Coronary Artery Disease Is Related to Methylation Disorders Caused by the c.1286A>C MTHFR Polymorphism and to Low Serum 5-MTHF and Folic Acid Concentrations—Preliminary Results
Abstract
1. Introduction
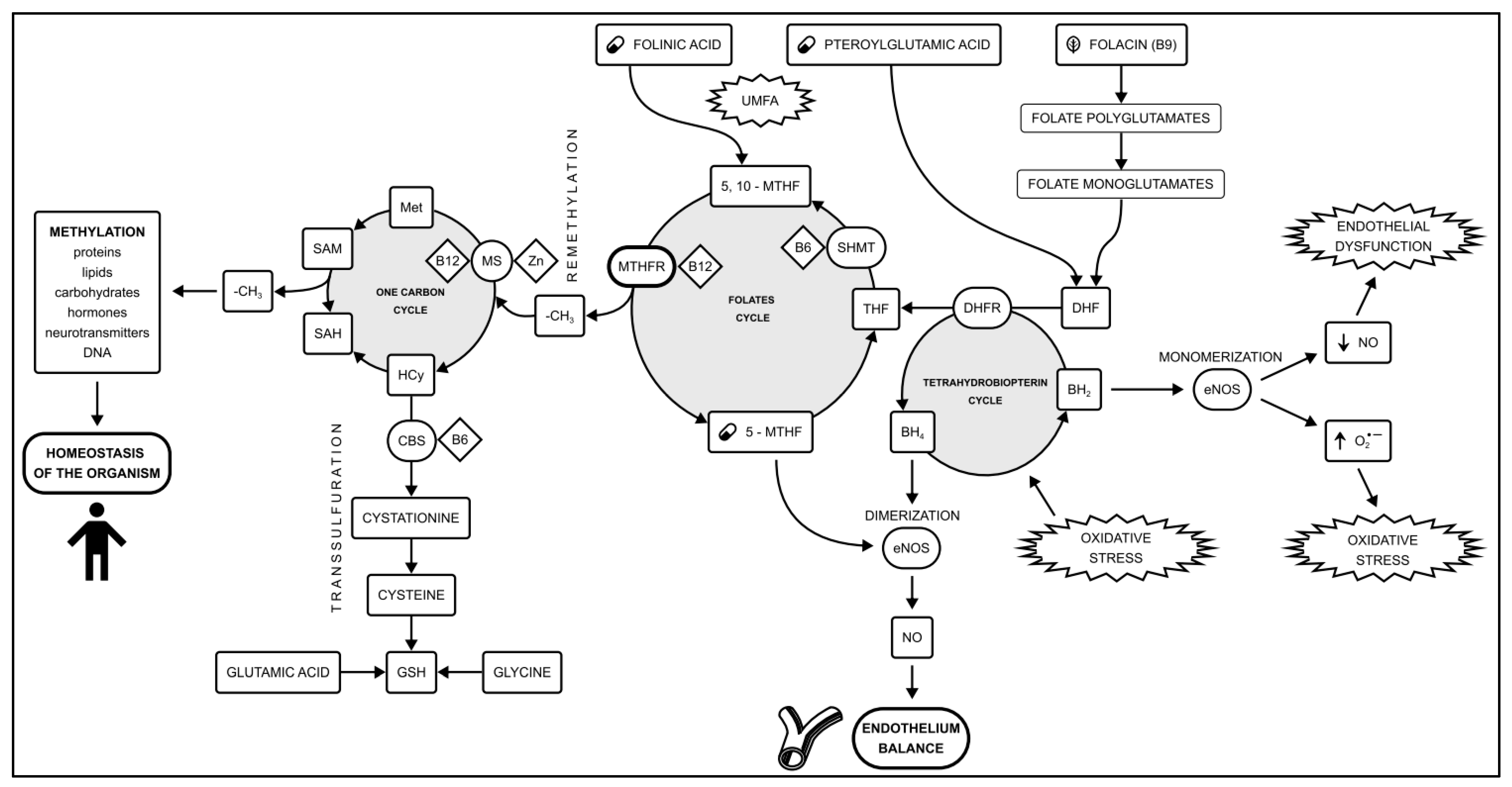
2. Materials and Methods
2.1. Study Design and Population
2.2. Inclusion/Exclusion Criteria
Blood Collection
2.3. Genetic Analysis
2.4. Folates Analysis
- Preparation of the sample for analysis: 1 mL of EtOH was added to the 0.5 mL of the sample, mixed together, and transferred to an Eppendorf (2 mL). Next, the sample was rinsed with an additional 0.5 mL of EtOH. Eppendorfs incubated at 37 °C for 20 min. After incubation, the probes were centrifuged. The supernatant was collected.
- LC analysis: gradient elution (component A—water and component B—acetonitrile, both solvents with the addition of 0.1% formic acid); reverse phase column by Kinetex C18 100 × 4.6 mm without thermostating, particle size—2.6 µm, pore size 100 Å; analysis program (B/A): 0–2 min 3/97, 2–31 min 95/5, 31–32 min 0/100, 32–33 min 0/100, 33–35 min 3/97, 35–37.5 min 3/97; analytical wavelengths of the DAD detector: 214, 220, 256 and 291 nm; sample injection volume per column—10 µL.
- MS analysis: electrospray ionization method, the flow rate of drying gas (nitrogen) 6.0 L/min, nebulizer pressure 2.4 Bar, capillary inlet temperature 250 °C, capillary voltage 4000 V; TOF (time of flight) detector. Solvents included in the eluent and used for sample preparation had the degree of purity required for LC-MS analysis. All obtained solutions were additionally centrifuged in order to remove possible impurities and residues of undissolved compounds.
2.5. Statistical Analysis
3. Results
3.1. Characteristics of the Groups
3.2. Frequencies of the Genotypes
3.2.1. Coronary Artery Disease Patients versus Controls Comparisons
3.2.2. Coronary Artery Disease Patients versus European Population Comparisons
3.3. Folates Concentrations
3.3.1. Folates Concentrations in Patients with MTHFR Polymorphism
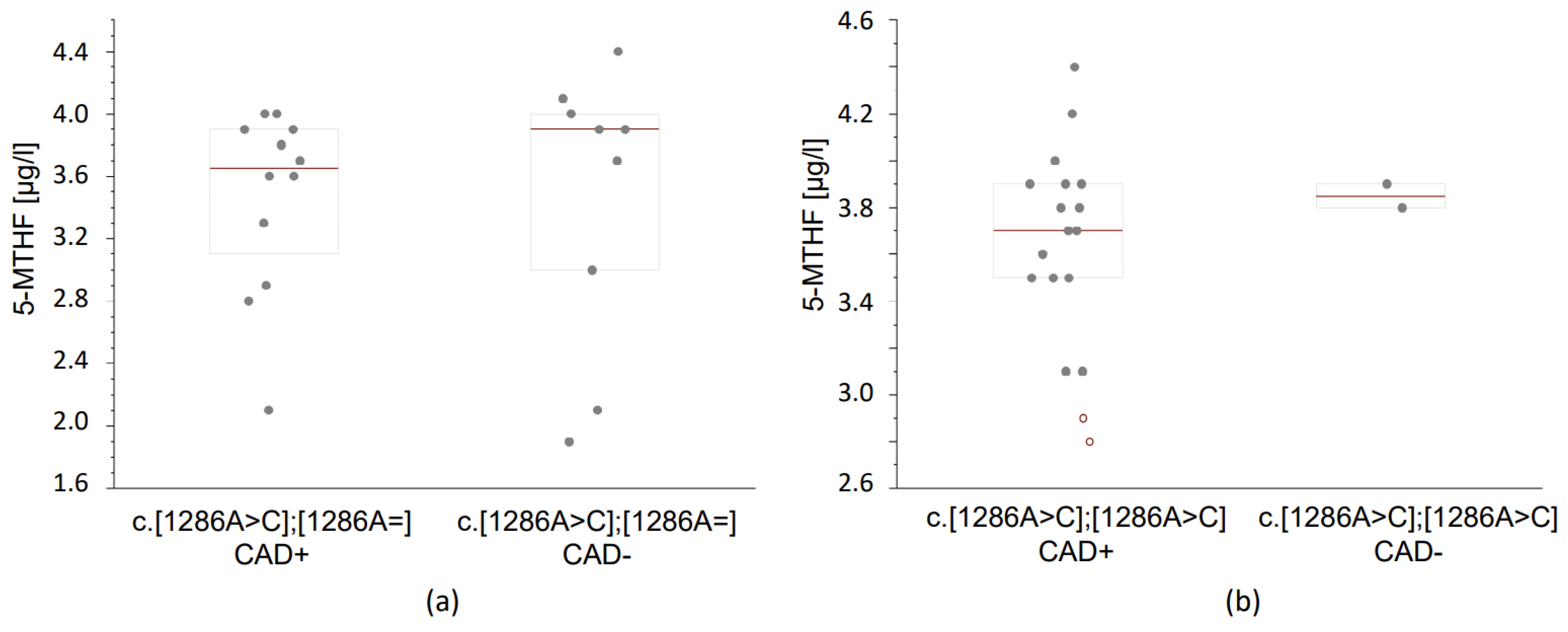
3.3.2. Folates Concentrations According to the MTHFR Genotype and the Occurrence of Coronary Artery Disease
4. Discussion
Practical Aspects of Results and Future Directions of Research
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- World Health Organization. Global Health Estimates: Life Expectancy and Leading Causes of Death and Disability. 2019. Available online: www.who.int/data/gho/data/themes/mortality-and-global-health-estimates (accessed on 18 October 2023).
- Flora, G.D.; Nayak, M.K. A Brief Review of Cardiovascular Diseases, Associated Risk Factors and Current Treatment Regimes. Curr. Pharm. Des. 2019, 25, 4063–4084. [Google Scholar] [CrossRef]
- Ferreira-Gonzalez, I. The epidemiology of coronary heart disease. Rev. Esp. Cardiol. (Engl. Ed.) 2014, 67, 139–144. [Google Scholar] [CrossRef] [PubMed]
- Szałtys, D. Umieralność w 2021 Roku. Zgony Według Przyczyn—Dane Wstępne; Główny Urząd Statystyczny: Warsaw, Poland, 2022. [Google Scholar]
- Fallahzadeh, A.; Mehraban, S.; Mahmoodi, T.; Sheikhy, A.; Naderian, M.; Afsaneh Aein, P.; Rafiee, H.; Mehrani, M.; Tajdini, M.; Masoud-kabir, F.; et al. Risk factor profile and outcomes of premature acute coronary syndrome after percutaneous coronary intervention: A 1-year prospective design. Clin. Cardiol. 2023, 47, e24170. [Google Scholar] [CrossRef] [PubMed]
- Roberts, R.; Chang, C.C.; Hadley, T. Genetic Risk Stratification: A Paradigm Shift in Prevention of Coronary Artery Disease. JACC Basic Transl. Sci. 2021, 6, 287–304. [Google Scholar] [CrossRef] [PubMed]
- Koyama, S.; Ito, K.; Terao, C.; Akiyama, M.; Horikoshi, M.; Momozawa, Y.; Matsunaga, H.; Ieki, H.; Ozaki, K.; Onouchi, Y.; et al. Population-specific and trans-ancestry genome-wide analyses identify distinct and shared genetic risk loci for coronary artery disease. Nat. Genet. 2020, 52, 1169–1177. [Google Scholar] [CrossRef]
- Kessler, T.; Schunkert, H. Coronary Artery Disease Genetics Enlightened by Genome-Wide Association Studies. JACC Basic Transl. Sci. 2021, 6, 610–623. [Google Scholar] [CrossRef]
- Erdmann, J.; Kessler, T.; Munoz Venegas, L.; Schunkert, H. A decade of genome-wide association studies for coronary artery disease: The challenges ahead. Cardiovasc. Res. 2018, 114, 1241–1257. [Google Scholar] [CrossRef] [PubMed]
- Askin, L.; Tanriverdi, O.; Tibilli, H.; Turkmen, S. Associations between Vaspin Levels and Coronary Artery Disease. Cardiovasc. Innov. Appl. 2020, 4, 211–216. [Google Scholar] [CrossRef]
- Mach, F.; Baigent, C.; Catapano, A.L.; Koskinas, K.C.; Casula, M.; Badimon, L.; Chapman, M.J.; De Backer, G.G.; Delgado, V.; Ference, B.A.; et al. 2019 ESC/EAS Guidelines for the management of dyslipidaemias: Lipid modification to reduce cardiovascular risk: The Task Force for the management of dyslipidaemias of the European Society of Cardiology (ESC) and European Atherosclerosis Society (EAS). Eur. Heart J. 2019, 41, 111–188. [Google Scholar] [CrossRef]
- David, S. A current guide to candidate gene association studies. Trends Genet. 2021, 37, 1056–1059. [Google Scholar] [CrossRef]
- De Clercq, E. The magic of a methyl group: Biochemistry at the service of medicine. Biochem. Pharmacol. 2023, 216, 115786. [Google Scholar] [CrossRef]
- Gladwin, R. Methylation. Encyclopedia Britannica. Available online: https://www.britannica.com/science/methylation (accessed on 18 October 2023).
- McMahon, A.; McNulty, H.; Hughes, C.F.; Strain, J.J.; Ward, M. Novel Approaches to Investigate One-Carbon Metabolism and Related B-Vitamins in Blood Pressure. Nutrients 2016, 8, 720. [Google Scholar] [CrossRef]
- Lee, W.E.; Genetzakis, E.; Figtree, G.A. Novel Strategies in the Early Detection and Treatment of Endothelial Cell-Specific Mitochondrial Dysfunction in Coronary Artery Disease. Antioxidants 2023, 12, 1359. [Google Scholar] [CrossRef] [PubMed]
- Karczewski, K.J.; Francioli, L.C.; Tiao, G.; Cummings, B.B.; Alföldi, J.; Wang, Q.; Collins, R.L.; Laricchia, K.M.; Ganna, A.; Birnbaum, D.P.; et al. The mutational constraint spectrum quantified from variation in 141,456 humans. Nature 2020, 581, 434–443. [Google Scholar] [CrossRef] [PubMed]
- Undas, A.; Chojnowski, K.; Klukowska, A.; Letowska, M.; Mital, A.; Mlynarski, W.; Musiał, J.; Podolak, M.; Sąsiadek, M.; Treliński, J.; et al. Determination and interpretation of MTHFR gene mutations in gynecology and internal medicine. Pol. Arch. Intern. Med. 2019, 129, 728–732. [Google Scholar] [CrossRef]
- Hickey, S.E.; Curry, C.J.; Toriello, H.V. ACMG Practice Guideline: Lack of evidence for MTHFR polymorphism testing. Genet. Med. 2013, 15, 153–156. [Google Scholar] [CrossRef]
- Greenland, P.; Alpert, J.S.; Beller, G.A.; Benjamin, E.J.; Budoff, M.J.; Fayad, Z.A.; Foster, E.; Hlatky, M.A.; Hodgson, J.M.; Kushner, F.G.; et al. 2010 ACCF/AHA guideline for assessment of cardiovascular risk in asymptomatic adults: A report of the American College of Cardiology Foundation/American Heart Association Task Force on Practice Guidelines. J. Am. Coll. Cardiol. 2010, 56, e50–e103. [Google Scholar] [CrossRef] [PubMed]
- Visseren, F.L.J.; Mach, F.; Smulders, Y.M.; Carballo, D.; Koskinas, K.C.; Back, M.; Benetos, A.; Biffi, A.; Boavida, J.-M.; Capodanno, D.; et al. 2021 ESC Guidelines on cardiovascular disease prevention in clinical practice: Developed by the Task Force for cardiovascular disease prevention in clinical practice with representatives of the European Society of Cardiology and 12 medical societies with the special contribution of the European Association of Preventive Cardiology (EAPC). Eur. Heart J. 2021, 42, 3227–3337. [Google Scholar]
- Hiraoka, M.; Kagawa, Y. Genetic polymorphisms and folate status. Congenit. Anom. 2017, 57, 142–149. [Google Scholar] [CrossRef]
- Crabtree, M.J.; Hale, A.B.; Channon, K.M. Dihydrofolate reductase protects endothelial nitric oxide synthase from uncoupling in tetrahydrobiopterin deficiency. Free Radic. Biol. Med. 2011, 50, 1639–1646. [Google Scholar] [CrossRef]
- Moens, A.L.; Vrints, C.J.; Claeys, M.J.; Timmermans, J.-P.; Champion, H.C.; Kass, D.A. Mechanisms and potential therapeutic targets for folic acid in cardiovascular disease. Am. J. Physiol.-Heart Circ. Physiol. 2008, 294, H1971–H1977. [Google Scholar] [CrossRef] [PubMed]
- Sobczyńska-Malefora, A.; Harrington, D.J. Laboratory assessment of folate (vitamin B9) status. J. Clin. Pathol. 2018, 71, 949–956. [Google Scholar] [CrossRef]
- Finer, S.; Saravanan, P.; Hitman, G.; Yajnik, C. The role of the one-carbon cycle in the developmental origins of Type 2 diabetes and obesity. Diabet. Med. 2014, 31, 263–272. [Google Scholar] [CrossRef] [PubMed]
- Population Detail. Available online: https://www.ncbi.nlm.nih.gov/projects/SNP/snp_viewTable.cgi?pop=4720 (accessed on 18 October 2023).
- Calderon-Larranaga, A.; Saadeh, M.; Hooshmand, B.; Refsum, H.; Smith, A.D.; Marengoni, A.; Vetrano, D.L. Association of Homocysteine, Methionine, and MTHFR 677C>T Polymorphism with Rate of Cardiovascular Multimorbidity Development in Older Adults in Sweden. JAMA Netw. Open 2020, 3, e205316. [Google Scholar] [CrossRef] [PubMed]
- Waśkiewicz, A.; Piotrowski, W.; Broda, G.; Sobczyk-Kopcioł, A.; Płoski, R. Impact of MTHFR C677T gene polymorphism and vitamins intake on homocysteine concentration in the Polish adult population. Kardiol. Pol. 2011, 69, 1259–1264. [Google Scholar]
- Luo, Z.; Lu, Z.; Muhammad, I.; Chen, Y.; Chen, Q.; Zhang, J.; Song, Y. Associations of the MTHFR rs1801133 polymorphism with coronary artery disease and lipid levels: A systematic review and updated meta-analysis. Lipids Health Dis. 2018, 17, 191. [Google Scholar] [CrossRef]
- Masud, R.; Qureshi, I.Z. Tetra primer ARMS-PCR relates folate/homocysteine pathway genes and ACE gene polymorphism with coronary artery disease. Mol. Cell. Biochem. 2011, 355, 289–297. [Google Scholar] [CrossRef] [PubMed]
- Masud, R.; Baqai, H.Z. The communal relation of MTHFR, MTR, ACE gene polymorphisms and hyperhomocysteinemia as conceivable risk of coronary artery disease. Appl. Physiol. Nutr. Metab. 2017, 42, 1009–1014. [Google Scholar] [CrossRef]
- Masud, R.; Anjum, A.F.; Anwar, M.Z.; Khan, W.U.; Shahzad, M.A.; Jawwad, G. The risk stratification of coronary vascular disease as linked to homocysteine, its modulating genes, genetic polymorphisms, conventional predictors, and with antihypertensive medicaments. Chin. J. Physiol. 2021, 64, 298–305. [Google Scholar] [CrossRef]
- Wrzosek, M.; Ślusarczyk, K. Methylenetetrahydrofolate Reductase C677T Gene Variant in Relation to Body Mass Index and Folate Concentration in a Polish Population. Biomedicines 2022, 10, 3140. [Google Scholar] [CrossRef]
- Cheng, T.-Y.D.; Ilozumba, M.N.; Balavarca, Y.; Neuhouser, M.L.; Miller, J.W.; Beresford, S.A.A.; Zheng, Y.; Song, X.; Duggan, D.J.; Toriola, A.T.; et al. Associations between Genetic Variants and Blood Biomarkers of One-Carbon Metabolism in Postmenopausal Women from the Women’s Health Initiative Observational Study. J. Nutr. 2022, 152, 1099–1106. [Google Scholar] [CrossRef]
- Tanaka, T.; Scheet, P.; Giusti, B.; Bandinelli, S.; Piras, M.G.; Usala, G.; Lai, S.; Mulas, A.; Corsi, A.M.; Vestrini, A.; et al. Genome-wide association study of vitamin B6, vitamin B12, folate, and homocysteine blood concentrations. Am. J. Hum. Genet. 2009, 84, 477–482. [Google Scholar] [CrossRef]
- Yang, S.; Ye, Z.; Liu, M.; Zhang, Y.; Wu, Q.; Zhou, C.; Zhang, Z.; He, P.; Zhang, Y.; Li, H.; et al. Associations of different serum folate forms with indices of nonalcoholic fatty liver disease and advanced fibrosis. Obes. Res. Clin. Pract. 2023, 17, 58–65. [Google Scholar] [CrossRef]
- Wei, Y.; Xu, B.; He, Q.; Chen, P.; Zhang, Q.; Zhang, X.; Yuan, H.; Duan, Y.; Wang, Z.; Zhou, Z.; et al. Serum total folate, 5-methyltetrahydrofolate and vitamin B12 concentrations on incident risk of lung cancer. Int. J. Cancer 2023, 152, 1095–1106. [Google Scholar] [CrossRef]
- Golbahar, J.; Mostafavi, E. Association between low red blood cell 5-methyltetrahydrofolate and hyperhomocysteinaemia with hypertension: A cross-sectional study. High Blood Press Cardiovasc. Prev. 2012, 19, 229–235. [Google Scholar] [CrossRef] [PubMed]
- Liu, M.; Zhang, Z.; Zhou, C.; Li, Q.; He, P.; Zhang, Y.; Li, H.; Liu, C.; Liang, M.; Wang, X.; et al. Relationship of several serum folate forms with the risk of mortality: A prospective cohort study. Clin. Nutr. 2021, 40, 4255–4262. [Google Scholar] [CrossRef] [PubMed]
- Golbahar, J.; Rezaian, G.; Fathi, Z.; Aminzadeh, M.A. Association of low red blood cell folate concentrations with coronary artery disease in Iranians: A matched case-control study. J. Vasc. Res. 2005, 42, 325–330. [Google Scholar] [CrossRef] [PubMed]
- Goracy, J.; Peregud-Pogorzelska, M.; Goracy, I.; Kaczmarczyk, M.; Kornacewicz-Jach, Z.; Naruszewicz, M.; Ciechanowicz, A. Allelic variants of genes: Angiotensin I-converting enzyme (ACE), angiotensin-II type 1 receptor (AT1R), methylenetetrahydrofolate reductase and left ventricular mass in patients with myocardial infarction. Pol. Arch. Med. Wewn. 2006, 115, 105–111. [Google Scholar]
- Moczulska, H.; Pesz, K.; Gach, A.; Borowiec, M.; Sieroszewski, P.; Sąsiadek, M.; Jakubowski, L.; Wielgoś, M. Stanowisko ekspertów Polskiego Towarzystwa Genetyki Człowieka i Polskiego Towarzystwa Ginekologów i Położników w sprawie zlecania i interpretacji wyników badań pod kątem wariantów genetycznych w genie, M.T.H.F.R. Ginekol. Perinatol. Prakt. 2017, 2, 234–238. [Google Scholar]
- Chen, P.; Tang, L.; Song, Y.; Wang, B.; Qin, X.; Zhang, N.; Wei, Y.; Xu, X.; Zhou, Z.; He, Q.; et al. Association of folic acid dosage with circulating unmetabolized folic acid in Chinese adults with H-type hypertension: A multicenter, double-blind, randomized controlled trial. Front. Nutr. 2023, 10, 1191610. [Google Scholar] [CrossRef]
- Lange, H.; Suryapranata, H.; De Luca, G.; Börner, C.; Dille, J.; Kallmayer, K.; Pasalary, N.; Scherer, E.; Dambrink, J.-H.E. Folate therapy and in-stent restenosis after coronary stenting. N. Engl. J. Med. 2004, 350, 2673–2681. [Google Scholar] [CrossRef] [PubMed]
- Centeno Tablante, E.; Pachon, H.; Guetterman, H.M.; Finkelstein, J.L. Fortification of wheat and maize flour with folic acid for population health outcomes. Cochrane Database Syst. Rev. 2019, 7, CD012150. [Google Scholar] [CrossRef] [PubMed]
| Parameters | CAD+ (n = 34) | CAD− (n = 14) | p-Value |
|---|---|---|---|
| Sex (% males) | 31/34 (91.2%) | 11/14 (78.6%) | 0.339 |
| Mean age (years) | 52 (47–58) | 59 (56–61) | 0.010 # |
| Mean age at CAD diagnosis | 47.1 + 5.6 | N/A | |
| Total cholesterol (mg/dL) | 195.5 + 48.2 | 179.3 + 39.9 | 0.366 |
| HDL Cholesterol (mg/dL) | 47.1 + 12.1 | 56.2 + 19.4 | 0.295 |
| LDL Cholesterol (mg/dL) | 116.8 + 38.8 | 101.7 + 31.9 | 0.096 |
| Triglycerides (mg/dL) | 148 (93–206) | 81 (71–127) | 0.015 # |
| non-HDL Cholesterol (mg/dL) | 148.4 + 46.8 | 123.3 + 29.6 | 0.136 |
| LVEF < 50% (TTE) | 15/33 (45.5%) | 4/11 (36.4%) | 0.731 |
| Hypertension | 20/34 (58.8%) | 4/14 (28.6%) | 0.111 |
| Obesity (BMI > 30 kg/m2) | 11/34 (32.4%) | 3/14 (21.4%) | 0.510 |
| Dyslipidemia | 19/34 (55.9%) | 4/14 (28.6%) | 0.193 |
| Diabetes | 5/34 (14.7%) | 2/14 (14.3%) | 0.998 |
| Family history (+) | 8/34 (23.5%) | 1/14 (7.1%) | 0.410 |
| Nicotinism | 19/34 (55.9%) | 2/14 (14.3%) | 0.020 |
| Past ACS | 18/34 (52.9%) | N/A | |
| Past PCI | 30/34 (88.2%) | N/A | |
| Past CABG | 7/34 (20.6%) | N/A | |
| ASA | 33/33 (97.1%) | 1/13 (7.7%) | <0.0001 |
| DAPT | 29/33 (87.9%) | 0/13 (0.0%) | <0.0001 |
| ACEI/ARB | 28/33 (84.8%) | 8/13 (61.5%) | 0.117 |
| β-blockers | 31/33 (93.9%) | 7/13 (53.8%) | 0.004 |
| Ca-blockers | 1/33 (2.9%) | 3/13 (21.4%) | 0.062 |
| Diuretics | 6/33 (18.2%) | 3/13 (23.1%) | 0.698 |
| Statins | 32/33 (97.0%) | 1/13 (7.7%) | <0.0001 |
| Genotype/Allele | CAD+ (n = 34) | CAD− (n = 14) | Crude OR (95% CI) | p | Adjusted OR (95% CI) * | p | Adjusted OR (95% CI) # | p | ||
|---|---|---|---|---|---|---|---|---|---|---|
| Number | Frequency | Number | Frequency | |||||||
| MTHFR c.665C>T | ||||||||||
| c.[665C=];[665C=] | 11 | 0.324 | 3 | 0.214 | 1.754 (0.405–7.588) & 1.844 (0.375–9.065) | 0.452 0.451 | 2.001 (0.370–10.823) & 2.149 (0.328–14.080) | 0.420 0.425 | 2.144 (0.385–11.929) & 2.748 (0.306–24.668) | 0.384 0.367 |
| c.[665C>T];[665C=] | 9 | 0.265 | 3 | 0.214 | 1.320 (0.298–5.838) & 1.322 (0.265–6.598) | 0.714 0.734 | 1.434 (0.267–7.697) & 1.397 (0.195–10.016) | 0.674 0.740 | 1.347 (0.246–7.378) & 1.536 (0.158–14.933) | 0.731 0.712 |
| c.[665C>T];[665C>T] | 14 | 0.411 | 8 | 0.571 | 0.525 (0.149–1.850) & 0.507 (0.133–1.933) | 0.316 0.320 | 0.462 (0.111–1.917) & 0.412 (0.080–2.118) | 0.288 0.288 | 0.466 (0.112–1.943) & 0.297 (0.040–2.190) | 0.294 0.234 |
| χ2 = 0.974; p = 0.324 | ||||||||||
| C | 31 | 0.456 | 9 | 0.321 | 1.769 (0.701–4.463) & 1.872 (0.689–5.086) | 0.227 0.219 | 1.994 (0.698–5.697) & 2.097 (0.665–6.610) | 0.198 0.206 | 2.039 (0.707–5.877) & 2.312 (0.635–8.417) | 0.187 0.204 |
| T | 37 | 0.544 | 19 | 0.679 | 0.565 (0.224–1.427) & 0.540 (0.196–1.484) | 0.227 0.232 | 0.502 (0.176–1.433) & 0.483 (0.158–1.471) | 0.198 0.200 | 0.490 (0.170–1.414) & 0.445 (0.132–1.503) | 0.187 0.192 |
| MTHFR c.1286A>C | ||||||||||
| c.[1286A=];[1286A=] | 3 | 0.088 | 3 | 0.214 | 0.355 (0.062–2.025) & 0.342 (0.050–2.328) | 0.244 0.273 | 0.460 (0.062–3.411) & 0.431 (0.039–4.729) | 0.448 0.491 | 0.491 (0.067–3.602) 0.338 (0.009–1.287 | 0.484 0.559 |
| c.[1286A>C];[1286A=] | 12 | 0.353 | 9 | 0.643 | 0.303 (0.083–1.112) & 0.287 (0.072–1.150) | 0.072 0.078 | 0.081 (0.011–0.576) & 0.052 (0.005–0.580) | 0.012 0.016 | 0.073 (0.010–0.548) & 0.039 (0.002–0.687) | 0.011 0.027 |
| c.[1286A>C];[1286A>C] | 19 | 0.559 | 2 | 0.143 | 7.600 (1.470–39.274) & 7.355 (1.322–40.930) | 0.016 0.023 | 24.652 (2.024–257.429) & 25.633 (2.222–295.696) | 0.007 0.009 | 25.883 (2.430–275.707) 40.327 (1.860–874.464) | 0.007 0.019 |
| χ2 = 5.986; p < 0.015 | ||||||||||
| A | 18 | 0.265 | 15 | 0.536 | 0.312 (0.125–0.781) & 0.303 (0.117–0.784) | 0.013 0.014 | 0.238 (0.082—0.691) & 0.218 (0.062–0.772) | 0.008 0.018 | 0.239 (0.082–0.700) & 0.209 (0.055–0.793) | 0.009 0.021 |
| C | 50 | 0.735 | 13 | 0.464 | 3.205 (1.280–8.023) & 3.354 (1.319–8.527) | 0.013 0.011 | 4.207 (1.447–12.229) & 4.437 (1.356–14.523) | 0.008 0.014 | 4.176 (1.429–12.202 & 4.930 (1.403–17.321) | 0.009 0.009 |
| Genotype | n | Folic Acid | p | 5-MTHF | p |
|---|---|---|---|---|---|
| MTHFR c.665C>T | |||||
| c.[665C=];[665C=] | 14 | 1.95 (1.625–2.075) #,& | 3.65 (3.350–3.875) #,& | ||
| c.[665C>T];[665C=] | 12 | 1.90 (1.700–2.125) & | 0.414 & | 3.80 (3.475–3.925) & | 0.357 & |
| c.[665C>T];[665C>T] | 22 | 1.90 (1.825–2.100) # | 0.364 # | 3.75 (3.525–3.900) # | 0.347 # |
| MTHFR c.1286A>C | |||||
| c.[1286A=];[1286A=] | 6 | 1.80 (1.650–2.025) #,& | 3.70 (3.700–3.775) #,& | ||
| c.[1286A>C];[1286A=] | 21 | 1.90 (1.600–2.100) & | 0.505 & | 3.70 (3.000–3.900) & | 0.448 & |
| c.[1286A>C];[1286A>C] | 21 | 2.00 (1.900–2.100) # | 0.181 # | 3.80 (3.500–3.900) # | 0.428 # |
| Genotype | Folic Acid | 5-MTHF | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| CAD+ | n | CAD− | n | p | CAD+ | n | CAD− | n | p | |
| MTHFR c.665C>T | ||||||||||
| c.[665C=];[665C=] | 1.9 (1.6–2.05) | 11 | 2.0 (1.8–3.65) | 3 | 0.316 | 3.5 (3.1–3.8) | 11 | 3.9 (3.8–4.15) | 3 | 0.079 |
| c.[665C>T];[665C=] | 2.0 (1.7–2.5) | 9 | 1.8 (1.55–1.9) | 3 | 0.205 | 3.9 (3.6–3.9) | 9 | 3.7 (2.8–3.9) | 3 | 0.391 |
| c.[665C>T];[665C>T] | 1.9 (1.825–2.1) | 14 | 1.95 (1.75–2.1) | 8 | 0.454 | 3.7 (3.525–3.9) | 14 | 3.8 (3.525–3.9) | 8 | 0.427 |
| MTHFR c.1286A>C | ||||||||||
| c.[1286A=];[1286A=] | 1.8 (1.55–4.5) | 3 | 1.8 (1.7–1.95) | 3 | 0.4 | 3.7 (3.1–4.25) | 3 | 3.7 (3.7–3.75) | 3 | 0.5 |
| c.[1286A>C];[1286A=] | 1.8 (1.675–2.05) | 12 | 2.0 (1.3–2.1) | 9 | 0.357 | 3.65 (3.2–3.9) | 12 | 3.9 (3.0–4.0) | 9 | 0.269 |
| c.[1286A>C];[1286A>C] | 2.0 (1.85–2.1) | 19 | 1.95 (1.925–1.975) | 2 | 0.433 | 3.7 (3.5–3.9) | 19 | 3.85 (3.825–3.875) | 2 | 0.286 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Pietruszyńska-Reszetarska, A.; Pietruszyński, R.; Majsterek, I.; Popławski, T.; Skrzypek, M.; Kolesińska, B.; Waśko, J.; Kapusta, J.; Watała, C.; Irzmański, R. Coronary Artery Disease Is Related to Methylation Disorders Caused by the c.1286A>C MTHFR Polymorphism and to Low Serum 5-MTHF and Folic Acid Concentrations—Preliminary Results. Reports 2024, 7, 6. https://doi.org/10.3390/reports7010006
Pietruszyńska-Reszetarska A, Pietruszyński R, Majsterek I, Popławski T, Skrzypek M, Kolesińska B, Waśko J, Kapusta J, Watała C, Irzmański R. Coronary Artery Disease Is Related to Methylation Disorders Caused by the c.1286A>C MTHFR Polymorphism and to Low Serum 5-MTHF and Folic Acid Concentrations—Preliminary Results. Reports. 2024; 7(1):6. https://doi.org/10.3390/reports7010006
Chicago/Turabian StylePietruszyńska-Reszetarska, Agnieszka, Robert Pietruszyński, Ireneusz Majsterek, Tomasz Popławski, Maciej Skrzypek, Beata Kolesińska, Joanna Waśko, Joanna Kapusta, Cezary Watała, and Robert Irzmański. 2024. "Coronary Artery Disease Is Related to Methylation Disorders Caused by the c.1286A>C MTHFR Polymorphism and to Low Serum 5-MTHF and Folic Acid Concentrations—Preliminary Results" Reports 7, no. 1: 6. https://doi.org/10.3390/reports7010006
APA StylePietruszyńska-Reszetarska, A., Pietruszyński, R., Majsterek, I., Popławski, T., Skrzypek, M., Kolesińska, B., Waśko, J., Kapusta, J., Watała, C., & Irzmański, R. (2024). Coronary Artery Disease Is Related to Methylation Disorders Caused by the c.1286A>C MTHFR Polymorphism and to Low Serum 5-MTHF and Folic Acid Concentrations—Preliminary Results. Reports, 7(1), 6. https://doi.org/10.3390/reports7010006







