Randomized Feature and Bootstrapped Naive Bayes Classification
Abstract
1. Introduction
2. Related Theory
2.1. Gaussian Naive Bayes
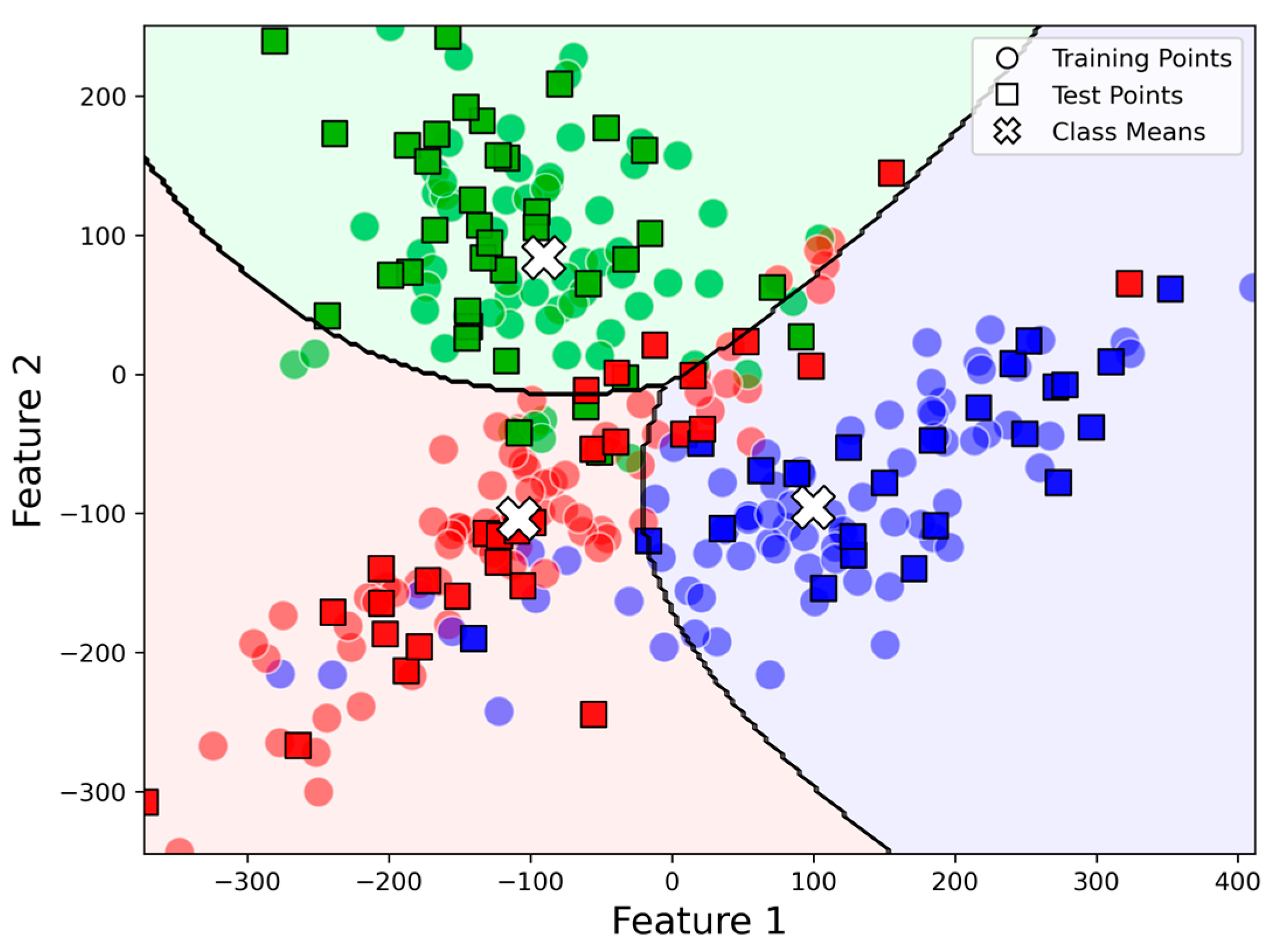
2.2. Decision Boundary
2.3. Parameter Estimation and Implementation
- Prior Probability: The proportion of training samples in class is used:where is the number of training samples in class and is the total size of the training set.
- Conditional Probabilities For continuous features, the Gaussian likelihood is parameterized using the estimated means and variances .
3. Proposed Methodology
3.1. Data Partitioning and Randomized Feature Selection
3.2. Bootstrap Sample Generation for Training
3.3. Gaussian Naive Bayes Training
3.4. Classification and Prediction Aggregation
3.5. Variance Reduction in RFB-NB
3.6. Cross-Validation and Model Assessment
3.7. Comparison of RFB-NB with TAN, WNB, and RF
4. Datasets
4.1. Dataset Description
4.2. Dataset Characteristics and Selection Rationale
- (1)
- Healthcare and Medical DiagnosticsThese datasets involve predicting health-related outcomes using clinical and physiological attributes.
- Breast Cancer Wisconsin [31]: Binary classification (malignant vs. benign tumors).
- Pima Indians Diabetes [32]: Binary classification (presence or absence of diabetes).
- Heart Disease [33]: Binary classification predicting the presence or absence of heart disease based on clinical parameters.
- Indian Liver Patients [34]: Binary classification (liver disease prediction).
- Hepatitis C Patients [35]: Classification of blood donors and Hepatitis C patients based on laboratory and demographic values.
- Heart Failure Clinical Records [36]: Binary classification (survival vs. death outcomes).
- (2)
- Financial and Business AnalyticsDatasets addressing customer behaviors, financial risks, and decision-making processes.
- Bank Marketing [37]: Predictive modeling of customer subscription behaviors.
- German Credit [38]: Binary classification of creditworthiness (good or bad credit).
- Telecom Churn Prediction [39]: Binary classification predicting customer churn behavior.
- Bike Sharing [40]: Ordinal classification predicting bike rental demand (categorized as low-, medium-, and high-demand days).
- (3)
- Signal Processing and Sensor-Based ClassificationDatasets involving an analysis of signals or sensor measurements for classification tasks.
- (4)
- Biological and Environmental ClassificationDatasets focused on classifying biological or agricultural samples and assessing environmental safety.
- Zoo [44]: Multi-class classification of animals into predefined categories.
- QSAR Bioconcentration Classes [45]: Classification of chemical compounds based on manually curated bioconcentration factors (BCF, fish) for QSAR modeling.
- Secondary Mushroom [46]: Binary classification (edible vs. poisonous mushrooms).
- Rice [47]: Binary classification identifying rice varieties (Cammeo vs. Osmancik).
- Seeds [48]: Multi-class classification of wheat kernel varieties based on geometrical and morphological features obtained using X-ray imaging techniques.
- (5)
- Product Quality AssessmentDatasets evaluating the quality or condition of consumer products.
4.3. Baseline Classification Methods
- (1)
- Random Forest
- (2)
- K-Nearest Neighbors
5. Results
5.1. Comparative Analysis by Domain
5.1.1. Healthcare and Medical Diagnostics
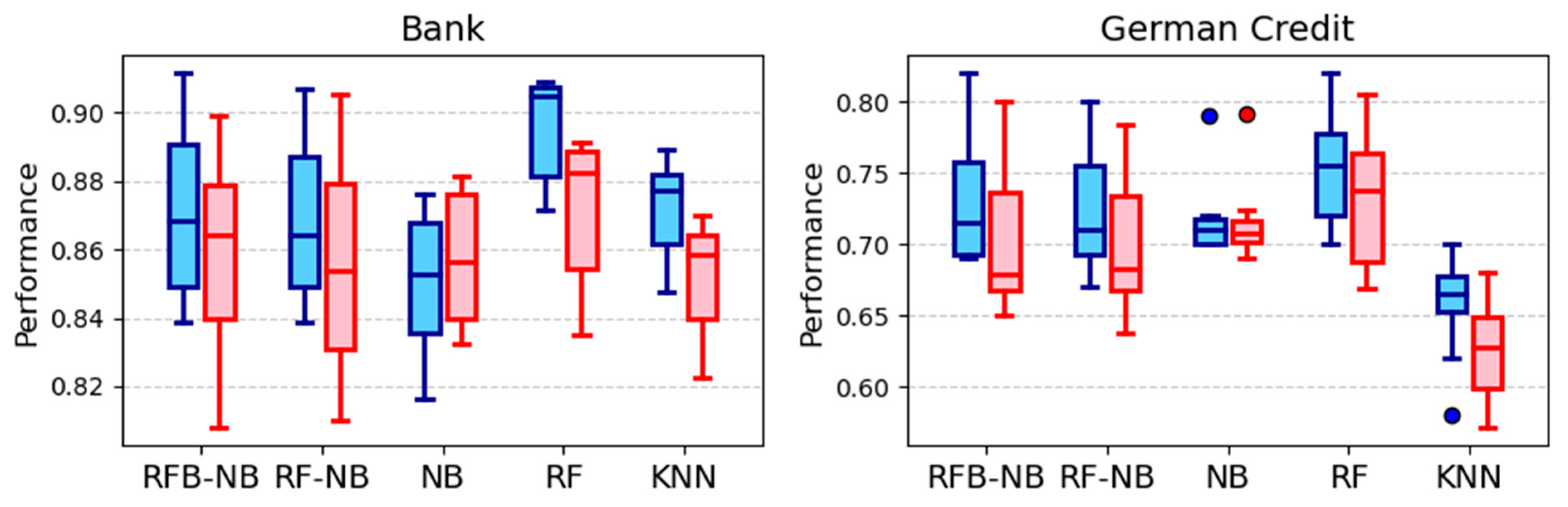
5.1.2. Financial and Business Analytics
5.1.3. Signal Processing and Sensor-Based Classification
5.1.4. Biological and Environmental Classification
5.1.5. Product Quality Assessment
5.2. Stability and Robustness Evaluation
5.3. Overall Comparative Performance
6. Conclusions
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Huang, Y.; Li, W.; Macheret, F.; Gabriel, R.A.; Ohno-Machado, L. A Tutorial on Calibration Measurements and Calibration Models for Clinical Prediction Models. J. Am. Med. Inform. Assoc. 2020, 27, 621–633. [Google Scholar] [CrossRef]
- Wang, Q.; Garrity, G.M.; Tiedje, J.M.; Cole, J.R. Naive Bayesian Classifier for Rapid Assignment of rRNA Sequences into the New Bacterial Taxonomy. Appl. Environ. Microbiol. 2007, 73, 5261–5267. [Google Scholar] [CrossRef] [PubMed]
- Jurafsky, D.; Martin, J.H. Naive Bayes and Sentiment Classification. In Speech and Language Processing, 3rd ed.; Prentice Hall: Upper Saddle River, NJ, USA, 2019. [Google Scholar]
- Zhang, H. The Optimality of Naive Bayes. In Proceedings of the 17th International Florida Artificial Intelligence Research Society Conference, Menlo Park, CA, USA, 12–14 May 2004; pp. 562–567. [Google Scholar]
- Domingos, P.; Pazzani, M. On the Optimality of the Simple Bayesian Classifier under Zero-One Loss. Mach. Learn. 1997, 29, 103–130. [Google Scholar] [CrossRef]
- Rennie, J.D.M.; Shih, L.; Teevan, J.; Karger, D.R. Tackling the Poor Assumptions of Naive Bayes Text Classifiers. In Proceedings of the Twentieth International Conference on Machine Learning (ICML-2003), Washington, DC, USA, 21–24 August 2003; Available online: https://api.semanticscholar.org/CorpusID:13606541 (accessed on 10 December 2024).
- Rish, I. An Empirical Study of the Naive Bayes Classifier. In Proceedings of the IJCAI 2001 Workshop on Empirical Methods in Artificial Intelligence, Seattle, WA, USA, 4 August 2001; pp. 41–46. [Google Scholar]
- Ratanamahatana, C.A.; Gunopulos, D. Feature Selection for the Naive Bayesian Classifier Using Decision Trees. Appl. Artif. Intell. 2003, 17, 475–487. [Google Scholar] [CrossRef]
- Askari, A.; d’Aspremont, A.; El Ghaoui, L. Naive Feature Selection: Sparsity in Naive Bayes. In Proceedings of the 23rd International Conference on Artificial Intelligence and Statistics, Online, 26–28 August 2020. [Google Scholar]
- Kohavi, R.; John, G.H. Wrappers for Feature Subset Selection. Artif. Intell. 1997, 97, 273–324. [Google Scholar] [CrossRef]
- Jiang, L.; Zhang, H.; Cai, Z.; Su, J. Learning Tree Augmented Naive Bayes for Ranking. In Proceedings of the 10th International Conference on Database Systems for Advanced Applications, Beijing, China, 17–20 April 2005; Lecture Notes in Computer Science; Springer: Berlin/Heidelberg, Germany, 2005; Volume 3453, pp. 688–698. [Google Scholar] [CrossRef]
- Hall, M.A. Correlation-Based Feature Selection for Machine Learning. Ph.D. Thesis, University of Waikato, Hamilton, New Zealand, 1999. [Google Scholar]
- May, S.S.; Isafiade, O.E.; Ajayi, O.O. An Enhanced Naïve Bayes Model for Crime Prediction Using Recursive Feature Elimination. In Proceedings of the 2021 Conference on Information Communications Technology and Society (ICTAS), Durban, South Africa, 10–11 March 2021. [Google Scholar]
- Pujianto, U.; Wibowo, A.; Widhiastuti, R.; Asma, D.A.I. Comparison of Naive Bayes and Random Forests Classifier in the Classification of News Article Popularity as Learning Material. J. Phys. Conf. Ser. 2021, 1808, 012028. [Google Scholar] [CrossRef]
- Aridas, C.K.; Kotsiantis, S.B.; Vrahatis, M.N. Increasing Diversity in Random Forests Using Naive Bayes. In Machine Learning and Knowledge Discovery in Databases; Springer: Cham, Switzerland, 2016; pp. 75–86. [Google Scholar] [CrossRef]
- Efron, B.; Tibshirani, R.J. An Introduction to the Bootstrap; Chapman and Hall/CRC: Boca Raton, FL, USA, 1994. [Google Scholar]
- Breiman, L. Random Forests. Mach. Learn. 2001, 45, 5–32. [Google Scholar] [CrossRef]
- Friedman, J.; Hastie, T.; Tibshirani, R. The Elements of Statistical Learning, 2nd ed.; Springer: New York, NY, USA, 2009. [Google Scholar]
- Liaw, A.; Wiener, M. Classification and Regression by randomForest. R News 2002, 2, 18–22. [Google Scholar]
- Zhang, H.; Sheng, S. Learning Weighted Naive Bayes with Accurate Ranking. In Proceedings of the 4th IEEE International Conference on Data Mining, Brighton, UK, 1–4 November 2004. [Google Scholar]
- Srisuradetchai, P.; Suksrikran, K. Random Kernel K-Nearest Neighbors Regression. Front. Big Data 2024, 7, 1402384. [Google Scholar] [CrossRef]
- Panichkitkosolkul, W.; Srisuradetchai, P. Bootstrap Confidence Intervals for the Parameter of Zero-Truncated Poisson-Ishita Distribution. Thail. Stat. 2022, 20, 918–927. Available online: https://ph02.tci-thaijo.org/index.php/thaistat/article/view/247474 (accessed on 10 January 2025).
- Jiang, L.; Zhang, H.; Cai, Z. A Novel Bayes Model: Hidden Naive Bayes. IEEE Trans. Knowl. Data Eng. 2009, 21, 1361–1371. [Google Scholar] [CrossRef]
- Guyon, I.; Elisseeff, A. An Introduction to Variable and Feature Selection. J. Mach. Learn. Res. 2003, 3, 1157–1182. [Google Scholar] [CrossRef][Green Version]
- Friedman, N.; Geiger, D.; Goldszmidt, M. Bayesian Network Classifiers. Mach. Learn. 1997, 29, 131–163. [Google Scholar] [CrossRef]
- Duan, W.; Lu, X. Weighted Naive Bayesian Classifier Model Based on Information Gain. In Proceedings of the 2010 International Conference on Intelligent System Design and Engineering Application, Changsha, China, 13–14 October 2010; pp. 819–822. [Google Scholar] [CrossRef]
- Foo, L.-K.; Chua, S.-L.; Ibrahim, N. Attribute Weighted Naïve Bayes Classifier. Comput. Mater. Contin. 2021, 71, 1945–1957. [Google Scholar] [CrossRef]
- McCallum, A.; Nigam, K. A Comparison of Event Models for Naive Bayes Text Classification. In Proceedings of the AAAI Workshop on Learning for Text Categorization, Madison, WI, USA, 26–27 July 1998. [Google Scholar]
- John, G.H.; Langley, P. Estimating Continuous Distributions in Bayesian Classifiers. In Proceedings of the Eleventh Conference on Uncertainty in Artificial Intelligence, Montréal, QC, Canada, 18–20th August 1995; pp. 338–345. [Google Scholar]
- Murphy, K.P. Machine Learning: A Probabilistic Perspective; MIT Press: Cambridge, MA, USA, 2012. [Google Scholar]
- Wolberg, W.; Mangasarian, O.; Street, N.; Street, W. Breast Cancer Wisconsin (Diagnostic) [Dataset]. UCI Machine Learning Repository. 1995. Available online: https://archive.ics.uci.edu/dataset/17/breast+cancer+wisconsin+diagnostic (accessed on 15 January 2025).
- Smith, J.W.; Everhart, J.E.; Dickson, W.C.; Knowler, W.C.; Johannes, R.S. Using the ADAP Learning Algorithm to Forecast the Onset of Diabetes Mellitus. In Proceedings of the Annual Symposium on Computer Applications in Medical Care, Washington, DC, USA, 9–11 November 1988; pp. 261–265. Available online: https://www.ncbi.nlm.nih.gov/pmc/articles/PMC2245318/ (accessed on 15 January 2025).
- Dua, D.; Graff, C. Heart Disease Dataset [Dataset]. UCI Machine Learning Repository. 2019. Available online: https://archive.ics.uci.edu/dataset/145/statlog+heart (accessed on 15 January 2025).
- Ramana, B.; Venkateswarlu, N. Indian Liver Patient Dataset (ILPD) [Dataset]. UCI Machine Learning Repository. 2022. Available online: https://archive.ics.uci.edu/dataset/225/ilpd+indian+liver+patient+dataset (accessed on 15 January 2025).
- Lichtinghagen, R.; Klawonn, F.; Hoffmann, G. Hepatitis C Virus Data (HCV) [Dataset]. UCI Machine Learning Repository. 2020. Available online: https://archive.ics.uci.edu/dataset/571/hcv+data (accessed on 15 January 2025).
- Chicco, D.; Jurman, G. Heart Failure Clinical Records [Dataset]. UCI Machine Learning Repository. 2020. Available online: https://archive.ics.uci.edu/dataset/519/heart+failure+clinical+records (accessed on 15 January 2025).
- Moro, S.; Rita, P.; Cortez, P. Bank Marketing Dataset [Dataset]. UCI Machine Learning Repository. 2014. Available online: https://archive.ics.uci.edu/dataset/222/bank+marketing (accessed on 15 January 2025).
- Hofmann, H. German Credit Data [Dataset]. UCI Machine Learning Repository. 1994. Available online: https://archive.ics.uci.edu/dataset/144/statlog+german+credit+data (accessed on 15 January 2025).
- Berson, A.; Smith, S.; Thearling, K. Building Data Mining Applications for CRM; McGraw-Hill: New York, NY, USA, 2000. [Google Scholar]
- Fanaee-T, H. Bike Sharing Dataset [Dataset]. UCI Machine Learning Repository. 2013. Available online: https://archive.ics.uci.edu/dataset/275/bike+sharing+dataset (accessed on 15 January 2025).
- Sigillito, V.; Wing, S.; Hutton, L.; Baker, K. Ionosphere [Dataset]. UCI Machine Learning Repository. 1989. Available online: https://archive.ics.uci.edu/dataset/52/ionosphere (accessed on 15 January 2025).
- Breiman, L.; Stone, C. Waveform Database Generator (Version 1) [Dataset]. UCI Machine Learning Repository. 1984. Available online: https://archive.ics.uci.edu/dataset/107/waveform+database+generator+version+1 (accessed on 15 January 2025).
- Singh, A.P.; Jain, V.; Chaudhari, S.; Kraemer, F.A.; Werner, S.; Garg, V. Room Occupancy Estimation [Dataset]. UCI Machine Learning Repository. 2018. Available online: https://archive.ics.uci.edu/dataset/864/room+occupancy+estimation (accessed on 15 January 2025).
- Forsyth, R. Zoo [Dataset]. UCI Machine Learning Repository. 1990. Available online: https://archive.ics.uci.edu/dataset/111/zoo (accessed on 15 January 2025).
- UCI Machine Learning Repository. QSAR Bioconcentration Classes Dataset [Dataset]. 2019. Available online: https://archive.ics.uci.edu/dataset/510/qsar+bioconcentration+classes+dataset (accessed on 15 January 2025).
- Wagner, D.; Heider, D.; Hattab, G. Secondary Mushroom [Dataset]. UCI Machine Learning Repository. 2021. Available online: https://archive.ics.uci.edu/dataset/848/secondary+mushroom+dataset (accessed on 15 January 2025).
- Cınar, I.; Koklu, M. Rice (Cammeo and Osmancik) [Dataset]. UCI Machine Learning Repository. 2019. Available online: https://archive.ics.uci.edu/dataset/545/rice+cammeo+and+osmancik (accessed on 15 January 2025).
- Charytanowicz, M.; Niewczas, J.; Kulczycki, P.; Kowalski, P.; Lukasik, S. Seeds [Dataset]. UCI Machine Learning Repository. 2010. Available online: https://archive.ics.uci.edu/dataset/236/seeds (accessed on 15 January 2025).
- Cortez, P.; Cerdeira, A.; Almeida, F.; Matos, T.; Reis, J. Wine Quality [Dataset]. UCI Machine Learning Repository. 2009. Available online: https://archive.ics.uci.edu/dataset/186/wine+quality (accessed on 15 January 2025).
- Bohanec, M. Car Evaluation [Dataset]. UCI Machine Learning Repository. 1988. Available online: https://archive.ics.uci.edu/dataset/19/car+evaluation (accessed on 15 January 2025).
- Vergni, L.; Todisco, F. A Random Forest Machine Learning Approach for the Identification and Quantification of Erosive Events. Water 2023, 15, 2225. [Google Scholar] [CrossRef]
- Svoboda, J.; Štych, P.; Laštovička, J.; Paluba, D.; Kobliuk, N. Random Forest Classification of Land Use, Land-Use Change and Forestry (LULUCF) Using Sentinel-2 Data—A Case Study of Czechia. Remote Sens. 2022, 14, 1189. [Google Scholar] [CrossRef]
- Purwanto, A.D.; Wikantika, K.; Deliar, A.; Darmawan, S. Decision Tree and Random Forest Classification Algorithms for Mangrove Forest Mapping in Sembilang National Park, Indonesia. Remote Sens. 2023, 15, 16. [Google Scholar] [CrossRef]
- Tian, S.; Zhang, X.; Tian, J.; Sun, Q. Random Forest Classification of Wetland Landcovers from Multi-Sensor Data in the Arid Region of Xinjiang, China. Remote Sens. 2016, 8, 954. [Google Scholar] [CrossRef]
- Kamlangdee, P.; Srisuradetchai, P. Circular Bootstrap on Residuals for Interval Forecasting in K-NN Regression: A Case Study on Durian Exports. Sci. Technol. Asia 2025, 30, 79–94. Available online: https://ph02.tci-thaijo.org/index.php/SciTechAsia/article/view/255306 (accessed on 28 March 2025).
- Srisuradetchai, P.; Panichkitkosolkul, W. Using Ensemble Machine Learning Methods to Forecast Particulate Matter (PM2.5) in Bangkok, Thailand. In Multi-Disciplinary Trends in Artificial Intelligence; Surinta, O., Kam, F., Yuen, K., Eds.; Lecture Notes in Computer Science; Springer: Cham, Switzerland, 2022; Volume 13651. [Google Scholar] [CrossRef]
- Srisuradetchai, P. A Novel Interval Forecast for K-Nearest Neighbor Time Series: A Case Study of Durian Export in Thailand. IEEE Access 2024, 12, 2032–2044. [Google Scholar] [CrossRef]
- Boateng, E.Y.; Otoo, J.; Abaye, D.A. Basic Tenets of Classification Algorithms K-Nearest-Neighbor, Support Vector Machine, Random Forest and Neural Network: A Review. J. Data Anal. Inf. Process. 2020, 8, 341–357. [Google Scholar] [CrossRef]
- Bellino, G.M.; Schiaffino, L.; Battisti, M.; Guerrero, J.; Rosado-Muñoz, A. Optimization of the KNN Supervised Classification Algorithm as a Support Tool for the Implantation of Deep Brain Stimulators in Patients with Parkinson’s Disease. Entropy 2019, 21, 346. [Google Scholar] [CrossRef] [PubMed]
- Florimbi, G.; Fabelo, H.; Torti, E.; Lazcano, R.; Madroñal, D.; Ortega, S.; Salvador, R.; Leporati, F.; Danese, G.; Báez-Quevedo, A.; et al. Accelerating the K-Nearest Neighbors Filtering Algorithm to Optimize the Real-Time Classification of Human Brain Tumor in Hyperspectral Images. Sensors 2018, 18, 2314. [Google Scholar] [CrossRef] [PubMed]





| Reference | Dataset | n | p | Response | Distribution Percentage (%) |
|---|---|---|---|---|---|
| [31] | Breast Cancer Wisconsin | 569 | 30 | Malignant/benign | 37.2/62.8 |
| [32] | Pima Indians Diabetes | 768 | 8 | Diabetic/non-diabetic | 34.9/65.1 |
| [33] | Heart Disease | 270 | 13 | Presence/absence of disease | 44.4/55.6 |
| [34] | Indian Liver Patients | 583 | 10 | Liver disease/no disease | 28.5/71.5 |
| [35] | Hepatitis C Patients | 612 | 12 | Blood donor/suspect blood donor/hepatitis C/fibrosis/cirrhosis | 91.4/3.2/2.5/2.1/0.8 |
| [36] | Heart Failure Clinical Records | 299 | 12 | Death/not death | 32.1/67.9 |
| [37] | Bank Marketing | 45,211 | 16 | Subscribed/not Subscribed | 11.5/88.5 |
| [38] | German Credit | 1000 | 20 | Good/bad credit | 70/30 |
| [39] | Telecom Churn Prediction | 7043 | 21 | Churn/no churn | 53.7/46.3 |
| [40] | Bike Sharing | 17,389 | 13 | Low-/medium-/high-demand bike days | 34.5/35.6/29.9 |
| [41] | Ionosphere | 351 | 34 | Good/bad radar return | 64.1/35.9 |
| [42] | Waveform Database Generator | 5000 | 21 | Class 1/2/3 | 33.2/32.9/33.9 |
| [43] | Room Occupancy | 10,129 | 18 | Number of occupants (0 to 3) | 81.2/4.5/7.4/6.9 |
| [44] | Zoo | 101 | 17 | Animal class (7 categories) | 40.5/19.8/5.0/12.8/4.0/ 8.0/9.9 |
| [45] | QSAR Bioconcentration Classes | 779 | 9 | Bioaccumulation classes (1 to 3) | 59.1/8.1/32.8 |
| [46] | Secondary Mushroom | 61,068 | 20 | Edible/poisonous | 44.5/55.5 |
| [47] | Rice | 3810 | 7 | Cammeo/Osmancik | 42.8/57.2 |
| [48] | Seeds | 210 | 7 | Three different varieties of wheat: Kama, Rosa, and Canadian | 33.7/32.7/33.6 |
| [49] | White Wine Quality | 4898 | 11 | Score quality (3 to 9) | 0.4/3.3/29.8/44.9/17.9/ 3.6/0.1 |
| [50] | Car Evaluation | 1728 | 6 | Evaluation level (unacceptable, acceptable, good, very good) | 22.2/4.0/70.0/3.8 |
| Dataset | Top Accuracy (Mean) | Top Accuracy (Mean) | Friedman p-Value | Significant Wilcoxon Post Hoc (vs. RFB-NB) |
|---|---|---|---|---|
| Breast Cancer Wisconsin | RF (0.958) | RF (0.957) | 0.0319 | RF better (p = 0.0494) |
| Pima Indians Diabetes | RFB-NB (0.753) | NB (0.748) | 0.0357 | RFB-NB better than KNN (p = 0.0199) |
| Heart Disease | RFB-NB (0.848) | RFB-NB (0.847) | <0.0001 | RFB-NB better than KNN |
| Indian Liver Patients | RF (0.701) | RF (0.677) | <0.0001 | RF (p = 0.0020), KNN (p = 0.0078) better than RFB-NB |
| Hepatitis C Patients | RF (0.947) | RF (0.938) | 0.2842 | None |
| Heart Failure Clinical Records | NB (0.796) | NB (0.784) | 0.0005 | RFB-NB better than KNN (p = 0.0076) |
| Bank Marketing | RF (0.896) | RF (0.872) | <0.0001 | RF better (p = 0.0039), RFB-NB better than NB (p = 0.0098) |
| German Credit | RF (0.751) | RF (0.732) | 0.0001 | RFB-NB better than KNN (p = 0.0020) |
| Telecom Churn Prediction | NB (0.550) | NB (0.538) | 0.1644 | None |
| Bike Sharing | RF (0.906) | RF (0.906) | 0.0001 | RFB-NB better than KNN (p = 0.0076) |
| Ionosphere | RF (0.929) | RF (0.928) | 0.0004 | RF better (p = 0.0391), RFB-NB better than KNN (p = 0.0117) |
| Waveform Database Generator | RF (0.841) | RF (0.840) | 0.0002 | RF better (p = 0.0020) |
| Room Occupancy | KNN (0.997) | KNN (0.997) | <0.0001 | RF (p = 0.0420), KNN (p = 0.0020) better than RFB-NB |
| Zoo | RFB-NB (0.960) | RFB-NB (0.961) | 0.1518 | None |
| QSAR Bioconcentration Classes | RF (0.680) | RF (0.665) | 0.0003 | RFB-NB better than NB (p = 0.0020) and RF-NB (p = 0.0176) |
| Secondary Mushroom | RFB-NB (0.583) | NB (0.628) | 0.0114 | RFB-NB better than RF (p = 0.0117) |
| Rice | RF (0.924) | RF (0.924) | <0.0001 | RFB-NB better than KNN (p = 0.0020) |
| Seeds | RF (0.909) | RF (0.909) | 0.1749 | None |
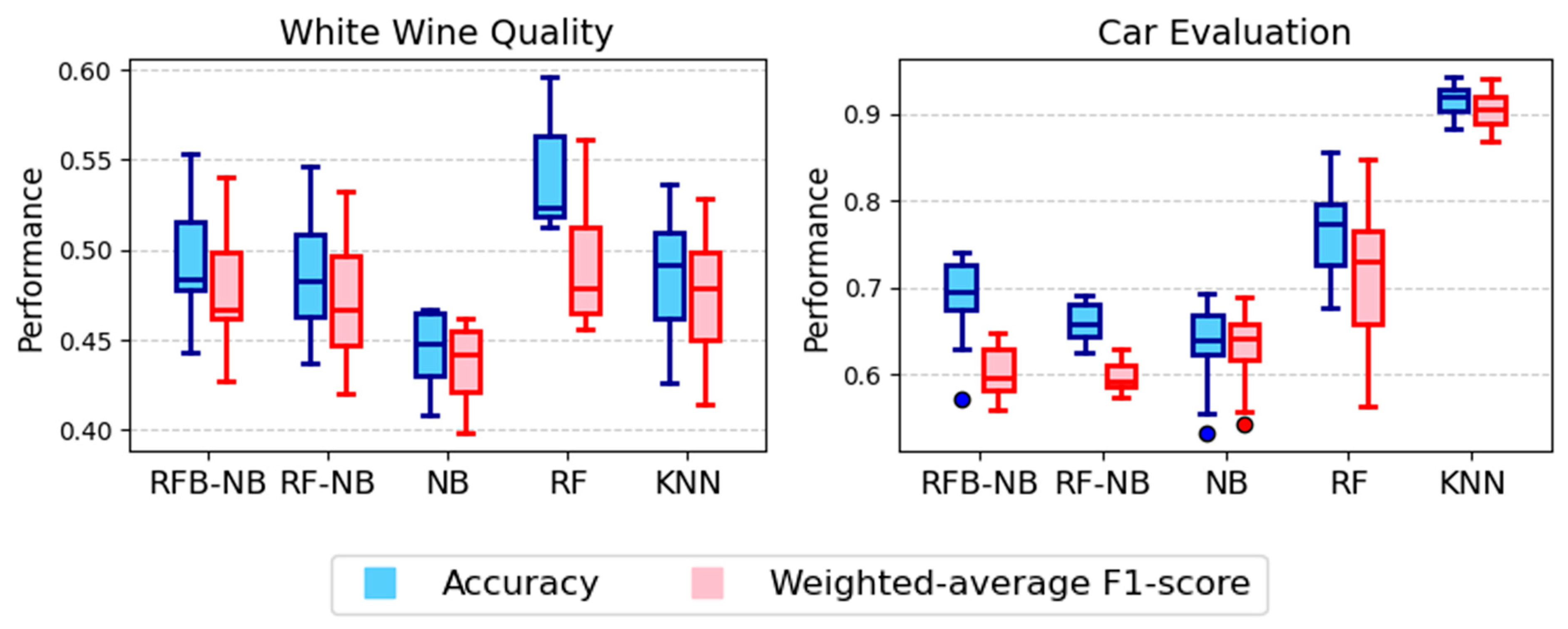
| White Wine Quality | RF (0.541) | RF (0.491) | <0.0001 | RFB-NB better than NB and RF-NB (p = 0.0020, 0.0215), RF better (p = 0.0020) |
| Car Evaluation | KNN (0.915) | KNN (0.905) | <0.0001 | KNN, RF, and NB better than RFB-NB (p = 0.0020, 0.0059, 0.0059) |
| Model | Mean Accuracy | Mean Std. Dev. | Average Rank |
|---|---|---|---|
| RF | 0.8061 | 0.0591 | 1.875 |
| RFB-NB | 0.7869 | 0.0447 | 2.375 |
| RF-NB | 0.7798 | 0.0449 | 3.000 |
| NB | 0.7672 | 0.0573 | 3.775 |
| KNN | 0.7494 | 0.0523 | 3.975 |
| Model | Mean Accuracy | Mean Std. Dev. | Average Rank |
| RF | 0.7979 | 0.0619 | 2.025 |
| RFB-NB | 0.7841 | 0.0471 | 2.600 |
| NB | 0.7758 | 0.0575 | 2.925 |
| RF-NB | 0.7726 | 0.0475 | 3.100 |
| KNN | 0.7579 | 0.0551 | 4.350 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Published by MDPI on behalf of the International Institute of Knowledge Innovation and Invention. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Phatcharathada, B.; Srisuradetchai, P. Randomized Feature and Bootstrapped Naive Bayes Classification. Appl. Syst. Innov. 2025, 8, 94. https://doi.org/10.3390/asi8040094
Phatcharathada B, Srisuradetchai P. Randomized Feature and Bootstrapped Naive Bayes Classification. Applied System Innovation. 2025; 8(4):94. https://doi.org/10.3390/asi8040094
Chicago/Turabian StylePhatcharathada, Bharameeporn, and Patchanok Srisuradetchai. 2025. "Randomized Feature and Bootstrapped Naive Bayes Classification" Applied System Innovation 8, no. 4: 94. https://doi.org/10.3390/asi8040094
APA StylePhatcharathada, B., & Srisuradetchai, P. (2025). Randomized Feature and Bootstrapped Naive Bayes Classification. Applied System Innovation, 8(4), 94. https://doi.org/10.3390/asi8040094








