Abstract
Introduction: Urological cancers account for a significant portion of cancer diagnoses and mortality rates worldwide. The traditional treatment options of surgery and chemoradiation can have significant morbidity and become ineffective in refractory disease. The discovery of the CRISPR system has opened up new avenues for cancer research by targeting specific genes or mutations that play a role in cancer development and progression. In this review, we summarise the current state of research on CRISPR in urology and discuss its potential for improving the diagnosis and treatment of urological cancers. Methods: A comprehensive literature search was conducted on databases including PubMed, Embase, and Cochrane Library. The keywords included CRISPR and urology OR prostate OR renal OR bladder OR testicular cancer. Results: CRISPR has been used extensively in a preclinical setting to identify and target genes in prostate cancer, including AR, NANOG, ERβ, TP53, PTEN, and PD-1. Targeting PRRX2 and PTEN has also been shown to overcome enzalutamide and docetaxel resistance in vitro. In bladder cancer, CBP, p300, hTERT, lncRNA SNGH3, SMAD7e, and FOXA1 have been targeted, with HNRNPU knockout demonstrating tumour inhibition, increased apoptosis and enhanced cisplatin sensitivity both in vitro and in vivo. Renal cancer has seen CRISPR target VHL, TWIST1, PTEN, and CD70, with the first in-human clinical trial of Anti-CD70 CAR T cell therapy showing an excellent safety profile and durable oncological results. Lastly, testicular cancer modelling has utilised CRISPR to knockout FLNA, ASH2L, HMGB4, CD24, and VIRMA, with NAE1 found to be over-expressed in cisplatin-resistant germ cell colonies. Conclusions: CRISPR is a cutting-edge technology that has been used extensively in the pre-clinical setting to identify new genetic targets, enhance drug sensitivity, and inhibit cancer progression in animal models. Although CAR T cell therapy has shown promising results in RCC, CRISPR-based therapeutics are far from mainstream, with further studies needed across all urological malignancies.
1. Introduction
In 2020, Doudna and Charpentier were awarded the Nobel Prize in chemistry for genome editing using Clustered Regularly Interspaced Short Palindromic Repeats (CRISPR) [1]. In 1978, Ishino et al. originally identified CRISPR and CRISPR-associated proteins (Cas) in the Escherichia coli genome when investigating genes involved in phosphate metabolism [2]. However, it was not until 2006 that advances in genomic analyses enabled the discovery that CRISPR and Cas proteins actually work together as an adaptive immune system in prokaryotic cells against viruses and plasmids, similar to the eukaryotic ribonucleic acid (RNA) interference system [3]. In 2012, Doudna and Charpentier fused crRNA and tracrRNA into a single guide RNA (sgRNA) and simplified the Cas9 protein for use in eukaryotic cells. They showcased CRISPR-Cas9’s ability to target and cleave specific DNA sequences in mammalian cells, opening the door to precise genome editing and revolutionising the field of genetic manipulation. Since then, the CRISPR-Cas system has been adapted for use in a wide range of applications, including genetic research, pharmaceutical development, and therapeutics for urological malignancies (Figure 1).
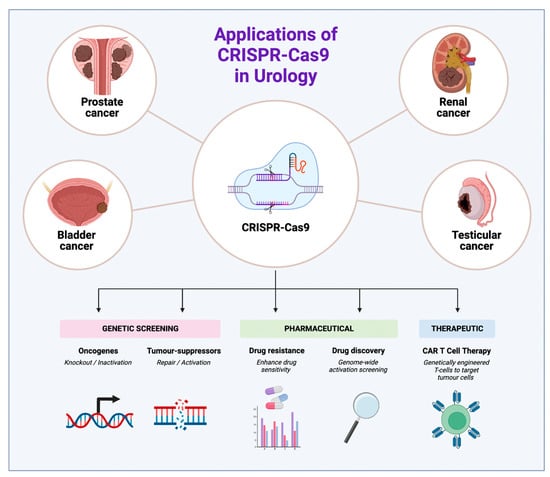
Figure 1.
Applications of CRISPR-Cas 9 in urology across four major urological cancers.
CRISPR-Cas systems have been found to act primarily as adaptive immune systems in many bacteria. They also play a role in regulating the expression of genes involved in virulence and metabolism. Three phases define the CRISPR-Cas-mediated immune response: adaptation, the production of CRISPR RNA, and targeting to destroy viral material [4]. Many CRISPR-Cas systems have been identified to date, but the CRISPR-Cas9 system is most popular for gene editing due to its ease of use, simplicity, and high efficacy [5]. The process of gene editing begins with designing an sgRNA specific to a target gene. It is then delivered, along with the Cas9 enzyme, into the cell. The sgRNA binds to the target DNA, and the Cas9 enzyme cuts the DNA at the target site near a protospacer adjacent motif (PAM) sequence. The PAM sequence is a critical component located immediately adjacent to the target DNA sequence that the CRISPR-Cas9 complex identifies and binds to. Whilst different Cas proteins have specific PAM requirements, the presence of the correct PAM sequence is necessary for the CRISPR-Cas system to function effectively [6]. After causing double-stranded DNA breaks (DSBs), the cell’s repair machinery will join the broken ends through either (A) non-homologous end joining (NHEJ) or (B) homology-directed repair (HDR) if a repair template is provided [7]. The former process is an error-prone repair mechanism that results in small insertions or deletions (indels) to occur, leading to disruptions in the target gene and commonly used to knockout genes. The latter process requires the presence of homologous DNA matrices to accurately repair the DSB and can be used to insert specific DNA sequences, allowing the directed insertion of desired modifications, such as the correction of disease-causing mutations (Figure 2).
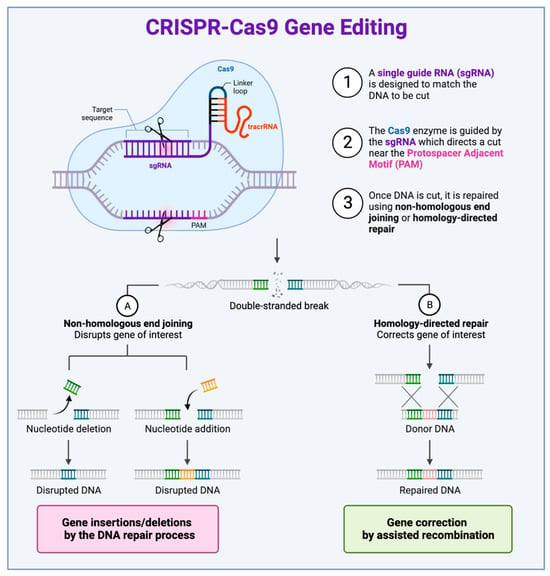
Figure 2.
The process of CRISPR-Cas9 genome editing. (1) Designing sgRNA: a single guide RNA (sgRNA) specific to the target gene is designed. This sgRNA binds to the complementary sequence of the target DNA. (2) Cas9 binding and cleavage: after designing sgRNA, it is delivered, along with the Cas9 enzyme, into the cell. The sgRNA binds to the target DNA, and the Cas9 enzyme cuts the DNA at the target site near a protospacer adjacent motif (PAM) sequence. (3) DNA repair: once the DNA is cut by the Cas9 enzyme, the cell’s repair machinery will either join the broken ends through (A) non-homologous end joining (NHEJ) or (B) repair the break via homology-directed repair (HDR) if a repair template is provided. The outcome of these repair mechanisms is the introduction of a specific mutation or insertion at the target site.
The versatility and accuracy of CRISPR-Cas have been well-recognised and bring new hope to cancer research. As cancer often involves multiple complex genetic mutations, translocations, and chromosomal losses and gains, the ability to identify and correct such mutations is an important goal in cancer treatment. In the context of this complex cancer genomic landscape, the CRISPR-Cas system has already proven to be a simple and flexible genetic tool that can effectively and rapidly identify oncogenes and tumour suppressor genes in urologic cancers [8]. By enabling the highly sensitive and specific detection of cancer-related genetic alterations, CRISPR-based diagnostic assays have the potential to identify these as biomarkers, facilitating early cancer detection and personalised treatment approaches. The ability to repair and correct these genetic events is increasingly being demonstrated in animal models and human tumour cell lines. This article provides a comprehensive review of CRISPR-Cas and its applications in four major urologic cancers, ranging from pre-clinical studies to clinical trials, with an emphasis on its advantages and directions for future uro-oncological research.
2. Methods
An online electronic database search was undertaken, using the platforms MEDLINE, Embase, and Cochrane Library. Databases were searched from their inception (MEDLINE 1946, Embase 1974, Cochrane 1996). Initial search utilised broad MeSH terms including ‘Clustered Regularly Interspaced Short Palindromic Repeats/’ and (Prostatic Neoplasms/OR Urinary Bladder Neoplasms/OR Kidney Neoplasms/OR Testicular Neoplasms/) and we extracted key terminology from reviews and a sample of potentially relevant primary data studies. The results of the literature search were downloaded to EndNote™ X9 software (Clarivate Analytics, London, UK). Exact article duplicates were removed using the duplicate tool in Endnote™ X9 software. Subsequently, a reference review of identified articles and reviews was conducted to identify any pertinent articles. The grey literature was searched via guidelines from EAU, AUA, and NICE and ongoing clinical trials through ClinicalTrials.gov, The ISRCTN registry, and the World Health Organisation International Clinical Trials Registry Platform portal. Authors of trials were contacted for preliminary or unpublished results for inclusion in the review.
3. Discussion
The CRISPR-Cas system has been extensively used for gene editing and has shown great potential in oncological research into the four major urological malignancies: prostate, bladder, kidney, and testicular cancer. A summary of CRISPR-driven research, including gene targets, study types, and overall findings, is provided in Appendix A.
3.1. Prostate Cancer
Prostate cancer (PCa) is the second-most-frequent cancer and the fifth-most-common cause of cancer death in men [9]. Androgen deprivation therapy (ADT) is the mainstay of treatment in advanced and metastatic hormone-sensitive PCa (mHSPC). However, over time, these men will inevitably progress to metastatic castration-resistant PCa (CRPC). This has led to the development of novel treatments including androgen receptor (AR)-targeted inhibitors and combination chemotherapy. However, there remains an ongoing challenge of identifying new therapeutic targets with a genetic emphasis in mCRPC. Given the move towards personalised medicine, this approach aims to avoid the ‘one size fits all’ treatment.
The early research utilising CRISPR-Cas9 included targeting the AR, which has been long-known to play a role in PCa carcinogenesis. Wei et al. (2017) designed three sgRNAs to target the AR gene in the LNCaP human cell line. This AR-sgRNA-guided CRISPR-Cas9 system was able to disrupt the AR at specific sites and inhibit the growth of androgen-sensitive PCa cells, demonstrating decreased cell proliferation due to apoptosis [10]. Similarly, Kawamura et al. (2015) used CRISPR-Cas9 to target the proteins NANOG and NANOGP8, which have been shown to be over-expressed in tissues with a higher Gleason score [11]. NANOG and NANOGP8 knockout in DU145 PCa cell lines reduced the malignant potential, including sphere formation, anchorage-independent growth, migration capability, and drug resistance [12]. This anti-tumourgenic effect was subsequently replicated in an in vivo murine model, thus demonstrating its potential as a therapeutic target in PCa.
In a recent study by Warner et al. (2020), CRISPR-Cas9 gene editing was used to demonstrate that estrogen receptor β (ERβ) may be a tumour suppressor gene for PCa [13]. Previous disagreements about the phenotype ERβ knockout mouse prompted researchers to use CRISPR-Cas9 technology to delete the entire ERβ gene from the mouse genome. They subsequently confirmed the role for ERβ in controlling growth of ventral prostate epithelium, where it opposes AR signalling. From a clinical perspective, this suggests ERβ can be targeted as a novel approach to treatment of PCa, which is an AR-driven disease. There are already ERβ agonists that have been synthesised and found to have a favourable safety profile [14,15].
Batir et al. (2019) evaluated the efficacy of CRISPR-Cas9 in repairing the dysfunctional mutant tumour protein p53 (TP53), a highly prevalent cancer-related mutation found in at least 50% of all human cancer cell lines [16]. These mutations of the TP53 gene result in impairment to, or loss of, p53 function, which is responsible for the transcriptional activation of apoptosis, cell cycle arrest, DNA repair, and senescence-related genes. By using the lentiviral delivery of sgRNA accompanied by single-stranded oligodeoxynucleotide (ssODN), the researchers were able to effectively repair the TP53 414delC gene region with an efficacy of 26%, resulting in increased apoptosis and reduced cell proliferation in the PC-3 cell line [16]. Although with a modest efficacy, this study represents a novel approach for the CRISPR-directed restoration of a mutant gene in a human PCa cell line and reveals the in vitro potential of targeting TP53.
PTEN (phosphatase and tensin homolog) is another tumour suppressor gene that has recently been inactivated using CRISPR-Cas9 by Takao et al. (2018) [17]. The knockout of PTEN from a murine prostate cell line leads to the activation of cyclin D1 expression and RAC-alpha serine/threonine-protein kinase phosphorylation; these are critical genes for cancer cell survival [17]. While knocking out a tumour suppressor gene could seem counterproductive, PTEN-CRISPR knockout has subsequently facilitated the discovery of genes such as TUBB3 and TCEAL1, which have been shown to overcome docetaxel resistance in laboratory settings [18,19]. TUBB3 knockdown enhanced PTEN expression, which, in turn, reversed docetaxel-resistance in cell lines. Interestingly, knockdown even re-sensitised docetaxel-resistant cells to cabazitaxel, indicating that TUBB3 mediates cross-resistance between both chemotherapeutic agents. The reverse was true, as PTEN knockout enhanced TUBB3 expression.
Whilst the above CRISPR knockout models have proven their value in oncological research by completely removing a target gene from the genome, this loss-of-function approach may not always reflect the true physiological effect of altering its expression levels in vivo. In contrast, a gain-of-function screen such as CRISPR activation (CRISPRa) allows for the targeted upregulation of specific genes without eliminating their function, enabling the identification of potential drug targets by determining genes that, when overexpressed, lead to therapeutic effects. Unlike traditional overexpression methods, CRISPRa can activate the expression of genes without the need for exogenous expression constructs or the potential for nonspecific effects [20]. CRISPRa provides immediate functional validation, which traditional whole-genome sequencing cannot do. More recently, Rodriguez et al. (2022) applied an in vitro genome-wide CRISPRa screen in the androgen-sensitive LNCaP cell line to identify genes that confer enzalutamide resistance. They identified the Paired Related Homeobox-2 (PRRX2) transcription factor as one of the top hits. Subsequently, they showed that PRRX2 is an oncogene in PCa and that PRRX2 overexpression mediates enzalutamide resistance, which can be overcome via BCL2 and CDK4/6 inhibition [21].
Chimeric antigen receptor (CAR) T cells, based on the genetic engineering of the patient’s own T cells for targeted tumour cell lysis, have great potential in immunotherapy for PCa [22]. A number of PCa-relevant antigens have been targeted by CAR T cell approaches, including prostate stem cell antigen (PSCA) and prostate-specific membrane antigen (PSMA). Although considerable clinical success has been seen in lymphoid malignancies, its use in solid tumours such as PCa has been limited by the hostile immunosuppressive tumour microenvironment. To overcome this, Ren et al. (2017) used CRISPR-Cas9 technology to eliminate PD-1 expression in PSCA-targeted CAR-T cells, which were subsequently infused into mice that had previously been injected with PC-3 tumour cells [23]. Whilst PSCA-CAR-T cell therapy alone reduced the tumour volume by 67%, the addition of CRISPR-mediated PD-1 knockout reduced the tumour volume by 88% compared to the control after 52 days. This CRISPR-enhanced anti-tumour activity of CAR-T cells during co-culture demonstrates the potential of combining both technologies.
3.2. Bladder Cancer
Given that close to a third of bladder cancer (BCa) presents as muscle-invasive with poor prognosis, there is an ongoing need for the development of therapeutic agents [24]. In the setting of metastatic disease, platinum-based therapy is the mainstay of treatment. Unfortunately, the overall response in clinical settings is less than 50% [25]. With the drive for personalised medicine, genetically emphasised treatments have become increasingly important.
Serving as key transcriptional co-activators, chromatin remodelling binding protein (CBP) and p300 are important in tumorigenesis, with recent genome-wide sequencing indicating that somatic mutations of these genes lead to multiple cancers, including BCa [26]. Using a CRISPR interference system, Li et al. (2019) selectively suppressed CBP and p300 expression, leading to BCa cell death in vitro; hence, this may be an attractive and novel strategy for prevention of BCa progression [27]. Similarly, engineered CRISPR-Cas13d sensing human telomerase reverse transcriptase (hTERT) selectively suppressed BCa progression in human BCa cell lines T24 and 5637 due to hTERT effects on maintaining cancer cell immortalisation, cancer growth, and metastases [28]. Unlike the more well-known Cas9 enzyme, which is commonly used in CRISPR gene editing, Cas13d is primarily known for its RNA-targeting capabilities rather than DNA editing.
Another popular target for the prevention of BCa progression is long non-coding RNA small nucleolar RNA host gene 3 (LncRNA SNGH3), which has been found to affect gene transcription, the post-transcriptional process, and, similarly, chromatin modification, with high expression in BCa cells. Hence, dysregulation and aberrant expression have been noted to promote tumorigenesis [29]. Unsurprisingly, the repression of the SNGH3 gene demonstrated reduced progression of BCa, similar to previous studies. However, the translation of such treatment targets remains to be seen.
Che et al. (2020) went one step further and utilised 38 BCa patients’ cells to determine in vivo the effects of SMAD enhancer RNA (SMAD7) knockdown in a mouse xenograft model [30]. SMAD7e is known to antagonise transforming growth factor β1 and facilitate cancer cell growth in colorectal, pancreatic, prostate, and lung cancer [31]. Hence, the authors examined SMAD7 enhancer’s significance in BCa and the effects of knockdown on the proliferation, apoptosis, migration, and invasion of BCa cells. The authors concluded that SMAD7e knockdown mediated by CRISPR-Cas13a reduced oestrogen’s cancer-promoting ability in vitro and in vivo in BCa cells and thus may represent an attractive target for treatment.
The cisplatin-based chemotherapy response in BCa is unsatisfactory due to genomic differences, pathological subtypes, and eventual drug resistance. Hence, Shi et al. (2022) attempted to elucidate the associated cisplatin resistance genes in BCa using high-throughput genome-wide CRISPR screening in human BCa cells and tumour xenograft mice models [32]. This method of CRISPR screening utilises large libraries of guide RNAs to target thousands of genes simultaneously and comprehensively explore gene functions and interactions on a genome-wide scale. Using this unbiased and systematic screening method, authors identified the Heterogenous Nuclear Ribonucleoprotein U (HNRNPU) gene and used in vitro and in vivo experiments to demonstrate HNRNPU function and depletion in cisplatin sensitivity. HNRNPU was highly expressed in tumour cells, and the subsequent knockout correlated with the inhibition of cell proliferation, invasion, and migration with apoptosis promotion in cisplatin-treated cells. Furthermore, HNRNPU knockout enhanced cisplatin sensitivity through the regulation of DNA damage repair genes. Hence, authors suggest that HNRNPU inhibition may a useful target in cisplatin-resistant BCa [32].
More recently, Neyret-Kahn et al. (2023) used CRISPR-Cas9 to establish a novel integrated epigenetic map for BCa and demonstrated a link between tumour subtypes. The group found that the long-term inactivation of FOXA1 alone through CRISPR mutation was sufficient to induce a shift from the luminal to basal subtype in luminal cells [33]. This finding is oncologically significant, as basal bladder tumours are typically of a high grade and stage, with a reduced response to chemotherapy and an overall poorer prognosis than luminal tumours. Not only did the study highlight the role of FOXA1 as a key transcription factor in subtype determination, it also induced ZBED2 overexpression, which plays a role in dampening the inflammatory response in cancer cells [33]. CRISPR has therefore demonstrated its ability to further our understanding of transcriptional regulation by identifying super-enhancer pathways providing potential targets for the treatment of aggressive disease.
3.3. Renal Cell Cancer
Patients presenting with metastatic renal cell cancer (mRCC) face a poor prognosis, with a five-year survival of less than 15%. Whilst mRCC tyrosine kinase inhibitors (TKIs) and checkpoint inhibitors (CPIs) have shown promising efficacy, treatment failure following use leads to a poor prognosis [34]. The lack of enduring interventions to combat mRCC underscores the need for better models to characterise this immunogenic malignancy and new insights into the mechanisms driving this condition.
The majority of RCCs are of the clear cell (ccRCC) subtype, much of our understanding of which is derived from studies investigating the von Hippel–Lindau (VHL) tumour suppressor gene. Schokrpur et al. (2016) used CRISPR-Cas9 to knockout VHL from RENCA mice. The RENCA murine model is a widely used xenograft model in which tumour cells from the RENCA cell line are implanted under the kidney capsule of immunocompetent mice and subsequently metastasise to sites seen in human ccRCC, including the lungs, liver, and lymph nodes, despite expressing wild-type VHL [35]. This study found that the loss of VHL led to morphologic and molecular changes indicative of the epithelial mesenchymal transition (EMT) phenotype, which, in turn, drives increased metastasis through stabilisation and therefore the oncogenic action of hypoxia-inducible factors-1α (HIF-1α) in mRCC [36]. A better understanding of this mechanism could lead to treatments to reduce the risk of progression to metastases in RCC in the first instance.
In a study by Yoshino et al. (2017), CRISPR-Cas9 was used to edit endogenous small non-coding RNAs, also known as microRNAs (miRNAs), that have been previously identified as highly upregulated in the RCC cell lines 786-O, A498, and Caki2 [37]. Upon deleting miR-210-3p from multiple RCC cell lines, the authors surprisingly found that its downregulation resulted in significantly increased cell invasiveness in vitro and promoted tumorigenesis in vivo in a mouse xenograft model. Although initially contradictory, these findings can be explained by earlier research showing miR-210-3p to be downregulated in high-grade late-stage ccRCC compared to low-grade early stage ccRCC [38]. Therefore, authors postulate that miR-210-3p expression has dual consequences in tumorigenesis and metastasis. Upregulation may be necessary to establish tumorigenesis in ccRCC. However, to then achieve EMT and metastasize, miR-210-3p needs to be downregulated in order to release the suppression of Twist-related protein 1 (TWIST1). These findings were supported by real-world patient data in the Cancer Genome Atlas database, where high TWIST1 and low miR-210-3p expression were associated with poorer overall and disease-free survival, suggesting that RCC progression is promoted by TWIST1 suppression mediated by miR-210-3p [39]. The success of these studies demonstrates that CRISPR-Cas9 gene-editing techniques can be applied to not only detect genes that cause RCC but also understand the complex mechanism by which they may progress to mRCC.
As with PCa, CRISPR-Cas9 has also been used to knockout the tumour suppressor gene PTEN from RCC cell lines. PTEN knockout was found to promote spheroid formation and decreased sensitivity to the commonly used TKIs sunitinib and sorafenib, suggesting that PTEN may be a biomarker and therapeutic target in patients with mRCC [40]. More recently, Makhov et al. (2020) used CRISPR-Cas9-based high-throughput loss-of-function screening to identify the cellular factors involved in the resistance to sunitinib [41]. In this type of screen, individual genes are targeted using CRISPR to disrupt their function, usually by creating small insertions or deletions (indels) that result in frameshift mutations and non-functional proteins. Cells with knocked-out genes are then subjected to a particular assay, and the changes in phenotype or behaviour are analysed. Farnesyltransferase was identified among the top hits contributing to the sunitinib-resistant phenotype in ccRCC. This was subsequently validated in cell and animal models of ccRCC by combining the farnesyltransferase inhibitor lonafarnib with sunitinib, and a significantly augmented anti-tumour efficacy was found both in vitro and in vivo mouse models [41]. This highlights the ability of CRISPR-Cas9 to identify and validate the druggable factors involved in resistance to targeted therapeutics.
The results from the first in-human clinical trial have recently been presented from the phase 1 COBALT-RCC trial (NCT04438083), which evaluates the effect of CTX130—an allogeneic CRISPR-Cas9 gene-edited, anti-CD70 CAR T cell therapy [42]. CD70 is a CD27 ligand that has transient expression on activated lymphocytes, and it is found to be highly expressed in ccRCC tumour samples. In this single-arm, open-label, multicentre dose-escalation and cohort-expansion study, 14 patients with unresectable/metastatic or refractory ccRCC were recruited after prior exposure to standard CPIs or VEGF-TKIs. Patients received standard lymphodepleting chemotherapy followed by CTX130 infusion, which was manufactured from healthy donor T cells that were then edited with CRISPR-Cas9 and cryopreserved for off-the-shelf use. The results showed an excellent safety profile with no dose-limiting or off-target toxicities. One patient (8%) had complete remission maintained at 18+ months, whilst nine patients (70%) had stable disease [42]. This encouraging anti-tumour activity and durable result is the first to be achieved with CAR T cell therapy in patients with solid tumours and represents a proof of concept for further CD70+ malignancies.
3.4. Testicular Cancer
In men aged 20–34, testicular cancer is the most common malignancy. It represents one of the most curable cancers when identified and treated promptly. Although rarer than the above cancers, the incidence has doubled over the past 40 years for unknown reasons, with increasing significance due to the long impact of both the disease and treatment on a younger age group [43]. In the setting of CRISPR, testicular cancer has been investigated less compared to the other common urologic cancers mentioned above, with the majority of the literature currently available focusing on the targeting of genes leading to chemotherapy resistance. However, given the younger age group, increasing life expectancy, and unexplained rising incidence, the development of screening tools and treatment efficacy is at the forefront of early diagnosis, the investigation into the rising incidence, disease monitoring, and personalised medicine.
Interestingly, filamin A (FLNA) has been found to be crucial in balancing stem cell characteristics and invasive properties in seminoma cells and possibly testicular germ cells [44]. Welter et al. (2020) investigated FLNA due to its abundance in seminoma TCam-2 cells using FLNA knockout via the CRIPSR-Cas9 system [44]. Given its importance in the mechanosensitive properties of cells, FLNA loss subjected the cell to actin cytoskeletal irregularity, leading to mechanical instability and impaired adhesive properties and ultimately disrupting migratory ability. FLNA knockout was able to reduce the invasive capacity of testicular tumorigenesis, thus demonstrating potential as a target in future therapeutics.
In an attempt to identify biomarkers to predict the efficacy of DNA-damaging drugs (genotoxins), Constantin et al. (2020) utilised the whole-genome CRISPR-Cas9 gene knockout screen to identify ASH2L [45]. As a core component of the H3K4 methyl transferase complex, which is required for bleomycin sensitivity, ASH2L knockdown rendered testicular cancer cells resistant to bleomycin, etoposide, and cisplatin. The authors also note that testicular cancer patients with ASH2L gene alterations are more likely to relapse than those without, based on the Tumour Cancer Genome Atlas. Hence, this study concluded that ASH2L levels may serve as a screening tool to predict response to genotoxins. Interestingly, the sensitivity toward ataxia-telangiectasia-mutated (ATM) and ataxia-telangiectasia- and Rad3-related (ATR) inhibitors was not affected in ASH2L knockdown cells, suggesting that its use in genotoxin-resistant patients may be more efficacious.
Several studies have similarly investigated the genetic basis of cisplatin resistance in testicular germ cell tumours (TGCT) [46,47,48,49]. Awuah et al. (2017) identified high-mobility group box protein 4 (HMGB4), a protein expressed preferentially in the testes, which blocks the excision repair of cisplatin-DNA adducts, thereby increasing the sensitivity of TGCT to cisplatin [46]. On the other hand, Skowron et al. (2022) focused on molecular and epigenetic mechanisms through the signalling molecule CD24, upregulated in several cancers, including embryonal carcinoma [47]. The authors demonstrated a bivalent role in differentiation for CD24 using a CRISPR-Cas9 deficiency model. CD24 is involved in the suppression of spermatogenesis and mesodermal/endodermal differentiation and prepares cells for ectodermal differentiation. Hence, the authors note that blocking CD24 improved cisplatin sensitivity in embryonal carcinoma and cisplatin-resistant subclones, highlighting the potential for combination therapy.
The emerging role of RNA modifications such as N6-methyladenosine (m6A) have also been studied in TGCT using CRISPR-Cas9 technology. Miranda-Gonçalves et al. (2021) found that the knockdown of the vir-like m6A methyltransferase-associated (VIRMA) gene reduced TGCT aggressiveness (with decreased cell viability, tumour cell proliferation, migration, and invasion) and increased cisplatin sensitivity both in vitro and in vivo via chorioallantoic membrane assay [48]. This study suggests that VIRMA plays an oncogenic role in TGCTs and contributes to cisplatin resistance by interfering with DNA repair mechanisms. These findings support the potential therapeutic targeting of m6A writer complex elements to combat cisplatin resistance in TGCTs. A genome-scale CRISPR-Cas9 activation screen has also been used to identify NEDD8-activating enzyme (NAE1) as a potential factor in cisplatin resistance [49]. Screening identified NAE1 overexpression in cisplatin-resistant cell colonies. This was further validated by applying MLN4924 (a novel NAE1 inhibitor) to TGCT cell lines, resulting in increased cisplatin toxicity, apoptosis, and G2/M-phase cycle arrest.
4. Conclusions
CRISPR-Cas technology has emerged as a revolutionary tool for gene editing and has opened new avenues for research and treatment of urological cancers. It has provided a more precise and efficient way to target specific genes involved in cancer development and progression. As highlighted in this review, the CRISPR-Cas system has been applied predominantly in a pre-clinical setting, with promising results seen in genome-wide screening, drug discovery, and tumour reduction in cell lines and animal models. CRISPR-based functional genomic screens continue to discover new mutations and expression patterns that may be used as highly sensitive biomarkers for early diagnosis or surveillance in the future. We are now entering an exciting new era where the first CRISPR-engineered therapeutics have become accessible to clinicians. They have the potential to be combined with chemotherapy and immunotherapy to enhance sensitivity and specificity and ultimately improve patient outcomes. Despite the progress made, several challenges remain in the clinical translation of CRISPR-based therapies in urology, including the need for more reliable delivery systems, off-target effects, and potential ethical concerns. However, the continued development of CRISPR technology and the ongoing research in urological applications provide a promising future for the use of CRISPR in the diagnosis and treatment of urological diseases.
Author Contributions
A.Y. and M.A. contributed to the design of the research and writing of the manuscript; J.I., D.W. and M.H. contributed to the analysis of the results and the revision of the manuscript; and D.B. conceived the original concept and supervised the project. All authors have read and agreed to the published version of the manuscript.
Funding
This research received no external funding.
Data Availability Statement
No new data were created or analyzed in this study. Data sharing is not applicable to this article.
Conflicts of Interest
The authors declare no conflicts of interest.
Appendix A

Table A1.
Summary of selected studies utilising CRISPR in uro-oncological research.
Table A1.
Summary of selected studies utilising CRISPR in uro-oncological research.
| Cancer Type | Target Choice | Study Type | CRISPR Effect | Findings/Mechanism | Reference |
|---|---|---|---|---|---|
| PROSTATE CANCER | AR | In vitro | Knockout of AR | Inhibited growth of androgen-sensitive LNCaP human cells | Wei et al. (2017) [10] |
| NANOG, NANOGP8 | In vitro, in vivo | Knockout of NANOG and NANOGP8 | Knockout in DU145 PCa cell lines reduced malignant potential, including sphere formation, anchorage-independent growth, migration capability, and drug resistance | Kawamura et al. (2015) [12] | |
| ERβ | In vitro | Knockout of ERβ | Confirmed the role of ERβ in controlling the growth of the ventral prostate epithelium, where it opposes AR signalling | Warner et al. (2020) [13] | |
| TP53 | In vitro | Repair of the TP53 414delC gene mutation | The repair of TP53 414delC gene mutation induces apoptosis and inhibits cell proliferation in the PC-3 cell line | Batir et al. (2019) [16] | |
| PTEN | In vitro | Knockout of PTEN | The activation of cyclin D1 expression and RAC-alpha serine/threonine-protein kinase phosphorylation, critical genes for cancer cell survival | Takao et al. (2018) [17] | |
| TUBB3, TCEAL1 | In vitro | Knockout of PTEN | Overcame docetaxel resistance | Rushworth et al. (2020), Sekino et al. (2019) [18,19] | |
| PRRX2 | In vitro | CRISPRa screening/PRRX2 over-expression | PRRX2 is an oncogene that mediates enzalutamide resistance, which can be overcome via BCL2 and CDK4/6 inhibition | Rodriguez et al. (2022) [21] | |
| PD-1 | In vitro | Knockout of PD-1 in PSCA targeted CAR-T cells | Enhanced the anti-tumour activity of CAR-T cells, reduction in tumour volume in murine model | Ren et al. (2017) [23] | |
| BLADDER CANCER | CBP, p300 | In vitro | Suppression of CBP and p300 expression | BCa cell death in vitro | Li et al. (2019) [27] |
| hTERT | In vitro | Engineered CRISPR/Cas13d sensing hTERT | hTERT maintains cancer cell immortalization and cancer growth and metastases | Zhuang (2021) [28] | |
| lncRNA SNGH3 | In vitro | Overexpression of SNGH3 | Increased gene transcription, chromatin modification, tumorigenesis | Cao et al. (2021) [29] | |
| SMAD7e | In vivo | Knockdown of SMAD7e | SMAD7 antagonises transforming growth factor β1 and facilitates cancer cell growth | Che et al. (2020) [30] | |
| HNRNPU | In vitro, in vivo | Knockout of HNRNPU | Enhanced cisplatin sensitivity. Inhibition of cell proliferation, invasion, and migration with apoptosis promotion in cisplatin-treated cells | Shi et al. (2022) [32] | |
| FOXA1 | In vitro, in vivo | Knockout of FOXA1 | Induces a shift from luminal to basal subgroup in luminal cells and induces ZBED2 overexpression to dampen inflammatory response | Neyret-Kahn et al. (2023) [33] | |
| RENAL CANCER | VHL | In vitro | Knockout of VHL | Morphological changes to epithelial mesenchymal transition phenotype, predisposition to metastases through the stabilisation of hypoxia-inducible factors-1α | Schokrpur et al. (2016) [36] |
| TWIST1 | In vitro | Deletion of miR-210-3p microRNA | Findings supported by the Cancer Genome Atlas database, where high TWIST1 and low miR-210-3p expression were associated with poorer disease-free survival | Yoshino et al. (2017) [37] | |
| PTEN | In vitro | Knockout of PTEN | Promoted spheroid formation and decreased sensitivity to sunitinib and sorafenib | Sekino et al. (2020) [40] | |
| Farnesyl-transferase | In vitro, in vivo | Loss of function screening | Screening identified farnesyltransferase as the top hit contributing to sunitinib resistance in ccRCC | Makhov et al. (2020) [41] | |
| CD70 | In vivo | Anti-CD70 CAR T cell therapy | First in-human clinical trial demonstrating an excellent safety profile. One patient had complete remission, and nine had stable disease | Pal et al. (2022) [42] | |
| TESTICULAR CANCER | FLNA | In vitro | Knockout of FLNA | Irregular actin cytoskeletal irregularity leading to mechanical instability and impaired adhesive properties, ultimately affecting migratory ability | Welter et al. (2020) [44] |
| ASH2L | In vitro | Knockout of ASH2L | Knockdown rendered testicular cancer cells resistant to bleomycin, etoposide, and cisplatin. | Constantin and Widmann (2020) [45] | |
| HMGB4 | In vitro | Knockout of HMGB4 | HMGB4 specifically inhibits the repair of the major cisplatin-DNA adducts in testicular germ cell tumours by using the human excision repair system | Awuah et al. (2017) [46] | |
| CD24 | In vitro | Knockout of CD24 | CD24 suppresses the germ cell program and promotes an ectodermal rather than mesodermal cell fate in embryonal carcinomas | Skowron et al. (2022) [47] | |
| VIRMA | In vitro, in vivo | Knockout of VIRMA | The knockdown of VIRMA led to the disruption of the methyltransferase complex and a decrease in m6A abundance, reduced tumour aggressiveness, and increased sensitivity to cisplatin | Miranda-Goncalves et al. (2021) [48] | |
| NAE1 | In vitro | Genome-scale CRISPR/Cas9 activation screen | Screening identified NAE1 overexpression in cisplatin-resistant cell colonies. This was further validated by applying MLN4924 (NAE1 inhibitor) to TGCT cell lines, resulting in increased cisplatin toxicity, apoptosis, and G2/M-phase cycle arrest | Funke et al. (2023) [49] |
Legend: AR, androgen receptor; LNCaP, lymph node carcinoma of the prostate; NANOGP8, nanog homeobox retrogene P8; PCa, prostate cancer; Erβ, estrogen receptor beta; TP53, tumour protein p53; PTEN, phosphatase and tensin homolog; RAC, Rho family; TUBB3, tubulin beta 3 class III; TCEAL1, transcription elongation factor A like 1; PRRX2, paired mesoderm homeobox protein 2; CRISPR, clustered regularly interspaced short palindromic repeats; CRISPRa, CRISPR-mediated gene activation; PD-1, programmed death-1; CBP, CREB-binding protein; BCa, bladder cancer; hTERT, human telomerase reverse transcriptase; Cas, CRISPR-associated systems; RNA, ribonucleic acid; lncRNAs, long noncoding RNAs; SNGH3, small nucleolar RNA host gene 3; SMAD7e, small mothers against decapentaplegic 7 enhancer; HNRNPU, heterogenous nuclear ribonucleoprotein U; FOXA1, forkhead box A1; VHL, von Hippel–Lindau; TWIST1, twist-related protein 1; ccRCC, clear-cell RCC; CD70, cluster of differentiation 70; FLNA, filamin A; ASH2L, Set1/Ash2 histone methyltransferase complex subunit ASH2; HMGB4, high-mobility group box protein 4; CD24, cluster of differentiation 24; VIRMA, vir-like m6A methyltransferase associated; m6A, N6-methyladenosine; NAE1, NEDD8-activating enzyme E1 subunit 1; TGCT, testicular germ cell tumour.
Abbreviations
| BCa | Bladder cancer |
| CAR | Chimeric antigen receptor |
| Cas | CRISPR-associated protein |
| CRISPR | Clustered Regularly Interspaced Short Palindromic Repeats |
| ERβ | Estrogen receptor βeta |
| PCa | Prostate cancer |
| PTEN | Phosphatase and tensin homolog |
| RCC | Renal cell cancer |
| RNA | Ribonucleic acid |
| TGCT | Testicular germ cell tumours |
References
- Jinek, M.; Chylinski, K.; Fonfara, I.; Hauer, M.; Doudna, J.A.; Charpentier, E. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science 2012, 337, 816–821. [Google Scholar] [CrossRef]
- Ishino, Y.; Shinagawa, H.; Makino, K.; Amemura, M.; Nakata, A. Nucleotide sequence of the iap gene, responsible for alkaline phosphatase isozyme conversion in Escherichia coli, and identification of the gene product. J. Bacteriol. 1987, 169, 5429–5433. [Google Scholar] [CrossRef] [PubMed]
- Makarova, K.S.; Grishin, N.V.; Shabalina, S.A.; Wolf, Y.I.; Koonin, E.V. A putative RNA-interference-based immune system in prokaryotes: Computational analysis of the predicted enzymatic machinery, functional analogies with eukaryotic RNAi, and hypothetical mechanisms of action. Biol. Direct 2006, 1, 7. [Google Scholar] [CrossRef]
- Koonin, E.V.; Makarova, K.S. Origins and evolution of CRISPR-Cas systems. Philos. Trans. R Soc. Lond. B Biol. Sci. 2019, 374, 20180087. [Google Scholar] [CrossRef]
- Liu, C.; Zhang, L.; Liu, H.; Cheng, K. Delivery strategies of the CRISPR-Cas9 gene-editing system for therapeutic applications. J. Control Release 2017, 266, 17–26. [Google Scholar] [CrossRef]
- Sánchez-Rivera, F.J.; Jacks, T. Applications of the CRISPR-Cas9 system in cancer biology. Nat. Rev. Cancer 2015, 15, 387–395. [Google Scholar] [CrossRef]
- Chen, X.Z.; Guo, R.; Zhao, C.; Xu, J.; Song, H.; Yu, H.; Pilarsky, C.; Nainu, F.; Li, J.-Q.; Zhou, X.-K.; et al. A Novel Anti-Cancer Therapy: CRISPR/Cas9 Gene Editing. Front. Pharmacol. 2022, 13, 939090. [Google Scholar] [CrossRef]
- Ratan, Z.A.; Son, Y.-J.; Haidere, M.F.; Uddin, B.M.M.; Yusuf, M.A.; Zaman, S.B.; Kim, J.-H.; Banu, L.A.; Cho, J.Y. CRISPR-Cas9: A promising genetic engineering approach in cancer research. Ther. Adv. Med. Oncol. 2018, 10, 1758834018755089. [Google Scholar] [CrossRef]
- Sung, H.; Ferlay, J.; Siegel, R.L.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J. Clin. 2021, 71, 209–249. [Google Scholar] [CrossRef]
- Wei, C.; Wang, F.; Liu, W.; Zhao, W.; Yang, Y.; Li, K.; Xiao, L.; Shen, J. CRISPR/Cas9 targeting of the androgen receptor suppresses the growth of LNCaP human prostate cancer cells. Mol. Med. Rep. 2018, 17, 2901–2906. [Google Scholar] [CrossRef]
- Miyazawa, K.; Tanaka, T.; Nakai, D.; Morita, N.; Suzuki, K. Immunohistochemical expression of four different stem cell markers in prostate cancer: High expression of NANOG in conjunction with hypoxia-inducible factor-1α expression is involved in prostate epithelial malignancy. Oncol. Lett. 2014, 8, 985–992. [Google Scholar] [CrossRef] [PubMed]
- Kawamura, N.; Nimura, K.; Nagano, H.; Yamaguchi, S.; Nonomura, N.; Kaneda, Y. CRISPR/Cas9-mediated gene knockout of NANOG and NANOGP8 decreases the malignant potential of prostate cancer cells. Oncotarget 2015, 6, 22361–22374. [Google Scholar] [CrossRef]
- Warner, M.; Wu, W.-F.; Montanholi, L.; Nalvarte, I.; Antonson, P.; Gustafsson, J.-A. Ventral prostate and mammary gland phenotype in mice with complete deletion of the ERβ gene. Proc. Natl. Acad. Sci. USA 2020, 117, 4902–4909. [Google Scholar] [CrossRef]
- Roehrborn, C.G.; Spann, M.E.; Myers, S.L.; Serviss, C.R.; Hu, L.; Jin, Y. Estrogen receptor beta agonist LY500307 fails to improve symptoms in men with enlarged prostate secondary to benign prostatic hypertrophy. Prostate Cancer Prostatic. Dis. 2015, 18, 43–48. [Google Scholar] [CrossRef]
- Norman, B.H.; Dodge, J.A.; Richardson, T.I.; Borromeo, P.S.; Lugar, C.W.; Jones, S.A.; Chen, K.; Wang, Y.; Durst, G.L.; Barr, R.J.; et al. Benzopyrans Are Selective Estrogen Receptor β Agonists with Novel Activity in Models of Benign Prostatic Hyperplasia. J. Med. Chem. 2006, 49, 6155–6157. [Google Scholar] [CrossRef]
- Batır, M.B.; Şahin, E.; Çam, F.S. Evaluation of the CRISPR/Cas9 directed mutant TP53 gene repairing effect in human prostate cancer cell line PC-3. Mol. Biol. Rep. 2019, 46, 6471–6484. [Google Scholar] [CrossRef]
- Takao, A.; Yoshikawa, K.; Karnan, S.; Ota, A.; Uemura, H.; De Velasco, M.A.; Kura, Y.; Suzuki, S.; Ueda, R.; Nishino, T.; et al. Generation of PTEN-knockout (−/−) murine prostate cancer cells using the CRISPR/Cas9 system and comprehensive gene expression profiling. Oncol. Rep. 2018, 40, 2455–2466. [Google Scholar] [CrossRef]
- Sekino, Y.; Han, X.; Kawaguchi, T.; Babasaki, T.; Goto, K.; Inoue, S.; Hayashi, T.; Teishima, J.; Shiota, M.; Yasui, W.; et al. TUBB3 Reverses Resistance to Docetaxel and Cabazitaxel in Prostate Cancer. Int. J. Mol. Sci. 2019, 20, 3936. [Google Scholar] [CrossRef]
- Rushworth, L.K.; Harle, V.; Repiscak, P.; Clark, W.; Shaw, R.; Hall, H.; Bushell, M.; Leung, H.Y.; Patel, R. In vivo CRISPR/Cas9 knockout screen: TCEAL1 silencing enhances docetaxel efficacy in prostate cancer. Life Sci. Alliance 2020, 3, e202000770. [Google Scholar] [CrossRef] [PubMed]
- Konermann, S.; Brigham, M.D.; Trevino, A.E.; Joung, J.; Abudayyeh, O.O.; Barcena, C.; Hsu, P.D.; Habib, N.; Gootenberg, J.S.; Nishimasu, H.; et al. Genome-scale transcriptional activation by an engineered CRISPR-Cas9 complex. Nature 2015, 517, 583–588. [Google Scholar] [CrossRef] [PubMed]
- Rodríguez, Y.; Unno, K.; Truica, M.I.; Chalmers, Z.R.; Yoo, Y.A.; Vatapalli, R.; Sagar, V.; Yu, J.; Lysy, B.; Hussain, M.; et al. A Genome-Wide CRISPR Activation Screen Identifies PRRX2 as a Regulator of Enzalutamide Resistance in Prostate Cancer. Cancer Res. 2022, 82, 2110–2123. [Google Scholar] [CrossRef]
- Wolf, P.; Alzubi, J.; Gratzke, C.; Cathomen, T. The potential of CAR T cell therapy for prostate cancer. Nat. Rev. Urol. 2021, 18, 556–571. [Google Scholar] [CrossRef]
- Ren, J.; Liu, X.; Fang, C.; Jiang, S.; June, C.H.; Zhao, Y. Multiplex Genome Editing to Generate Universal CAR T Cells Resistant to PD1 Inhibition. Clin. Cancer Res. 2017, 23, 2255–2266. [Google Scholar] [CrossRef]
- Sylvester, R.J.; van der Meijden, A.P.; Oosterlinck, W.; Witjes, J.A.; Bouffioux, C.; Denis, L.; Newling, D.W.; Kurth, K. Predicting recurrence and progression in individual patients with stage Ta T1 bladder cancer using EORTC risk tables: A combined analysis of 2596 patients from seven EORTC trials. Eur. Urol. 2006, 49, 466–477, discussion 475–477. [Google Scholar] [CrossRef]
- Kamat, A.M.; Hahn, N.M.; Efstathiou, J.A.; Lerner, S.P.; Malmstrom, P.U.; Choi, W.; Guo, C.C.; Lotan, Y.; Kassouf, W. Bladder cancer. Lancet 2016, 388, 2796–2810. [Google Scholar] [CrossRef]
- Iyer, N.G.; Ozdag, H.; Caldas, C. p300/CBP and cancer. Oncogene 2004, 23, 4225–4231. [Google Scholar] [CrossRef]
- Li, J.; Huang, C.; Xiong, T.; Zhuang, C.; Zhuang, C.; Li, Y.; Ye, J.; Gui, Y. A CRISPR Interference of CBP and p300 Selectively Induced Synthetic Lethality in Bladder Cancer Cells In Vitro. Int. J. Biol. Sci. 2019, 15, 1276–1286. [Google Scholar] [CrossRef]
- Zhuang, C.; Zhuang, C.; Zhou, Q.; Huang, X.; Gui, Y.; Lai, Y.; Yang, S. Engineered CRISPR/Cas13d Sensing hTERT Selectively Inhibits the Progression of Bladder Cancer In Vitro. Front. Mol. Biosci. 2021, 8, 646412. [Google Scholar] [CrossRef]
- Cao, Y.; Hu, Q.; Zhang, R.; Li, L.; Guo, M.; Wei, H.; Zhang, L.; Wang, J.; Li, C. Knockdown of Long Non-coding RNA SNGH3 by CRISPR-dCas9 Inhibits the Progression of Bladder Cancer. Front. Mol. Biosci. 2021, 8, 657145. [Google Scholar] [CrossRef]
- Che, W.; Ye, S.; Cai, A.; Cui, X.; Sun, Y. CRISPR-Cas13a Targeting the Enhancer RNA-SMAD7e Inhibits Bladder Cancer Development Both in vitro and in vivo. Front. Mol. Biosci. 2020, 7, 607740. [Google Scholar] [CrossRef]
- Briones-Orta, M.A.; Tecalco-Cruz, A.C.; Sosa-Garrocho, M.; Caligaris, C.; Macias-Silva, M. Inhibitory Smad7: Emerging roles in health and disease. Curr. Mol. Pharmacol. 2011, 4, 141–153. [Google Scholar] [CrossRef]
- Shi, Z.D.; Hao, L.; Han, X.X.; Wu, Z.X.; Pang, K.; Dong, Y.; Qin, J.; Wang, G.; Zhang, X.; Xia, T.; et al. Targeting HNRNPU to overcome cisplatin resistance in bladder cancer. Mol. Cancer 2022, 21, 37. [Google Scholar] [CrossRef]
- Neyret-Kahn, H.; Fontugne, J.; Meng, X.Y.; Groeneveld, C.S.; Cabel, L.; Ye, T.; Guyon, E.; Krucker, C.; Dufour, F.; Chapeaublanc, E.; et al. Epigenomic mapping identifies an enhancer repertoire that regulates cell identity in bladder cancer through distinct transcription factor networks. Oncogene 2023, 42, 1524–1542. [Google Scholar] [CrossRef]
- Sharma, R.; Kadife, E.; Myers, M.; Kannourakis, G.; Prithviraj, P.; Ahmed, N. Determinants of resistance to VEGF-TKI and immune checkpoint inhibitors in metastatic renal cell carcinoma. J. Exp. Clin. Cancer Res. 2021, 40, 186. [Google Scholar] [CrossRef]
- Murphy, G.P.; Hrushesky, W.J. A Murine Renal Cell Carcinoma. JNCI J. Natl. Cancer Inst. 1973, 50, 1013–1025. [Google Scholar] [CrossRef]
- Schokrpur, S.; Hu, J.; Moughon, D.L.; Liu, P.; Lin, L.C.; Hermann, K.; Mangul, S.; Guan, W.; Pellegrini, M.; Xu, H.; et al. CRISPR-Mediated VHL Knockout Generates an Improved Model for Metastatic Renal Cell Carcinoma. Sci. Rep. 2016, 6, 29032. [Google Scholar] [CrossRef]
- Yoshino, H.; Yonemori, M.; Miyamoto, K.; Tatarano, S.; Kofuji, S.; Nohata, N.; Nakagawa, M.; Enokida, H. microRNA-210-3p depletion by CRISPR/Cas9 promoted tumorigenesis through revival of TWIST1 in renal cell carcinoma. Oncotarget 2017, 8, 20881–20894. [Google Scholar] [CrossRef]
- McCormick, R.I.; Blick, C.; Ragoussis, J.; Schoedel, J.; Mole, D.R.; Young, A.C.; Selby, P.J.; Banks, R.E.; Harris, A.L. miR-210 is a target of hypoxia-inducible factors 1 and 2 in renal cancer, regulates ISCU and correlates with good prognosis. Br. J. Cancer 2013, 108, 1133–1142. [Google Scholar] [CrossRef]
- The Cancer Genome Atlas Research Network. Comprehensive molecular characterization of clear cell renal cell carcinoma. Nature 2013, 499, 43–49. [Google Scholar] [CrossRef]
- Sekino, Y.; Hagura, T.; Han, X.; Babasaki, T.; Goto, K.; Inoue, S.; Hayashi, T.; Teishima, J.; Shigeta, M.; Taniyama, D.; et al. PTEN Is Involved in Sunitinib and Sorafenib Resistance in Renal Cell Carcinoma. Anticancer Res. 2020, 40, 1943–1951. [Google Scholar] [CrossRef]
- Makhov, P.; Sohn, J.A.; Serebriiskii, I.G.; Fazliyeva, R.; Khazak, V.; Boumber, Y.; Uzzo, R.G.; Kolenko, V.M. CRISPR/Cas9 genome-wide loss-of-function screening identifies druggable cellular factors involved in sunitinib resistance in renal cell carcinoma. Br. J. Cancer 2020, 123, 1749–1756. [Google Scholar] [CrossRef]
- Pal, S.; Tran, B.; Haanen, J.; Hurwitz, M.; Sacher, A.; Agarwal, N.; Tannir, N.; Budde, E.; Harrison, S.; Klobuch, S.; et al. 558 CTX130 allogeneic CRISPR-Cas9–engineered chimeric antigen receptor (CAR) T cells in patients with advanced clear cell renal cell carcinoma: Results from the phase 1 COBALT-RCC study. J. ImmunoTherapy Cancer 2022, 10 (Suppl. S2), A584. [Google Scholar]
- Boccellino, M.; Vanacore, D.; Zappavigna, S.; Cavaliere, C.; Rossetti, S.; D’aniello, C.; Chieffi, P.; Amler, E.; Buonerba, C.; Di Lorenzo, G.; et al. Testicular cancer from diagnosis to epigenetic factors. Oncotarget 2017, 8, 104654–104663. [Google Scholar] [CrossRef]
- Welter, H.; Herrmann, C.; Fröhlich, T.; Flenkenthaler, F.; Eubler, K.; Schorle, H.; Nettersheim, D.; Mayerhofer, A.; Müller-Taubenberger, A. Filamin A Orchestrates Cytoskeletal Structure, Cell Migration and Stem Cell Characteristics in Human Seminoma TCam-2 Cells. Cells 2020, 9, 2563. [Google Scholar] [CrossRef]
- Constantin, D.; Widmann, C. ASH2L drives proliferation and sensitivity to bleomycin and other genotoxins in Hodgkin’s lymphoma and testicular cancer cells. Cell Death Dis. 2020, 11, 1019. [Google Scholar] [CrossRef]
- Awuah, S.G.; Riddell, I.A.; Lippard, S.J. Repair shielding of platinum-DNA lesions in testicular germ cell tumors by high-mobility group box protein 4 imparts cisplatin hypersensitivity. Proc. Natl. Acad. Sci. USA 2017, 114, 950–955. [Google Scholar] [CrossRef]
- Skowron, M.A.; Becker, T.K.; Kurz, L.; Jostes, S.; Bremmer, F.; Fronhoffs, F.; Funke, K.; Wakileh, G.A.; Müller, M.R.; Burmeister, A.; et al. The signal transducer CD24 suppresses the germ cell program and promotes an ectodermal rather than mesodermal cell fate in embryonal carcinomas. Mol. Oncol. 2022, 16, 982–1008. [Google Scholar] [CrossRef]
- Miranda-Gonçalves, V.; Lobo, J.; Guimarães-Teixeira, C.; Barros-Silva, D.; Guimarães, R.; Cantante, M.; Braga, I.; Maurício, J.; Oing, C.; Honecker, F.; et al. The component of the m(6)A writer complex VIRMA is implicated in aggressive tumor phenotype, DNA damage response and cisplatin resistance in germ cell tumors. J. Exp. Clin. Cancer Res. 2021, 40, 268. [Google Scholar] [CrossRef]
- Funke, K.; Einsfelder, U.; Hansen, A.; Arévalo, L.; Schneider, S.; Nettersheim, D.; Stein, V.; Schorle, H. Genome-scale CRISPR screen reveals neddylation to contribute to cisplatin resistance of testicular germ cell tumours. Br. J. Cancer 2023, 128, 2270–2282. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).