Untargeted Exometabolomics Provides a Powerful Approach to Investigate Biogeochemical Hotspots with Vegetation and Polygon Type in Arctic Tundra Soils
Abstract
1. Introduction
2. Materials and Methods
2.1. Study Site and Sample Description
2.2. Soil Extraction and Sample Preparation
2.3. Instrumentation and LC/MS Data Collection
2.4. Untargeted LC/MS Data Processing
2.5. Data Filtering, Normalization, and Statistical Analyses
2.6. Feature Annotation
3. Results
3.1. Evaluation of Analytical Performance Across Multiple Sites
3.2. Impacts of Polygon Type and Vegetation on LMW DOM Abundance
3.3. Molecular Characterization of Differentially Abundant LMW DOM Features
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Disclaimer
References
- Tarnocai, C.; Canadell, J.G.; Schuur, E.A.G.; Kuhry, P.; Mazhitova, G.; Zimov, S. Soil organic carbon pools in the northern circumpolar permafrost region. Glob. Biogeochem. Cycles 2009, 23. [Google Scholar] [CrossRef]
- IPCC. Climate Change 2013: The Physical Science Basis. Contribution of Working Group I to the Fifth Assessment Report of the Intergovernmental Panel on Climate Change; Cambridge University Press: Cambridge, UK; New York, NY, USA, 2013; p. 1535. [CrossRef]
- Hugelius, G.; Strauss, J.; Zubrzycki, S.; Harden, J.W.; Schuur, E.A.G.; Ping, C.L.; Schirrmeister, L.; Grosse, G.; Michaelson, G.J.; Koven, C.D.; et al. Estimated stocks of circumpolar permafrost carbon with quantified uncertainty ranges and identified data gaps. Biogeosciences 2014, 11, 6573–6593. [Google Scholar] [CrossRef]
- Abbott, B.W.; Larouche, J.R.; Jones, J.B.; Bowden, W.B.; Balser, A.W. Elevated dissolved organic carbon biodegradability from thawing and collapsing permafrost. J. Geophys. Res. Biogeosci. 2014, 119, 2049–2063. [Google Scholar] [CrossRef]
- Hopkins, F.M.; Torn, M.S.; Trumbore, S.E. Warming accelerates decomposition of decades-old carbon in forest soils. Proc. Natl. Acad. Sci. USA 2012, 109, E1753–E1761. [Google Scholar] [CrossRef]
- Romanovsky, V.E.; Smith, S.L.; Christiansen, H.H. Permafrost Thermal State in the Polar Northern Hemisphere during the International Polar Year 2007–2009: A Synthesis. Permafrost Periglac. Process. 2010, 21, 106–116. [Google Scholar] [CrossRef]
- Schuur, E.A.G.; McGuire, A.D.; Schadel, C.; Grosse, G.; Harden, J.W.; Hayes, D.J.; Hugelius, G.; Koven, C.D.; Kuhry, P.; Lawrence, D.M.; et al. Climate change and the permafrost carbon feedback. Nature 2015, 520, 171–179. [Google Scholar] [CrossRef]
- Schuur, E.A.G.; Bockheim, J.; Canadell, J.G.; Euskirchen, E.; Field, C.B.; Goryachkin, S.V.; Hagemann, S.; Kuhry, P.; Lafleur, P.M.; Lee, H.; et al. Vulnerability of permafrost carbon to climate change: Implications for the global carbon cycle. Bioscience 2008, 58, 701–714. [Google Scholar] [CrossRef]
- Hinzman, L.D.; Deal, C.J.; McGuire, A.D.; Mernild, S.H.; Polyakov, I.V.; Walsh, J.E. Trajectory of the Arctic as an integrated system. Ecol. Appl. 2013, 23, 1837–1868. [Google Scholar] [CrossRef]
- Voigt, C.; Lamprecht, R.E.; Marushchak, M.E.; Lind, S.E.; Novakovskiy, A.; Aurela, M.; Martikainen, P.J.; Biasi, C. Warming of subarctic tundra increases emissions of all three important greenhouse gases—Carbon dioxide, methane, and nitrous oxide. Glob. Chang. Biol. 2017, 23, 3121–3138. [Google Scholar] [CrossRef]
- Lara, M.J.; Nitze, I.; Grosse, G.; Martin, P.; McGuire, A.D. Reduced arctic tundra productivity linked with landform and climate change interactions. Sci. Rep. 2018, 8. [Google Scholar] [CrossRef]
- Wookey, P.A.; Aerts, R.; Bardgett, R.D.; Baptist, F.; Brathen, K.A.; Cornelissen, J.H.C.; Gough, L.; Hartley, I.P.; Hopkins, D.W.; Lavorel, S.; et al. Ecosystem feedbacks and cascade processes: Understanding their role in the responses of Arctic and alpine ecosystems to environmental change. Glob. Chang. Biol. 2009, 15, 1153–1172. [Google Scholar] [CrossRef]
- Olefeldt, D.; Goswami, S.; Grosse, G.; Hayes, D.; Hugelius, G.; Kuhry, P.; McGuire, A.D.; Romanovsky, V.E.; Sannel, A.B.K.; Schuur, E.A.G.; et al. Circumpolar distribution and carbon storage of thermokarst landscapes. Nat. Commun. 2016, 7, 13043. [Google Scholar] [CrossRef]
- Lipson, D.A.; Zona, D.; Raab, T.K.; Bozzolo, F.; Mauritz, M.; Oechel, W.C. Water-table height and microtopography control biogeochemical cycling in an Arctic coastal tundra ecosystem. Biogeosciences 2012, 9, 577–591. [Google Scholar] [CrossRef]
- Zona, D.; Lipson, D.A.; Zulueta, R.C.; Oberbauer, S.F.; Oechel, W.C. Microtopographic controls on ecosystem functioning in the Arctic Coastal Plain. J. Geophys. Res. Biogeosci. 2011, 116. [Google Scholar] [CrossRef]
- Lara, M.J.; Nitze, I.; Grosse, G.; McGuire, A.D. Tundra landform and vegetation productivity trend maps for the Arctic Coastal Plain of northern Alaska. Sci. Data 2018, 5. [Google Scholar] [CrossRef]
- Iversen, C.M.; Sloan, V.L.; Sullivan, P.F.; Euskirchen, E.S.; McGuire, A.D.; Norby, R.J.; Walker, A.P.; Warren, J.M.; Wullschleger, S.D. The unseen iceberg: Plant roots in arctic tundra. New Phytol. 2015, 205, 34–58. [Google Scholar] [CrossRef]
- Tas, N.; Prestat, E.; Wang, S.; Wu, Y.X.; Ulrich, C.; Kneafsey, T.; Tringe, S.G.; Torn, M.S.; Hubbard, S.S.; Jansson, J.K. Landscape topography structures the soil microbiome in arctic polygonal tundra. Nature Commun. 2018, 9. [Google Scholar] [CrossRef]
- Hodgkins, S.B.; Tfaily, M.M.; McCalley, C.K.; Logan, T.A.; Crill, P.M.; Saleska, S.R.; Rich, V.I.; Chanton, J.P. Changes in peat chemistry associated with permafrost thaw increase greenhouse gas production. Proc. Natl. Acad. Sci. USA 2014, 111, 5819–5824. [Google Scholar] [CrossRef]
- Sjogersten, S.; Caul, S.; Daniell, T.J.; Jurd, A.P.S.; O’Sullivan, O.S.; Stapleton, C.S.; Titman, J.J. Organic matter chemistry controls greenhouse gas emissions from permafrost peatlands. Soil Biol. Biochem. 2016, 98, 42–53. [Google Scholar] [CrossRef]
- Pare, M.C.; Bedard-Haughn, A. Soil organic matter quality influences mineralization and GHG emissions in cryosols: A field-based study of sub- to high Arctic. Glob. Chang. Biol. 2013, 19, 1126–1140. [Google Scholar] [CrossRef]
- Xu, X.F.; Schimel, J.P.; Thornton, P.E.; Song, X.; Yuan, F.M.; Goswami, S. Substrate and environmental controls on microbial assimilation of soil organic carbon: A framework for Earth system models. Ecol. Lett. 2014, 17, 547–555. [Google Scholar] [CrossRef]
- Riley, W.J.; Subin, Z.M.; Lawrence, D.M.; Swenson, S.C.; Torn, M.S.; Meng, L.; Mahowald, N.M.; Hess, P. Barriers to predicting changes in global terrestrial methane fluxes: Analyses using CLM4Me, a methane biogeochemistry model integrated in CESM. Biogeosciences 2011, 8, 1925–1953. [Google Scholar] [CrossRef]
- Tang, J.Y.; Riley, W.J. Weaker soil carbon-climate feedbacks resulting from microbial and abiotic interactions. Nat. Clim. Chang. 2015, 5, 56–60. [Google Scholar] [CrossRef]
- Liljedahl, A.K.; Boike, J.; Daanen, R.P.; Fedorov, A.N.; Frost, G.V.; Grosse, G.; Hinzman, L.D.; Iijma, Y.; Jorgenson, J.C.; Matveyeva, N.; et al. Pan-Arctic ice-wedge degradation in warming permafrost and its influence on tundra hydrology. Nat. Geosci. 2016, 9, 312–318. [Google Scholar] [CrossRef]
- Wainwright, H.M.; Dafflon, B.; Smith, L.J.; Hahn, M.S.; Curtis, J.B.; Wu, Y.X.; Ulrich, C.; Peterson, J.E.; Torn, M.S.; Hubbard, S.S. Identifying multiscale zonation and assessing the relative importance of polygon geomorphology on carbon fluxes in an Arctic tundra ecosystem. J. Geophys. Res. Biogeosci. 2015, 120, 788–808. [Google Scholar] [CrossRef]
- Hubbard, S.S.; Gangodagamage, C.; Dafflon, B.; Wainwright, H.; Peterson, J.; Gusmeroli, A.; Ulrich, C.; Wu, Y.; Wilson, C.; Rowland, J.; et al. Quantifying and relating land-surface and subsurface variability in permafrost environments using LiDAR and surface geophysical datasets. Hydrogeol. J. 2013, 21, 149–169. [Google Scholar] [CrossRef]
- Norby, R.J.; Sloan, V.L.; Iversen, C.M.; Childs, J. Controls on Fine-Scale Spatial and Temporal Variability of Plant-Available Inorganic Nitrogen in a Polygonal Tundra Landscape. Ecosystems 2018, 22, 528–543. [Google Scholar] [CrossRef]
- Newman, B.D.; Throckmorton, H.M.; Graham, D.E.; Gu, B.; Hubbard, S.S.; Liang, L.; Wu, Y.; Heikoop, J.M.; Herndon, E.M.; Phelps, T.J.; et al. Microtopographic and depth controls on active layer chemistry in Arctic polygonal ground. Geophys. Res. Lett. 2015, 42, 1808–1817. [Google Scholar] [CrossRef]
- Lehmann, J.; Kleber, M. The contentious nature of soil organic matter. Nature 2015, 528, 60–68. [Google Scholar] [CrossRef]
- Yang, Z.; Wullschleger, S.D.; Liang, L.; Graham, D.E.; Gu, B. Effects of warming on the degradation and production of low-molecular-weight labile organic carbon in an Arctic tundra soil. Soil Biol. Biochem. 2016, 95, 202–211. [Google Scholar] [CrossRef]
- Chen, H.M.; Yang, Z.M.; Chu, R.K.; Tolic, N.; Liang, L.Y.; Graham, D.E.; Wullschleger, S.D.; Gu, B.H. Molecular Insights into Arctic Soil Organic Matter Degradation under Warming. Environ. Sci. Technol. 2018, 52, 4555–4564. [Google Scholar] [CrossRef] [PubMed]
- Boddy, E.; Roberts, P.; Hill, P.W.; Farrar, J.; Jones, D.L. Turnover of low molecular weight dissolved organic C (DOC) and microbial C exhibit different temperature sensitivities in Arctic tundra soils. Soil Biol. Biochem. 2008, 40, 1557–1566. [Google Scholar] [CrossRef]
- Herndon, E.M.; Yang, Z.M.; Bargar, J.; Janot, N.; Regier, T.; Graham, D.; Wullschleger, S.; Gu, B.H.; Liang, L.Y. Geochemical drivers of organic matter decomposition in arctic tundra soils. Biogeochemistry 2015, 126, 397–414. [Google Scholar] [CrossRef]
- Judd, K.E.; Crump, B.C.; Kling, G.W. Variation in dissolved organic matter controls bacterial production and community composition. Ecology 2006, 87, 2068–2079. [Google Scholar] [CrossRef]
- Guigue, J.; Leveque, J.; Mathieu, O.; Schmitt-Kopplin, P.; Lucio, M.; Arrouays, D.; Jolivet, C.; Dequiedt, S.; Prevost-Boure, N.C.; Ranjard, L. Water-extractable organic matter linked to soil physico-chemistry and microbiology, at the regional scale. Soil Biol. Biochem. 2015, 84, 158–167. [Google Scholar] [CrossRef]
- Swenson, T.L.; Karaoz, U.; Swenson, J.M.; Bowen, B.P.; Northen, T.R. Linking soil biology and chemistry in biological soil crust using isolate exometabolomics. Nat. Commun. 2018, 9. [Google Scholar] [CrossRef]
- Swenson, T.L.; Jenkins, S.; Bowen, B.P.; Northen, T.R. Untargeted soil metabolomics methods for analysis of extractable organic matter. Soil Biol. Biochem. 2015, 80, 189–198. [Google Scholar] [CrossRef]
- Vorkas, P.A.; Isaac, G.; Anwar, M.A.; Davies, A.H.; Want, E.J.; Nicholson, J.K.; Holmes, E. Untargeted UPLC-MS Profiling Pipeline to Expand Tissue Metabolome Coverage: Application to Cardiovascular Disease. Anal. Chem. 2015, 87, 4184–4193. [Google Scholar] [CrossRef]
- Zhou, B.; Xiao, J.F.; Tuli, L.; Ressom, H.W. LC-MS-based metabolomics. Mol. Biosyst. 2012, 8, 470–481. [Google Scholar] [CrossRef] [PubMed]
- Evans, A.M.; DeHaven, C.D.; Barrett, T.; Mitchell, M.; Milgram, E. Integrated, Nontargeted Ultrahigh Performance Liquid Chromatography/Electrospray Ionization Tandem Mass Spectrometry Platform for the Identification and Relative Quantification of the Small-Molecule Complement of Biological Systems. Anal. Chem. 2009, 81, 6656–6667. [Google Scholar] [CrossRef] [PubMed]
- Warren, C.R. Development of liquid chromatography mass spectrometry method for analysis of organic N monomers in soil. Soil Biol. Biochem. 2014, 78, 233–242. [Google Scholar] [CrossRef]
- Ladd, M.P.; Giannone, R.J.; Abraham, P.E.; Wullschleger, S.D.; Hettich, R.L. Evaluation of an untargeted nano-liquid chromatography-mass spectrometry approach to expand coverage of low molecular weight dissolved organic matter in Arctic soil. Sci. Rep. 2019, 9, 5810. [Google Scholar] [CrossRef] [PubMed]
- Sloan, V.L.; Brooks, J.D.; Wood, S.J.; Liebig, J.A.; Siegrist, J.; Iversen, C.M.; Norby, R.J. Plant Community Composition and Vegetation Height, Barrow, Alaska, Ver. 1; Oak Ridge National Laboratory, Carbon Dioxide Information Analysis Center: Oak Ridge, TN, USA, 2014. [Google Scholar] [CrossRef]
- Simmons, K.; Deatrick, J.; Lewis, B. Operating Procedure: Soil Sampling. Science and Ecosystem Support Division; EPA: Washington, DC, USA, 2014.
- Tfaily, M.M.; Chu, R.K.; Tolic, N.; Roscioli, K.M.; Anderton, C.R.; Pasa-Tolic, L.; Robinson, E.W.; Hess, N.J. Advanced Solvent Based Methods for Molecular Characterization of Soil Organic Matter by High-Resolution Mass Spectrometry. Anal. Chem. 2015, 87, 5206–5215. [Google Scholar] [CrossRef] [PubMed]
- De Livera, A.M.; Sysi-Aho, M.; Jacob, L.; Gagnon-Bartsch, J.A.; Castillo, S.; Simpson, J.A.; Speed, T.P. Statistical Methods for Handling Unwanted Variation in Metabolomics Data. Anal. Chem. 2015, 87, 3606–3615. [Google Scholar] [CrossRef]
- Pluskal, T.; Castillo, S.; Villar-Briones, A.; Oresic, M. MZmine 2: Modular framework for processing, visualizing, and analyzing mass spectrometry-based molecular profile data. BMC Bioinform. 2010, 11. [Google Scholar] [CrossRef]
- Olivon, F.; Grelier, G.; Roussi, F.; Litaudon, M.; Touboul, D. MZmine 2 Data-Preprocessing To Enhance Molecular Networking Reliability. Anal. Chem. 2017, 89, 7836–7840. [Google Scholar] [CrossRef]
- Gika, H.G.; Theodoridis, G.A.; Plumb, R.S.; Wilson, I.D. Current practice of liquid chromatography-mass spectrometry in metabolomics and metabonomics. J. Pharm. Biomed. Anal. 2014, 87, 12–25. [Google Scholar] [CrossRef]
- Koch, B.P.; Dittmar, T.; Witt, M.; Kattner, G. Fundamentals of molecular formula assignment to ultrahigh resolution mass data of natural organic matter. Anal. Chem. 2007, 79, 1758–1763. [Google Scholar] [CrossRef]
- Kind, T.; Fiehn, O. Seven Golden Rules for heuristic filtering of molecular formulas obtained by accurate mass spectrometry. BMC Bioinform. 2007, 8. [Google Scholar] [CrossRef]
- Kujawinski, E.B.; Behn, M.D. Automated analysis of electrospray ionization Fourier transform ion cyclotron resonance mass spectra of natural organic matter. Anal. Chem. 2006, 78, 4363–4373. [Google Scholar] [CrossRef]
- Kanehisa, M.; Furumichi, M.; Tanabe, M.; Sato, Y.; Morishima, K. KEGG: New perspectives on genomes, pathways, diseases and drugs. Nucleic Acids Res. 2017, 45, D353–D361. [Google Scholar] [CrossRef]
- Smith, C.A.; O’Maille, G.; Want, E.J.; Qin, C.; Trauger, S.A.; Brandon, T.R.; Custodio, D.E.; Abagyan, R.; Siuzdak, G. METLIN—A metabolite mass spectral database. Ther. Drug Monit. 2005, 27, 747–751. [Google Scholar] [CrossRef]
- Cui, Q.; Lewis, I.A.; Hegeman, A.D.; Anderson, M.E.; Li, J.; Schulte, C.F.; Westler, W.M.; Eghbalnia, H.R.; Sussman, M.R.; Markley, J.L. Metabolite identification via the Madison Metabolomics Consortium Database. Nat. Biotechnol. 2008, 26, 162–164. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.; Thiessen, P.A.; Bolton, E.E.; Chen, J.; Fu, G.; Gindulyte, A.; Han, L.Y.; He, J.E.; He, S.Q.; Shoemaker, B.A.; et al. PubChem Substance and Compound databases. Nucleic Acids Res. 2016, 44, D1202–D1213. [Google Scholar] [CrossRef] [PubMed]
- Wishart, D.S.; Jewison, T.; Guo, A.C.; Wilson, M.; Knox, C.; Liu, Y.F.; Djoumbou, Y.; Mandal, R.; Aziat, F.; Dong, E.; et al. HMDB 3.0-The Human Metabolome Database in 2013. Nucleic Acids Res. 2013, 41, D801–D807. [Google Scholar] [CrossRef] [PubMed]
- Fahy, E.; Sud, M.; Cotter, D.; Subramaniam, S. LIPID MAPS online tools for lipid research. Nucleic Acids Res. 2007, 35, W606–W612. [Google Scholar] [CrossRef] [PubMed]
- Schlapfer, P.; Zhang, P.; Wang, C.; Kim, T.; Banf, M.; Chae, L.; Dreher, K.; Chavali, A.K.; Nilo-Pyanco, R.; Bernard, T.; et al. Genome-Wide Prediction of Metabolic Enzymes, Pathways, and Gene Clusters in Plants. Plant Physiol. 2017, 173, 2041–2059. [Google Scholar] [CrossRef]
- Darrouzet-Nardi, A.; Weintraub, M.N. Evidence for spatially inaccessible labile N from a comparison of soil core extractions and soil pore water lysimetry. Soil Biol. Biochem. 2014, 73, 22–32. [Google Scholar] [CrossRef]
- Verchot, L.V.; Dutaur, L.; Shepherd, K.D.; Albrecht, A. Organic matter stabilization in soil aggregates: Understanding the biogeochemical mechanisms that determine the fate of carbon inputs in soils. Geoderma 2011, 161, 182–193. [Google Scholar] [CrossRef]
- Kujawinski, E.B.; Freitas, M.A.; Zang, X.; Hatcher, P.G.; Greenchurch, K.B.; Jones, R.B. The application of electrospray ionization mass spectrometry (ESI MS) to the structural characterization of natural organic matter. Org. Geochem. 2002, 33, 171. [Google Scholar] [CrossRef]
- Brown, T.L.; Rice, J.A. Effect of experimental parameters on the ESI FT-ICR mass spectrum of fulvic acid. Anal. Chem. 2000, 72, 384. [Google Scholar] [CrossRef] [PubMed]
- Lipson, D.A.; Raab, T.K.; Goria, D.; Zlamal, J. The contribution of Fe(III) and humic acid reduction to ecosystem respiration in drained thaw lake basins of the Arctic Coastal Plain. Glob. Biogeochem. Cycles 2013, 27, 399–409. [Google Scholar] [CrossRef]
- Lipson, D.A.; Jha, M.; Raab, T.K.; Oechel, W.C. Reduction of iron (III) and humic substances plays a major role in anaerobic respiration in an Arctic peat soil. J. Geophys. Res. Biogeosci. 2010, 115. [Google Scholar] [CrossRef]
- Herndon, E.M.; AlBashaireh, A.; Singer, D.; Roy Chowdhury, T.; Gu, B.; Graham, D. Influence of iron redox cycling on organo-mineral associations in Arctic tundra soil. Geochim. Cosmochim. Acta 2017, 207, 210–231. [Google Scholar] [CrossRef]
- Ladd, M.P.; Wullschleger, S.; Iversen, C. Soil Organic Horizon Exometabolomics Extracts and Soil Characteristics with Depth, Barrow, Alaska, 2014; Oak Ridge National Laboratory, U.S. Department of Energy, Next Generation Ecosystem Experiments Arctic Data Collection: Oak Ridge, TN, USA, 2018. [CrossRef]
- Kim, S.; Kramer, R.W.; Hatcher, P.G. Graphical method for analysis of ultrahigh-resolution broadband mass spectra of natural organic matter, the van Krevelen diagram. Anal. Chem. 2003, 75, 5336–5344. [Google Scholar] [CrossRef]
- Mann, B.F.; Chen, H.M.; Herndon, E.M.; Chu, R.K.; Tolic, N.; Portier, E.F.; Chowdhury, T.R.; Robinson, E.W.; Callister, S.J.; Wullschleger, S.D.; et al. Indexing Permafrost Soil Organic Matter Degradation Using High-Resolution Mass Spectrometry. PLoS ONE 2015, 10. [Google Scholar] [CrossRef]
- Rivas-Ubach, A.; Liu, Y.; Bianchi, T.S.; Tolic, N.; Jansson, C.; Pasa-Tolic, L. Moving beyond the van Krevelen Diagram: A New Stoichiometric Approach for Compound Classification in Organisms. Anal. Chem. 2018, 90, 6152–6160. [Google Scholar] [CrossRef]
- Gerhard, M.; Dietmar, S. Biochemical Pathways: An Atlas of Biochemistry and Molecular Biology, 2nd ed.; Wiley: Hoboken, NJ, USA, 2012. [Google Scholar]
- Kostiainen, R.; Kauppila, T.J. Effect of eluent on the ionization process in liquid chromatography-mass spectrometry. J. Chromatogr. A 2009, 1216, 685–699. [Google Scholar] [CrossRef]
- Kaiser, K.; Kalbitz, K. Cycling downwards—Dissolved organic matter in soils. Soil Biol. Biochem. 2012, 52, 29–32. [Google Scholar] [CrossRef]
- Tfaily, M.M.; Wilson, R.M.; Cooper, W.T.; Kostka, J.E.; Hanson, P.; Chanton, J.P. Vertical Stratification of Peat Pore Water Dissolved Organic Matter Composition in a Peat Bog in Northern Minnesota. J. Geophys. Res. Biogeosci. 2018, 123, 479–494. [Google Scholar] [CrossRef]
- Strom, L.; Tagesson, T.; Mastepanov, M.; Christensen, T.R. Presence of Eriophorum scheuchzeri enhances substrate availability and methane emission in an Arctic wetland. Soil Biol. Biochem. 2012, 45, 61–70. [Google Scholar] [CrossRef]
- Kielland, K. Short-Circuiting the Nitrogen Cycle: Ecophysiological Strategies of Nitrogen Uptake in Plants from Marginal Environments. In Plant Nutrient Acquisition; Springer: Berlin, Germany, 2001; pp. 376–398. [Google Scholar]
- Kielland, K.; McFarland, J.; Olson, K. Amino acid uptake in deciduous and coniferous taiga ecosystems. Plant Soil 2006, 288, 297–307. [Google Scholar] [CrossRef]
- Nashölm, T.; Kielland, K.; Ganeteg, U. Uptake of organic nitrogen by plants. New Phytol. 2009, 182, 31–48. [Google Scholar] [CrossRef]
- Mendez-Millan, M.; Dignac, M.F.; Rumpel, C.; Rasse, D.P.; Derenne, S. Molecular dynamics of shoot vs. root biomarkers in an agricultural soil estimated by natural abundance 13C labeling. Soil Biol. Biochem. 2010, 42, 169–177. [Google Scholar] [CrossRef]
- Miller, P.C.; Mangan, R.; Kummerow, J. Vertical distribution of organic matter in eight vegetation types near Eagle Summit, Alaska. Holarct. Ecol. 1982, 5, 117–124. [Google Scholar] [CrossRef]
- Chowdhury, T.R.; Herndon, E.M.; Phelps, T.J.; Elias, D.A.; Gu, B.H.; Liang, L.Y.; Wullschleger, S.D.; Graham, D.E. Stoichiometry and temperature sensitivity of methanogenesis and CO2 production from saturated polygonal tundra in Barrow, Alaska. Glob. Chang. Biol. 2015, 21, 722–737. [Google Scholar] [CrossRef]
- Eisenhauer, N.; Lanoue, A.; Strecker, T.; Scheu, S.; Steinauer, K.; Thakur, M.; Mommer, L. Root biomass and exudates link plant diversity with soil bacterial and fungal biomass. Sci. Rep. 2017, 7. [Google Scholar] [CrossRef]
- Sloan, V.L.; Liebig, J.; Hahn, M.; Curtis, B.; Brooks, J.; Rogers, A.; Iversen, C.; Norby, R. Soil Temperature, Soil Moisture, and Thaw Depth, Barrow, Alaska Ver. 1; Oak Ridge National Laboratory, U.S. Department of Energy, Next Generation Ecosystem Experiments Arctic Data Collection: Oak Ridge, TN, USA, 2014. [CrossRef]
- Gardeñas, A.I.; Agren, G.I.; Bird, J.A.; Clarholm, M.; Hallin, S.; Ineson, P.; Katterer, T.; Knicker, H.; Nilsson, S.I.; Nashölm, T.; et al. Knowledge gaps in soil carbon and nitrogen interactions—From molecular to global scale. Soil Biol. Biochem. 2011, 43, 702–717. [Google Scholar] [CrossRef]
- Kuzyakov, Y. Review: Factors affecting rhizosphere priming. J. Plant Nutr. Soil Sci. 2002, 165, 382–396. [Google Scholar] [CrossRef]
- Reemtsma, T.; These, A.; Venkatachari, P.; Xia, X.Y.; Hopke, P.K.; Springer, A.; Linscheid, M. Identification of fulvic acids and sulfated and nitrated analogues in atmospheric aerosol by electrospray ionization Fourier transform ion cyclotron resonance mass spectrometry. Anal. Chem. 2006, 78, 8299–8304. [Google Scholar] [CrossRef]
- EPA. Secondary Organic Aerosol (SOAs) Research. Available online: https://www.epa.gov/air-research/secondary-organic-aerosol-soas-research (accessed on 20 October 2018).
- Xie, M.; Mladenov, N.; Williams, M.W.; Neff, J.C.; Wasswa, J.; Hannigan, M.P. Water soluble organic aerosols in the Colorado Rocky Mountains, USA: Composition, sources and optical properties. Sci. Rep. 2016, 6, 39339. [Google Scholar] [CrossRef] [PubMed]
- Kirpes, R.M.; Bondy, A.L.; Bonanno, D.; Moffet, R.C.; Wang, B.B.; Laskin, A.; Ault, A.P.; Pratt, K.A. Secondary sulfate is internally mixed with sea spray aerosol and organic aerosol in the winter Arctic. Atmos. Chem. Phys. 2018, 18, 3937–3949. [Google Scholar] [CrossRef]
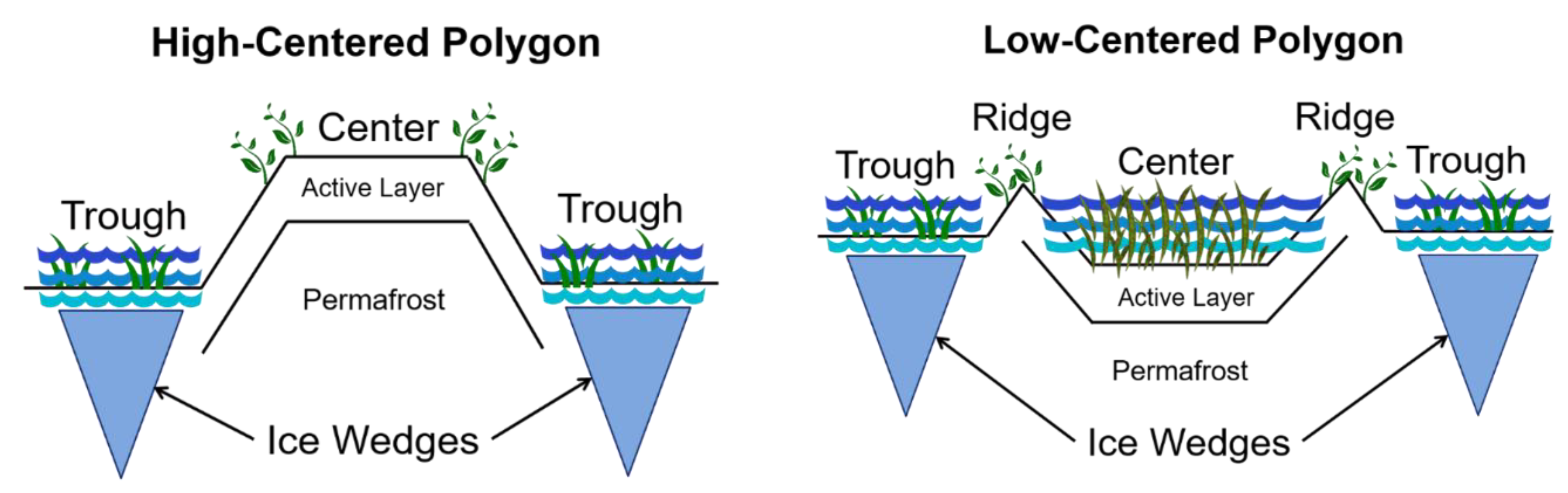

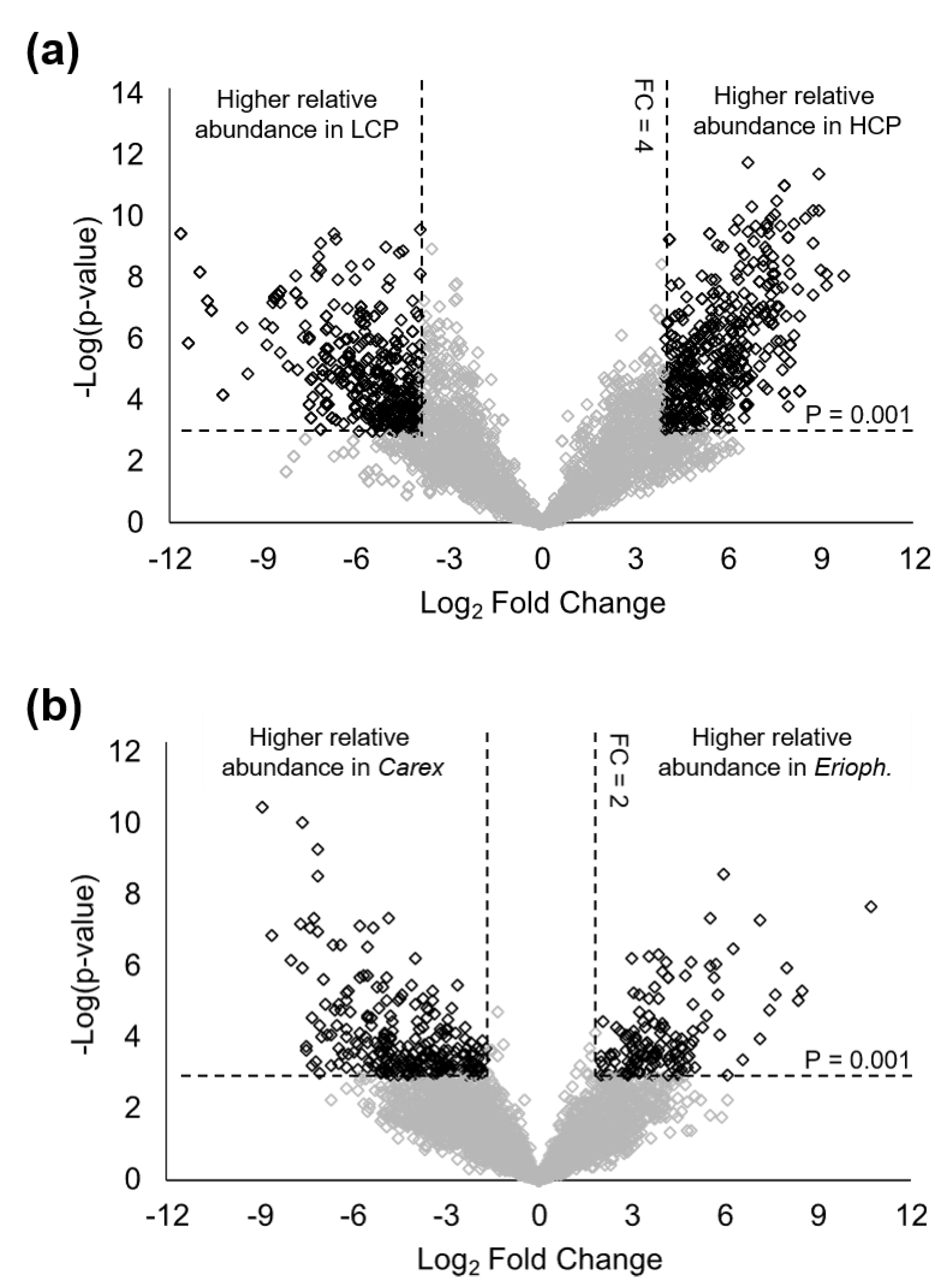

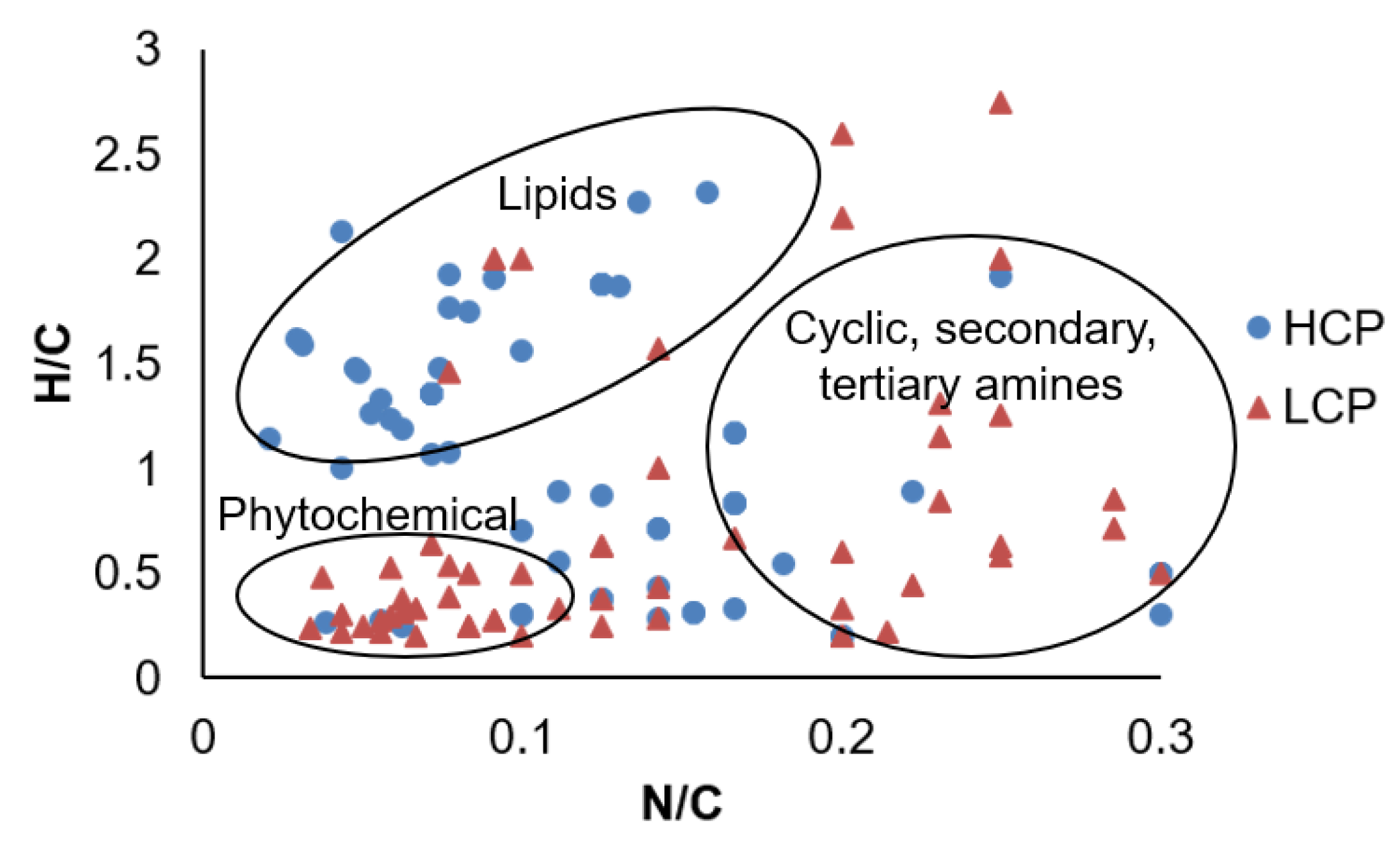

| HILIC (+) | HILIC (−) | RP (+) | RP (−) | |
|---|---|---|---|---|
| Aligned peaks a | 4686 | 2853 | 4213 | 1921 |
| Features b | 4352 | 2249 | 3655 | 1762 |
| High-Quality Features c | 3929 | 2170 | 3618 | 1541 |
| Unique HQFs d | 3414 | 1942 | 3494 | 1287 |
| Abundant HQFs e | 1966 | 776 | 1259 | 99 |
| Differentially abundant f | 322 | 76 | 122 | 1 |
| Annotated g | 283 | 74 | 117 | 1 |
| HCP | LCP | Due to Vegetation | All Differentially Abundant Features | |
|---|---|---|---|---|
| Number of features | 92 | 95 | 30 | 217 |
| Average formula | C17H21.5O4.9N1.2S2.0P0.2 | C11.9H7.8O6.5N1.1S2.3P0.5 | C20.3H20.6O6N0.9S0.5P0.2 | C15.2H15.4O5.7N1.1S1.9P0.3 |
| m/z | 393.0249 | 361.4926 | 398.1955 | 379.9353 |
| DBE | 7.88 | 9.51 | 11.5 | 9.09 |
| AI | 0.37 | 0.75 | 0.42 | 0.54 |
| H/C | 1.12 | 0.75 | 1.12 | 0.81 |
| O/C | 0.45 | 3.40 | 0.38 | 0.62 |
| N/C | 0.15 | 0.17 | 0.08 | 0.20 |
| O/S | 2.6 | 3.4 | 5.13 | 3.26 |
| DBE/C | 0.59 | 0.79 | 0.56 | 0.67 |
| DBE/H | 1.15 | 2.10 | 0.76 | 1.51 |
| DBE/O | 3.2 | 2.35 | 3.62 | 2.89 |
| C:N | 14.4 | 10.9 | 23.4 | 13.9 |
| % lipid | 16.3 | 2.11 | 6.67 | 8.75 |
| % protein | 7.61 | 6.32 | 6.67 | 6.91 |
| % lignin | 13.0 | 4.21 | 30.0 | 11.5 |
| % carbohydrate | 5.43 | 6.32 | 6.67 | 5.99 |
| % unsaturated | 12.0 | 4.21 | 10.0 | 8.29 |
| % aromatic | 30.4 | 51.6 | 33.3 | 40.1 |
| % tannin | 15.2 | 25.3 | 6.67 | 18.4 |
| Site with Higher Relative Abundance | LC/MS Condition | m/z | Predicted Formula | ppm Error | Class | Fold Change (Erioph Cores) | Fold Change (Carex Cores) |
|---|---|---|---|---|---|---|---|
| LCP | HILIC (−) | 347.8884 | C9H2O9P1S2 | −4.7 | Tannin | −11.63 | −7.645 |
| LCP | HILIC (−) | 400.8711 | C7H2N2O12S3 | 3.5 | Tannin | −11.01 | −6.884 |
| LCP | HILIC (−) | 367.8842 | C8H3NO10S3 | −1.1 | Tannin | −10.61 | −8.179 |
| LCP | HILIC (−) | 400.8702 | C7H2N2O12S3 | 1.3 | Tannin | −10.16 | −6.893 |
| LCP | HILIC (−) | 192.0527 | C7H7N5O2 | 0.1 | Lignin | - | −5.111 |
| HCP | HILIC (+) | 702.5350 | C50H68O2 | 0.2 | Unsaturated Hydrocarbon | 9.189 | 8.014 |
| HCP | HILIC (+) | 506.8323 | C13H4N2O5S7 | −1.9 | Condensed Hydrocarbon | 7.311 | 8.501 |
| HCP | RP (+) | 273.2535 | C15H32N2O2 | −0.7 | Lipid | 5.685 | 7.433 |
| HCP | RP (+) | 453.3682 | C26H48N2O4 | −1.1 | Lipid | 6.616 | 9.623 |
| HCP | RP (+) | 104.0705 | C4H9NO2 | −1.2 | Protein | 8.020 | 8.594 |
| Site with Higher Relative Abundance | LC/MS Condition | m/z | Predicted Formula | ppm Error | Class | Fold Change (HCP Cores) | Fold Change (LCP Cores) |
| Erioph | HILIC (+) | 286.1138 | C9H19NO9 | 1.8 | Carbohydrate | −5.562 | −7.98873 |
| Erioph | HILIC (+) | 363.0902 | C10H14N6O9 | 1.9 | Tannin | - | −7.38981 |
| Erioph | HILIC (+) | 251.0764 | C9H14O8 | 0.9 | Carbohydrate | −6.575 | −6.22072 |
| Erioph | RP (+) | 459.1955 | C32H26O3 | 0 | Unsaturated Hydrocarbon | −7.075 | - |
| Erioph | RP (+) | 548.2498 | C11H21N27O | 0.5 | Lipid | −7.523 | - |
| Erioph | RP (+) | 550.2340 | C18H27N15O6 | −0.3 | Lignin | −8.613 | - |
| Erioph | RP (+) | 226.1285 | C8H19NO6 | −0.2 | Carbohydrate | −7.708 | - |
| Carex | HILIC (+) | 191.0233 | C12H2N2O | −3.8 | Aromatic | 7.139 | - |
| Carex | HILIC (−) | 236.8647 | no hit | - | - | 4.198 | - |
| Carex | HILIC (−) | 414.7706 | no hit | - | - | 4.127 | - |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ladd, M.P.; Reeves, D.T.; Poudel, S.; Iversen, C.M.; Wullschleger, S.D.; Hettich, R.L. Untargeted Exometabolomics Provides a Powerful Approach to Investigate Biogeochemical Hotspots with Vegetation and Polygon Type in Arctic Tundra Soils. Soil Syst. 2021, 5, 10. https://doi.org/10.3390/soilsystems5010010
Ladd MP, Reeves DT, Poudel S, Iversen CM, Wullschleger SD, Hettich RL. Untargeted Exometabolomics Provides a Powerful Approach to Investigate Biogeochemical Hotspots with Vegetation and Polygon Type in Arctic Tundra Soils. Soil Systems. 2021; 5(1):10. https://doi.org/10.3390/soilsystems5010010
Chicago/Turabian StyleLadd, Mallory P., David T. Reeves, Suresh Poudel, Colleen M. Iversen, Stan D. Wullschleger, and Robert L. Hettich. 2021. "Untargeted Exometabolomics Provides a Powerful Approach to Investigate Biogeochemical Hotspots with Vegetation and Polygon Type in Arctic Tundra Soils" Soil Systems 5, no. 1: 10. https://doi.org/10.3390/soilsystems5010010
APA StyleLadd, M. P., Reeves, D. T., Poudel, S., Iversen, C. M., Wullschleger, S. D., & Hettich, R. L. (2021). Untargeted Exometabolomics Provides a Powerful Approach to Investigate Biogeochemical Hotspots with Vegetation and Polygon Type in Arctic Tundra Soils. Soil Systems, 5(1), 10. https://doi.org/10.3390/soilsystems5010010






