1. Introduction
The wide-spread utilisation of capacity and digitising advancements, specifically the digitisation of clinical records, presents numerous information examination chances. Notwithstanding, to arrive at their maximum capacity, such investigation frameworks need to remove organised information from unstructured content reports. An expanding volume of unstructured clinical information about patients is put away electronically by clinics and medical services. Organised data is fundamental for applications, for example, reporting, reasoning, and retrieving, for instance, malignancy observations from medical reports and death certificates [
1], checking radiology reports to forestall missed fractures [
2], and clinical data retrieval [
3]. Late advancements of Natural Language Processing (NLP) and information extraction (IE) have confronted fundamental difficulties in adequately catching valuable data from this free-text resources [
4]. IE is a nontrivial interaction for extricating helpful, organised data like examples and different connections from unstructured info text.
One of the challenges is distinguishing cases of ideas that are alluded to in manners not captured inside current lexical assets and tackle uncertainty, polysemy, synonymy, and word order varieties. Moreover, the data introduced in clinical narratives are frequently unstructured, ungrammatical, and divided. Along these lines, standard NLP advances and frameworks cannot be straightforwardly applied to the clinical domain [
5].
ML-based algorithms, rule-based and existing dictionary-based methods can be utilised to identify and extract the concepts from raw text corpus in finance, medical, and various other domains [
6,
7,
8,
9,
10]. In the clinical domain, the ShARe/CLEF 2013 eHealth Evaluation Lab and the i2b2/VA challenge methodologies have been applied in shared tasks [
11,
12,
13]. The results demonstrated that ML-based algorithms are scalable and usually beats the rule-based approaches.
A critical challenge is a clinical text contains domain expert words which requires domain expert efforts to presented rule-based methods or label huge corpora as training data for supervised ML-based methods. Usually, rule-based approaches are expensive because it needs domain experts and is a challenging task itself that can create error [
14] and not adaptable or transferable to other tasks. The results of the supervised ML-based approached increases as the set of labelled data is used for training. Using crowdsourcing for labelling clinical data is not useful in the general domain; manual labelling is an expensive and labour-intensive task.
AL [
15] and semi-supervised learning [
16] are viable options in contrast to standard supervised ML methods and can reduce labelling costs. AL can prepare to accomplish an automated system with high adequacy and less labelling cost. Training an ML-based approach using a small subset of labelled data, selected randomly, leads to reduced effectiveness compared to when the model uses complete labelled data, while in AL, the aim is to reach high viability and effectiveness by training a small chunk of data.
AL is a human-in-the-loop technique with the capacity to radically decrease human inclusion contrasted with the conventional supervised ML techniques that require a massive amount of labelled data at the start.
Figure 1 presents the overall general cycle of AL for extracting information from text. It is an iterative cycle, where informative samples from raw and unstructured text documents are chosen utilising a query strategy. A human annotator then labels these samples to extricate data and construct a supervised ML-based model at every iteration. The viability of AL techniques has been shown and decisively demonstrated in numerous spaces, for example, text classification, IE, and speech recognition [
15].
Regardless of comparative findings on various tasks and domains, AL is not thoroughly investigated in biomedical tasks. Our research is based on the following research questions. RQ1: How AL can be used to reduce the labelling cost while maintaining the good quality of extracted information? RQ2: Which existing AL techniques perform well compared to other AL methods to reduce the labelling time?
RQ3: How can other ML approaches (i.e., representation learning and unsupervised learning) can produce effective information extraction while maintaining the quality and minimising the labelling effort?
Despite similar findings, the aim of our research is to provide a framework to the research community for extracting information from large amounts of unstructured biomedical documents by developing an AL-based framework that extracts high-quality concepts and reduces the burden of manual annotation.
2. Related Work
Expanding volumes of clinical information that can be presently digitised and put away in electronic medical records makes the extraction of information from clinical text progressively basic, especially in the region of NLP and ML. While numerous clinical assets and advances are presently accessible to encourage the preparing of clinical information, clinical data extraction remains challenging.
Recent studies focus on IE from biomedical literature, for example, books and scientific articles [
17,
18,
19,
20] and the subsequent gathering centres around IE from free-text clinical narratives delivered by clinical staff, for example, radiology and pathology reports or release synopses. Besides, other studies represent a more troublesome errand on account of the unstructured idea of the free content and the simple language used [
6]. IE is a significant essential advance in extricating essential data from clinical records. The fundamental challenge is to create cost-productive methodologies that help automatic idea extraction from clinical free-text assets while guaranteeing the extracted ideas’ high quality. Automatic handling of such volumes of information could incredibly profit clinical information systems.
2.1. Information Extraction from Biomedical Corpus
Extracting information from biomedical documents involves capturing words of natural language from raw and unstructured document which express the significant information within a given domain [
14]. NLP-based techniques cannot be directly used to extraction information from biomedical corpus due to its ubiquitous, raw, and unstructured nature. Current methods can be divided into following techniques.
2.1.1. Dictionary-Based Methods
Dictionary-based methods consist of matching a provided list of terms in a text and use patterns to extract structures like entities and text strings from a pre-defined dictionary. A large number of domain specific dictionaries are largely available which can be used to extract biomedical information. These include SNOMED CT [
21] and UMLS [
22]. Bashyam et al. [
23] presented and demonstrated a lexical lookup approach for radiology reports to detect UMLS concepts. They showed that their method is 7 times faster than MetaMap in identifying the same concepts.
Dictionary-based techniques can be helpful in extracting information from free text with the help of dictionaries, and they can also normalise entities and be useful for both the syntactical and semantic level of information by associating the entities with terms in the dictionaries. These dictionary-based methods are useful but suffer from coverage issues, which makes their use limited in this domain.
2.1.2. Rule-Based Methods
Rule-based methods have been generally evolved to extract entities in the biomedical domain [
24]. Rule-based methods contain manually created rules to extract biomedical information from the corpus. Various techniques are used to define these rules, which are used to capture patterns within natural language [
25].
Current databases have coverage issues and do not cover recently discovered elements; some helpful objective substances and biomedical-related data covered up in non-important settings may be missed and not extracted utilising a word reference-based methodology. Hamon and Graber [
26] presented a rule-based method to extract biomedical information using existing terms, rules, and shallow parsing methods. Mack et al. [
27] proposed BioTeks, a rule-based approach to capture biomedical information from biomedical corpus. These methods are widely used in the biomedical domain; however, implementation requires domain expertise and they are not adaptable nor transferable to other domains [
28].
2.2. Machine Learning (ML)
Machine learning (ML)-based methods are presented to address the shortcomings of the abovementioned techniques by making the machine learn and improve the performance [
29]. Biomedical/clinical extraction can be classified as a labelling task sequence, referred to as a classification task in supervised learning of ML algorithms. Both support vector machines (SVMs) [
30] and conditional random fields (CRFs) [
31] are the methods mostly used in the classification for sequence labelling tasks. For another high level of sophisticated tasks, a large number and high quality of training data are required to train the models. Although a huge amount of data is available, the labelling cost is high and the task cumbersome. The AL technique is proposed to limit the required high volume of manual labelling of data. AL’s main idea is to query and label those samples that carry useful information for the learning model compared to other available samples. It can attain better performance with less-annotated training data [
15,
32].
Semi-supervised learning is another approach to address annotated corpus [
33]. It has been effectively applied to some real-world applications. An abundance of unlabelled examples is effortlessly accessible, while physically naming them is an escalated and costly errand. Self-training is a customarily utilised technique where unannotated text is annotated in an iterative interaction. The updated labelled set is utilised to retrain and refresh the fundamental classifier at every emphasis. This examination researches how to increase the learning model at every point by consolidating self-preparing into the AL process.
Representation learning refers to learning data representations to facilitate information extraction. The IE is fed into the training of the machine learning model [
34]. Mikolov et al. [
35] introduced a novel word embedding concept where words are represented in continuous vector representations of words based on their various dimensions of difference.
2.3. Natural Language Processing (NLP)
NLP is the intersection of computing science and linguistics that includes dissecting and understanding common human language from both speech and written texts. Over the years, NLP has been used in various applications such as email filtering [
36], irony and sarcasm detection [
37] document organisation [
38], sentiment and opinion mining prediction [
39,
40,
41], hate speech detection [
42,
43,
44], question answering [
45], content mining [
46], biomedical text mining [
47,
48], and many more [
8,
49,
50].
In biomedical named entity recognition (BioNER), Yao et al. [
51] initially created embeddings of words on unlabelled texts of biological topics using neural networks, going on to establish a multi-layer neural network to obtain cutting edge output. Li et al. [
52] mixed sentence vectors and twin word embeddings and utilised the BiLSTM on biomedical texts to identify domain-relevant entities. To identify drug entities, Zeng et al. [
53] developed their model, BiLSTM-CRF. CNN was utilised to get the representation of features on a character level. This was done with representations on a word level and used as data to be fed to the BiLSTM-CRF for BioNER. In biomedical literature, many words can cause information redundancy whilst neural network models are trained for feature capture, preventing critical information from being obtained. This may cause the crucial areas not to focus on the BioNER models, and loss of information could occur. It is a salient focus to make models of neural networks attentive to areas of importance. In machine translation, Bahdanau et al. [
54] suggest the attention focusing mechanism. Taking the decoder model into account, the focus can be made on the initial text’s essential bits as it is decoded, reducing information loss. An attention-based BiLSTM-CRF model is used by Luo et al. [
55] for BioNER on a document level. They optimise the tagging inconsistency problem by using, between various sentences, mechanisms that are attention-focused. The best results are obtained on CHEMDNER and CDR corpora using this approach.
Several other works have investigated the benefit of contextual models in biomedical and clinical areas. Several researchers trained ELMo on biomedical corpora and presented BioELMo and found that BioELMo beats ELMo on BioNER tasks [
56,
57]. Along with their work, a pre-trained BioELMo model was published, enabling further clinical research. Beltagy et al. [
58] released Scientific BERT (SciBERT), where BERT was trained on the scientific texts. In non-contextual embedding, BERT has been usually superior and better than ELMo. Similarly, innovative wireless connectivity techniques could be applied to the remote execution of these techniques [
59,
60,
61,
62]
Si et al. [
63], trained the BERT on clinical notes corpora, using complex task-specific models to improve both traditional embedding and ELMo embedding i2b2 2010 and 2012 BioNER. Similarly, in another study, a new domain-specific language model, BioBERT [
64], trained a BERT model on biomedical documents from PMC abstracts and articles from PubMed that resulted in improved BioNER results. Peng et al. [
65] introduced Biomedical Language Understanding Evaluation (BLUE), a collection of resources for evaluating and analysing natural biomedical language representation models.
2.4. Active Learning (AL)
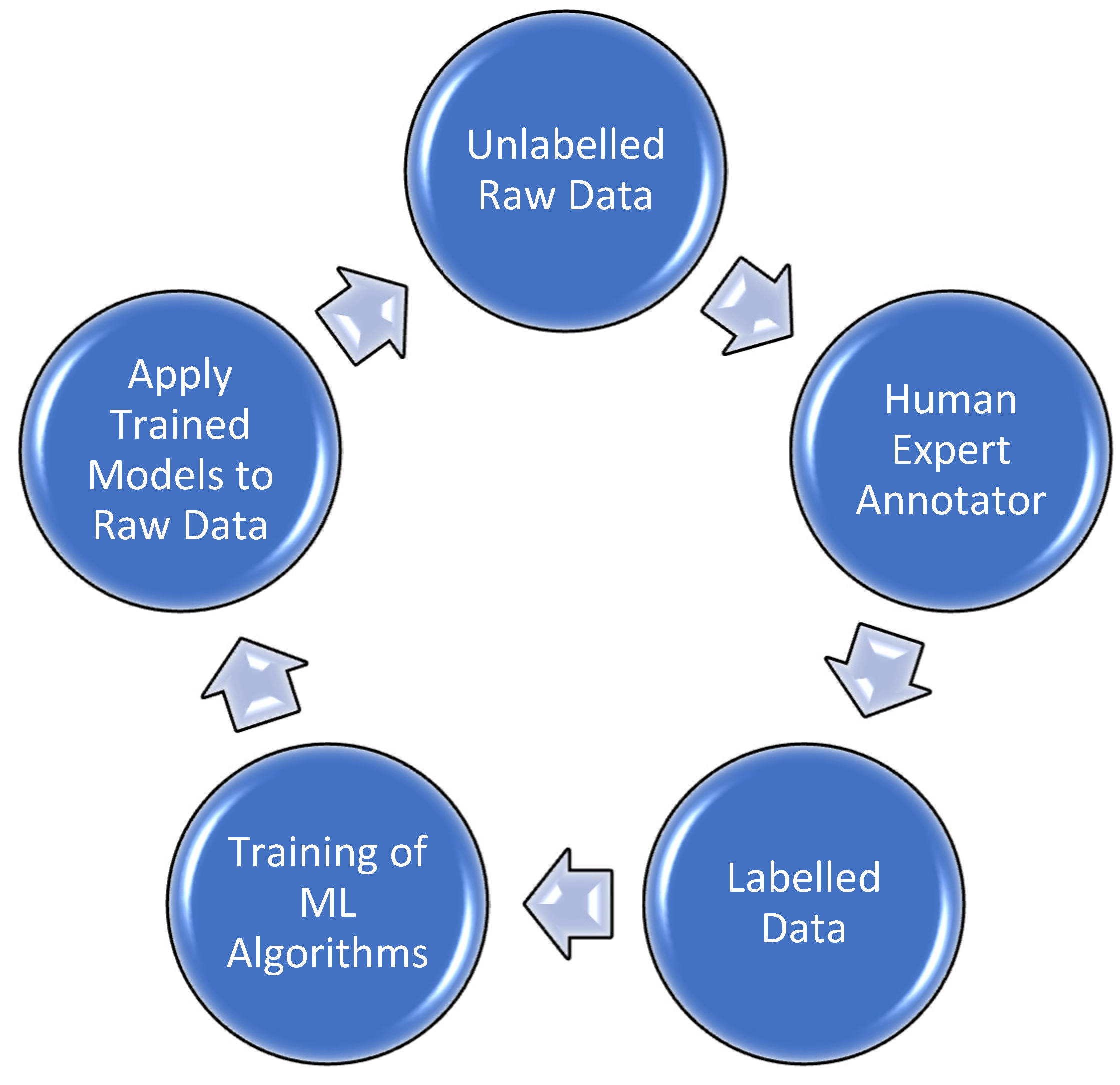
AL algorithms are beneficial in ML, especially when we have large amounts of unannotated data. AL techniques use supervised ML methods in an iterative way. A human annotator is involved in the learning process and can drastically decrease the human involvement as demonstrated in
Figure 2. Despite its strength, AL has not been fully explored for biomedical information extraction. AL’s primary goal is to maximise the model’s effectiveness by reducing the number of samples that need manual labelling. The main challenge is to find informative samples that are available to train a model, achieving the better performance and high effectiveness.
2.5. Active Learning in Clinical Domain
AL aims to reduce the costs and issues related to the manual annotation step in supervised ML methods. Decreasing the manual annotation burden becomes highly critical in the clinical domain because of qualified experts’ high costs to annotate the clinical free text. AL is used for various biomedical tasks [
66], de-identifying clinical records [
67], clinical text classification [
68], and clinical named entity recognition [
69]. Random sampling (RS), where samples are chosen randomly, is a commonly used AL technique.
Rosales et al. [
70] presented an AL method to identify biomedical information to two groups. Their method outperformed the traditional methods. Chen et al. [
66] presented a sampling technique established on the changes appearing in different learning models during AL. Another study on de-identifying of Swedish biomedical samples as a classification task was presented by Boström and Dalianis [
67]. They presented the comparison on the performance of two AL methods against RS baseline methods. Recently, Chen et al. [
69] proposed new AL query strategies that belong to uncertainty-based approaches and diversity-based approaches. Authors presented a comprehensive evaluation of current and new AL methods on biomedical tasks and found that uncertainty sample-based methods resulted in less effort being required to label the corpus as compared to diversity-based methods.
Considering the basic need of having cost-effective AL approaches for biomedical tasks, the highlighted limitations need to be addressed. Therefore, in this research, our aim is to address the cost needed for manual annotation using AL and representation learning.
3. Methodology
3.1. Dataset
In this study, we used the following datasets.
DDI extraction 2013 corpus is a collection of 792 texts selected from the DrugBank database and other 233 Medline abstracts [
71]. The drug-drug interactions, including both pharmacokinetics and pharmacodynamic interactions, were annotated by two expert pharmacists with a substantial pharmacovigilance background. In our benchmark, we use 624 train files and 191 test files to evaluate the performance and report the micro-average F1-score of the four DDI types.
ChemProt consists of 1820 PubMed abstracts with chemical-protein interactions annotated by domain experts and was used in the BioCreative VI text mining chemical-protein interactions shared task [
72]. We use the standard training, and test sets in the ChemProt shared task and evaluate the same five classes: CPR:3, CPR:4, CPR:5, CPR:6, and CPR:9.
HoC (the Hallmarks of Cancers corpus) consists of 1580 PubMed abstracts annotated with ten currently known hallmarks of cancer [
73]. Annotation was performed at the sentence level by an expert with 15+ years of experience in cancer research. We used 315 (20%) abstracts for testing and the remaining abstracts for training.
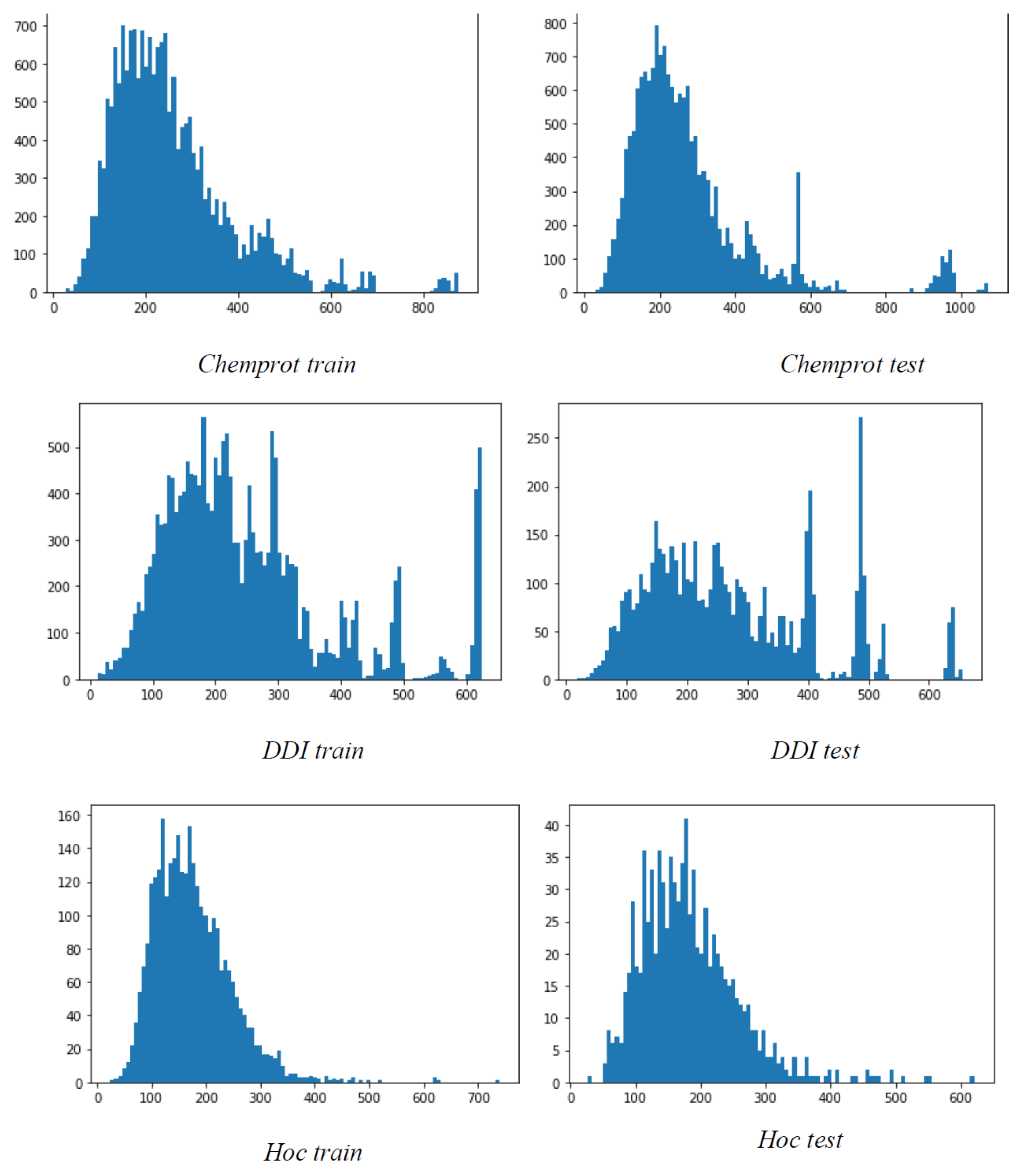
Table 1 shows the name along with the task description of the dataset used in this study. Further,
Figure 3 depicts the data analysis of the dataset used in our study.
3.2. Active Learning Query Strategies
3.2.1. Random Sampling (RS)
The key idea for random sampling of AL is that it takes a small, random portion of the entire dataset to represent the entire dataset. Each member has an equal probability. During the AL application, random sampling is quite the most straightforward algorithm compared to other query strategies. It applies the random state and shuffles to achieve the random selection of the training and testing pools.
3.2.2. Least Confidence (LC)
Least confidence is one of the methods belonging to uncertainty sampling, a query strategy that tries to determine the word’s values by calculating the real uncertainty of the word.
3.2.3. Informative Diversity and Density (IDD)
IDD is a method used to calculate the information density of an instance x. Unlike uncertainty sampling, IDD can lead us to take the structure of data into account.
3.2.4. Margin
Margin is also belonging to uncertainty sampling; unlike LC, the margin is designed to measure the difference in probability of the first and second most likely prediction.
3.2.5. Maximum Representativeness-Diversity (MRD)
Maximum representativeness diversity is a method that relies only on the similarity between samples and all other samples in unlabelled sets. The most representative is to mark various samples in the current batch and then add them to the training set. This method could prevent experts from waiting until the learning model is on the current set of tags, and then the next batch of samples selects tags using one of the above query strategies.
3.3. AL Query Strategies
There are many query strategies in AL; however, not all query strategies are invented for all situations. We pick up LC and margin because they are the most popular query strategies in other areas. We pick up RS because it is different from other algorithms as it picks up pools randomly. Then we choose IDD because IDD uses a different measure way compared with LC and margin. For the same reason, we pick up MRD to increase the variety of our query strategies schemes to get a better and reliable result for analysing.
3.4. Feature Extraction Methods
For feature extraction methods, we pick TF-IDF for feature extraction method in many areas. Then, we add FastText to compare with TF-IDF because TF-IDF only considers the frequency of a word in a document. FastText, consider more than that which can give our study result analysis some other aspect to analyse the performance. In the end, we decided to add BERT and ELMo and their extension into our study. Because BERT and ELMo are heavy methods compared to others and perform well, especially with NLP tasks in other areas than other methods. Therefore, we decided also to include this to analyse its performance with clinical datasets.
3.5. Machine Learning Methods
For ML methods, first of all, we determined to choose some the widely used method as the basic ML methods for our study is why we pick SVM, KNN, and NB; they are widely used methods in many different aspects of the dataset. Then, to make some comparison with SVM, KNN, and NB, we pick up some algorithms with different schemes compared to SVM, KNN, and NB. XGBoost and CatBoost are both gradient boosts based on decision trees. Random forest (RF) and AdaBoost are both ensemble functions. Furthermore, each of them is the most popular method in their area. Therefore, we finally pick these 7 methods as our ML methods to make our results more reliable by analysing different schemes’ performance.
4. Results and Discussion
The results (
Table 2,
Table 3,
Table 4,
Table 5,
Table 6,
Table 7,
Table 8,
Table 9,
Table 10,
Table 11,
Table 12 and
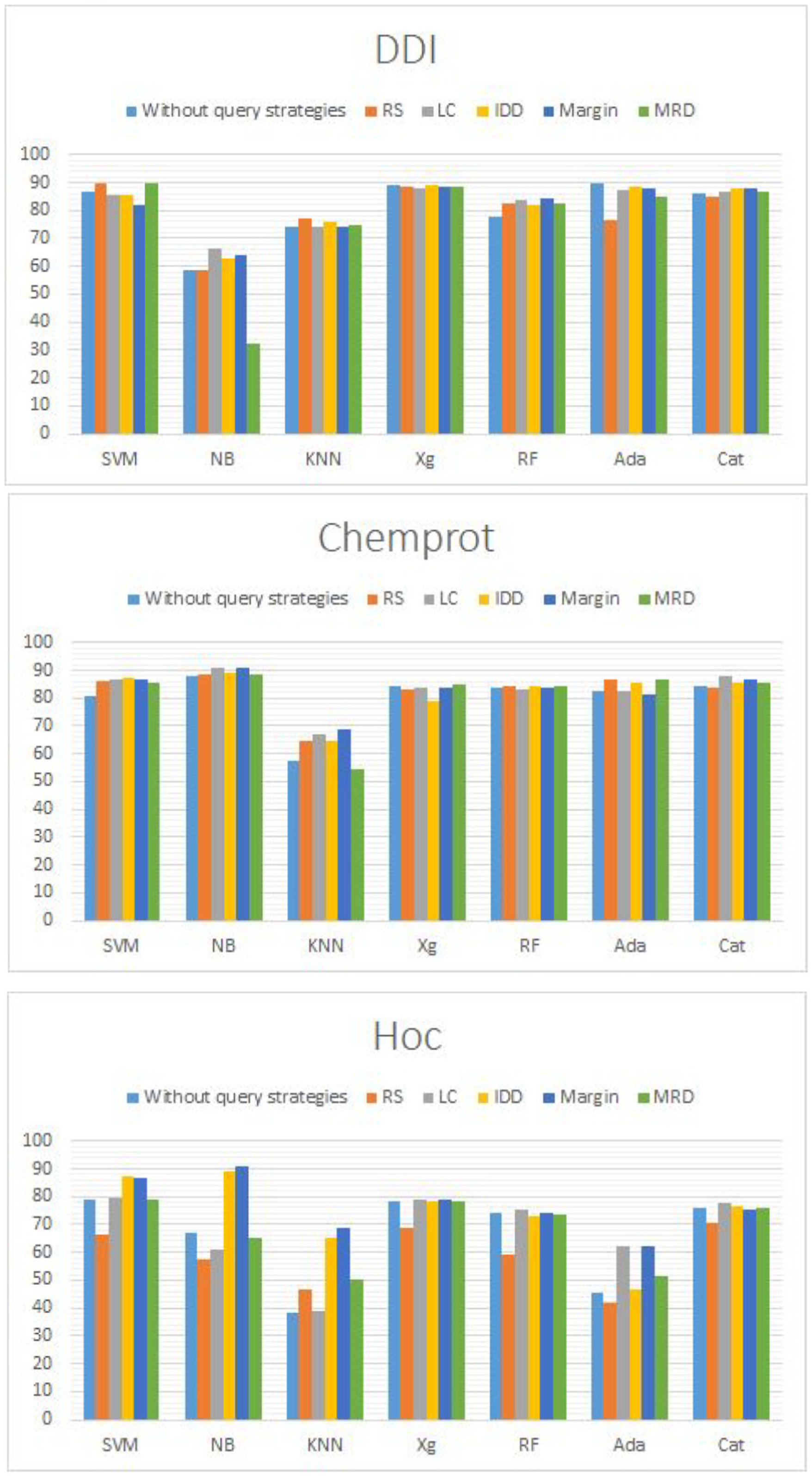
Table 13) show that the DDI dataset, which applies BERT for feature extraction, has the best performance in accuracy when we apply an SVM algorithm with an AL framework which builds based on MRD query strategies.
The result shows a table DDI dataset, which applies BERT for feature extraction and has the best performance in accuracy when applying an SVM algorithm with an AL framework that builds based on MRD query strategies.
Almost all ML methods have good performance except KNN algorithms. Further- more, in general, AL algorithms have slightly better performance than passive learning algorithms.
HoC datasets have a much clearer difference between different methods applied. In general, AL performs better than passive learning algorithms. For ML algorithms, we can see that XGBoost and CatBoost have relatively better performance than others. Furthermore, for query strategies, margins have overall better performance than others.
The following are answers to our research questions.
CatBoost performs better than others in most situations after we summarise all results tables.
In general, LC and margin have better performance than other query strategies after we summarise all result tables.
Overall, AL performance is better; therefore, AL is more recommended than passive learning.
In addition to the above results, we also notice that CatBoost always performs stably in every situation where other classifiers somehow have some bad performance. The judgement of LC and margin performance is challenging since they still have similar performance in almost all cases.
For the first results, CatBoost, as described in the methodology, is part of the gradient boost based on DT. This structure gives CatBoost the ability to get more chances to recover the errors during the implementation of the entire CatBoost structure since the later tree will fix the error that occurred by the previous tree. At the same time, the CatBoost boosting scheme is modified to be more efficient than other gradient boost algorithms, such as XGBoost, which gives CatBoost stability when changing the hyperparameters, especially with extensive data. All these advantages make CatBoost, overall, have a better ability to perform better in our study. Besides other algorithms such as AdaBoost, SVM is very sensitive with the correlation between data and data, which lead them to make more mistakes during the training and prediction than other algorithms.
For the second result, both LC and margin belong to uncertainty sampling, which calculates the uncertainty between data to measure the value of the word to decide the query order. Therefore, we can consider these two methods as a similar scheme used for AL. Then, uncertainty sampling was invented to reduce classification errors, making them more able to reduce classification errors than other query strategies, which is also what our study aims for. At the same time, IDD and MRD focused more on one word to decide the values. This could be better than uncertainty sampling with efficiently pre-computed; however, we cannot develop an efficiently pre-computed IDD and MRD algorithm to test the performance due to the tight time for implementation. This lead IDD and MRD cannot perform better than LC and Margin.
For the third result, the main reason why we can achieve better performance since with fewer data trained by using AL than using passive learning is the unbalance of the dataset. More data does not mean more accuracy for text classification. There exist iterations for the classification, even with all valuable data. In this time, insufficient data can immediately cause errors and reduce the accuracy of the classification. Therefore, the most important thing for classification is not the number of training pools. The most important thing is, can you find out which data is valuable enough to train the classifier. The AL algorithm is invented to achieve this goal by applying different query strategies. Therefore, AL can perform better than passive learning. Graphical representation of results is shown in
Figure 4.
5. Conclusions
We conducted a simulated study to compare different AL algorithms for a clinical task. Our results showed that most AL algorithms outperformed the passive learning method when we assume equal annotation cost for each sentence. However, savings of annotation by AL were reduced when the length of sentences was considered. We suggest that the effectiveness of AL for clinical NER needs to be further evaluated by developing AL enabled annotation systems and conducting user studies.
We can conclude that AL is more recommended to test a clinical dataset classification with unlabelled data than passive learning. Compared to nowadays techniques to generate the health care outcomes, it will provide at least the same accuracy as before and even with less training dataset, which will significantly decrease the cost of collecting and labelling the dataset. Also, we can see that CatBoost makes a great performance combined with the uncertainty sampling AL framework. This also gives more options to choose when people want to implement the AL to text classification. Furthermore, the domain knowledge is not so hard to understand since AL is still one part of ML; therefore, the required knowledge is only ML; once master this knowledge, the rest part is not hard to implement.