; Casentini, B.; Marani, D. The reduction of selenium (IV) by hydrogen sulfide in aqueous solutions. Geochim. Cosmochim. Acta 2012, 83, 37–47. [Google Scholar] [CrossRef]
- Yeager, C.M.; Amachi, S.; Grandbois, R.; Kaplan, D.I.; Xu, C.; Schwehr, K.A.; Santschi, P.H. Microbial Transformation of Iodine: From Radioisotopes to Iodine Deficiency. Adv. Appl. Microbiol. 2017. [Google Scholar] [CrossRef]
- Fernández-Calviño, D.; Bååth, E. Interaction between pH and Cu toxicity on fungal and bacterial performance in soil. Soil Biol. Biochem. 2016, 96, 20–29. [Google Scholar] [CrossRef]
- Starr, M.; Lindroos, A.J.; Ukonmaanaho, L.; Tarvainen, T.; Tanskanen, H. Weathering release of heavy metals from soil in comparison to deposition, litterfall and leaching fluxes in a remote, boreal coniferous forest. Appl. Geochem. 2003, 18, 607–613. [Google Scholar] [CrossRef]
- Moffett, B.F.; Nicholson, F.A.; Uwakwe, N.C.; Chambers, B.J.; Harris, J.A.; Hill, T.C. Zinc contamination decreases the bacterial diversity of agricultural soil. FEMS Microbiol. Ecol. 2003, 43, 13–19. [Google Scholar] [CrossRef]
- Kour, R.; Jain, D.; Bhojiya, A.A.; Sukhwal, A.; Sanadhya, S.; Saheewala, H.; Jat, G.; Singh, A.; Mohanty, S.R. Zinc biosorption, biochemical and molecular characterization of plant growth-promoting zinc-tolerant bacteria. 3 Biotech 2019, 9, 1–17. [Google Scholar] [CrossRef]
- Myers, B.; Webster, K.L.; Mclaughlin, J.W.; Basiliko, N. Microbial activity across a boreal peatland nutrient gradient: The role of fungi and bacteria. Wetl. Ecol. Manag. 2012, 20, 77–88. [Google Scholar] [CrossRef]
- Song, N.; Xu, H.; Yan, Z.; Yang, T.; Wang, C.; Jiang, H.L. Improved lignin degradation through distinct microbial community in subsurface sediments of one eutrophic lake. Renew. Energy 2019, 138, 861–869. [Google Scholar] [CrossRef]
- Lin, X.; Tfaily, M.M.; Steinweg, J.M.; Chanton, P.; Esson, K.; Yang, Z.K.; Chanton, J.P.; Cooper, W.T.; Schadt, C.W.; Kostka, J.E. Microbial community stratification linked to utilization of carbohydrates and phosphorus limitation in a boreal peatland at Marcell Experimental Forest, Minnesota, USA. Appl. Environ. Microbiol. 2014, 80, 3518–3530. [Google Scholar] [CrossRef] [PubMed]
- Coolen, M.J.; van de Giessen, J.; Zhu, E.Y.; Wuchter, C. Bioavailability of soil organic matter and microbial community dynamics upon permafrost thaw. Environ. Microbiol. 2011, 13, 2299–2314. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).