Primary Adult Retroperitoneal Sarcoma: A Comprehensive Genomic Profiling Study
Abstract
:1. Introduction
2. Methods
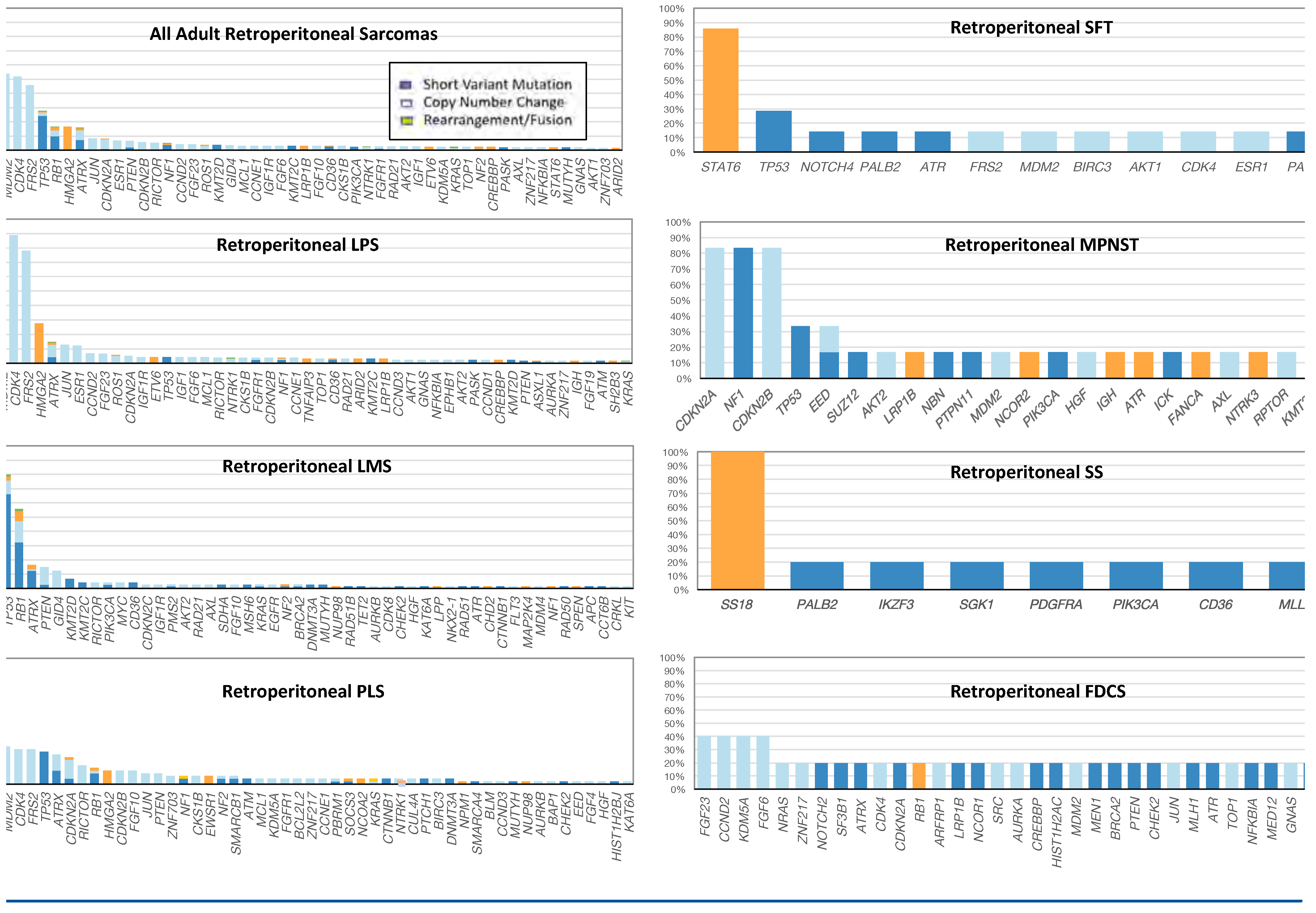
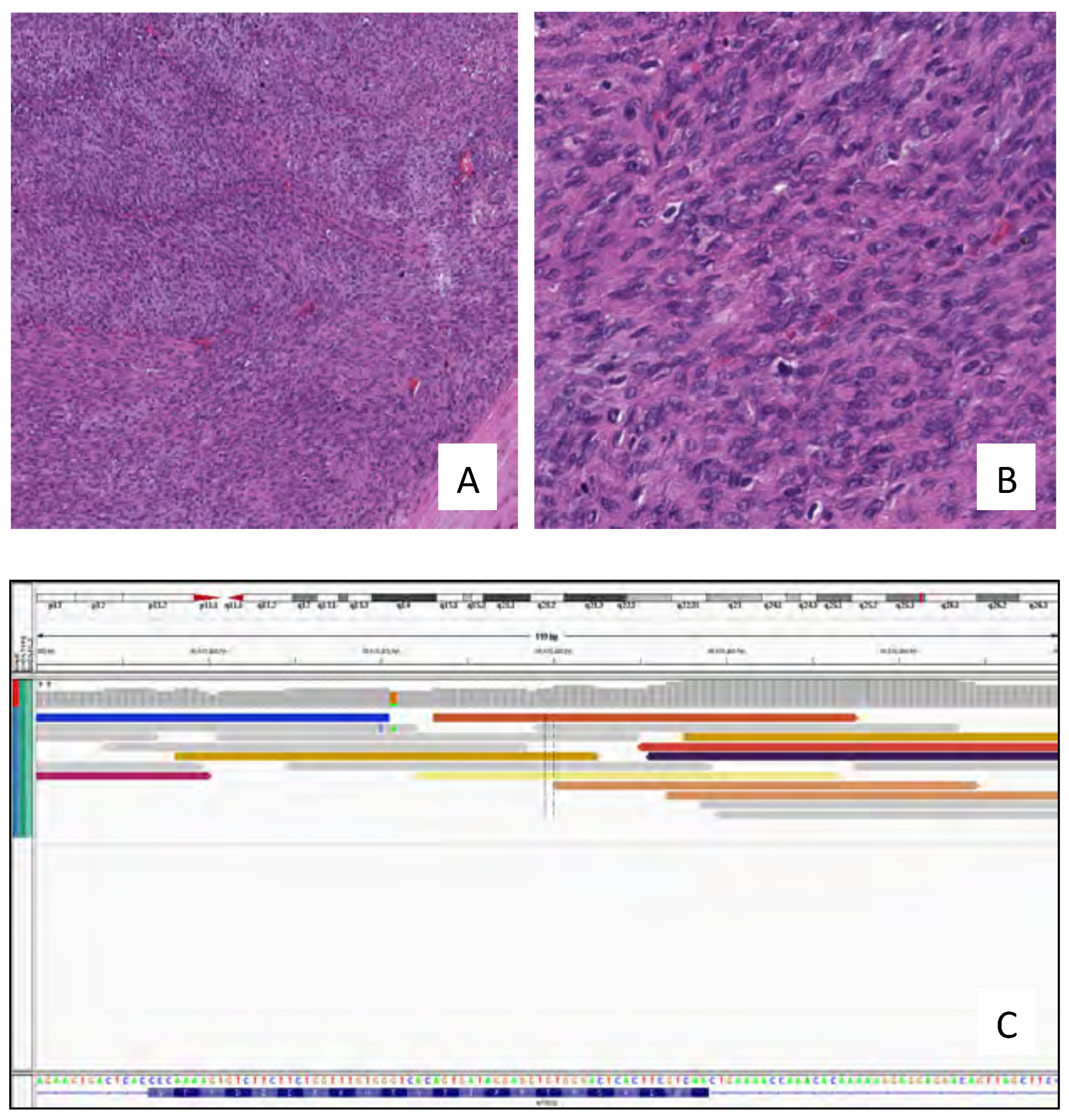
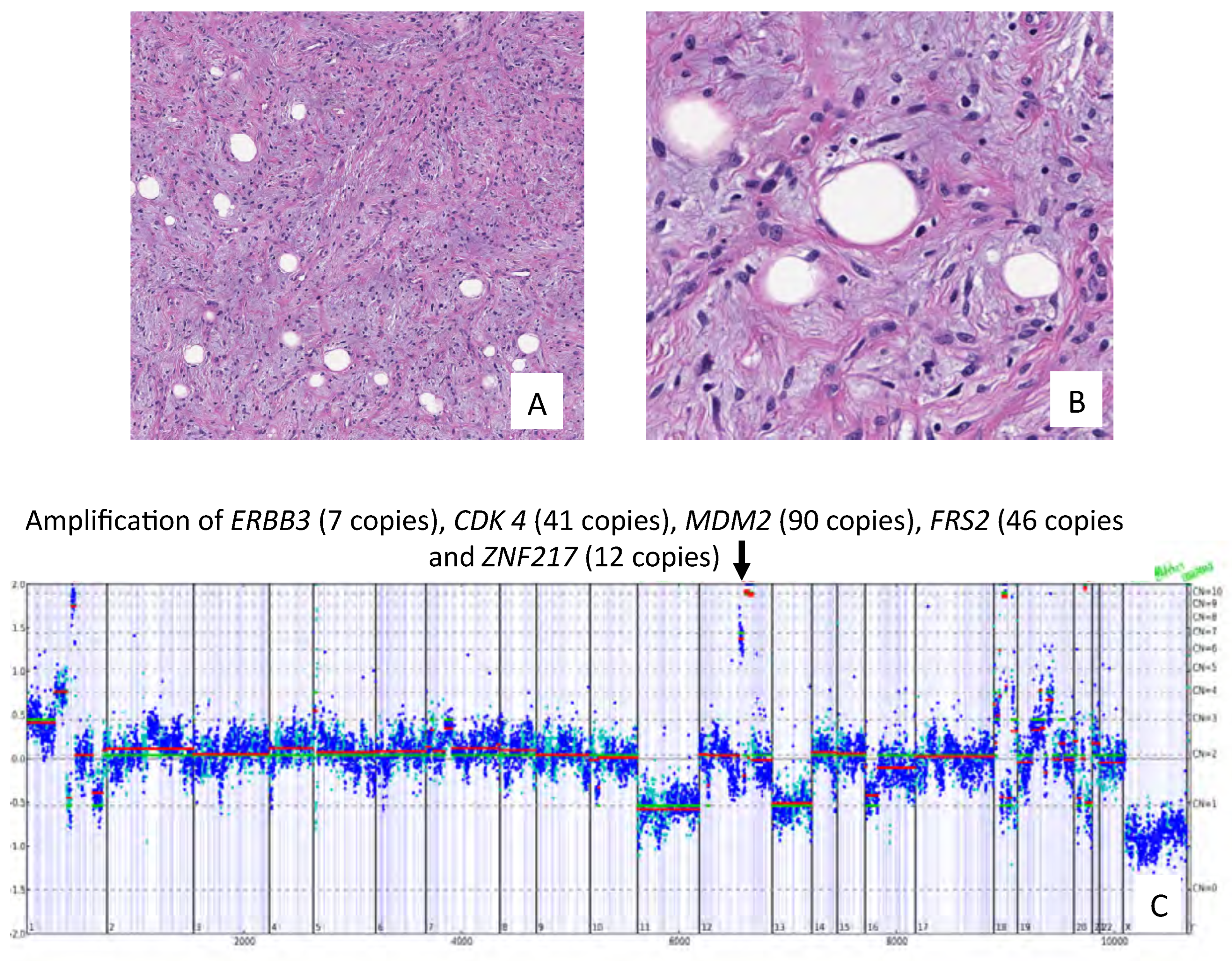
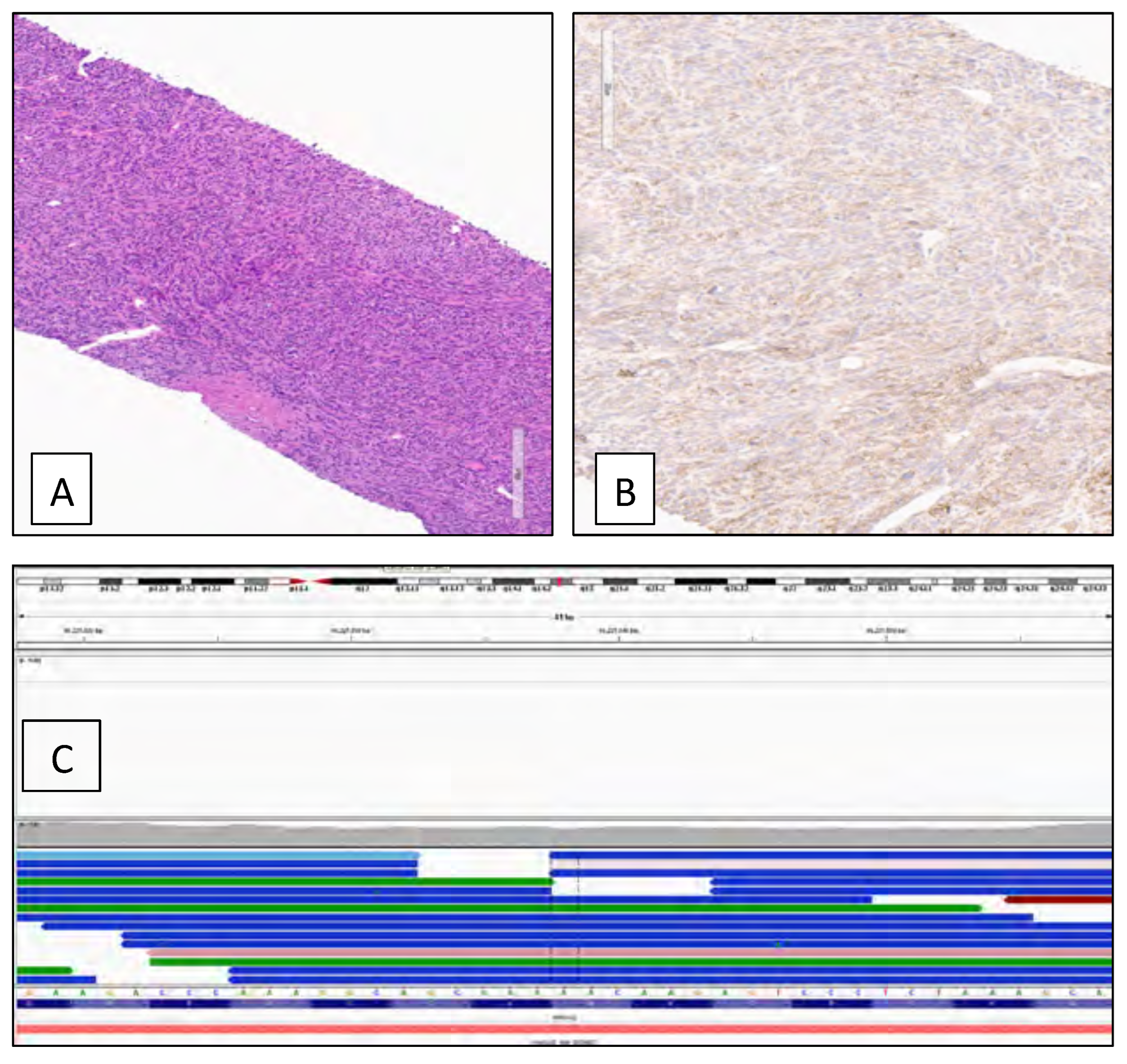
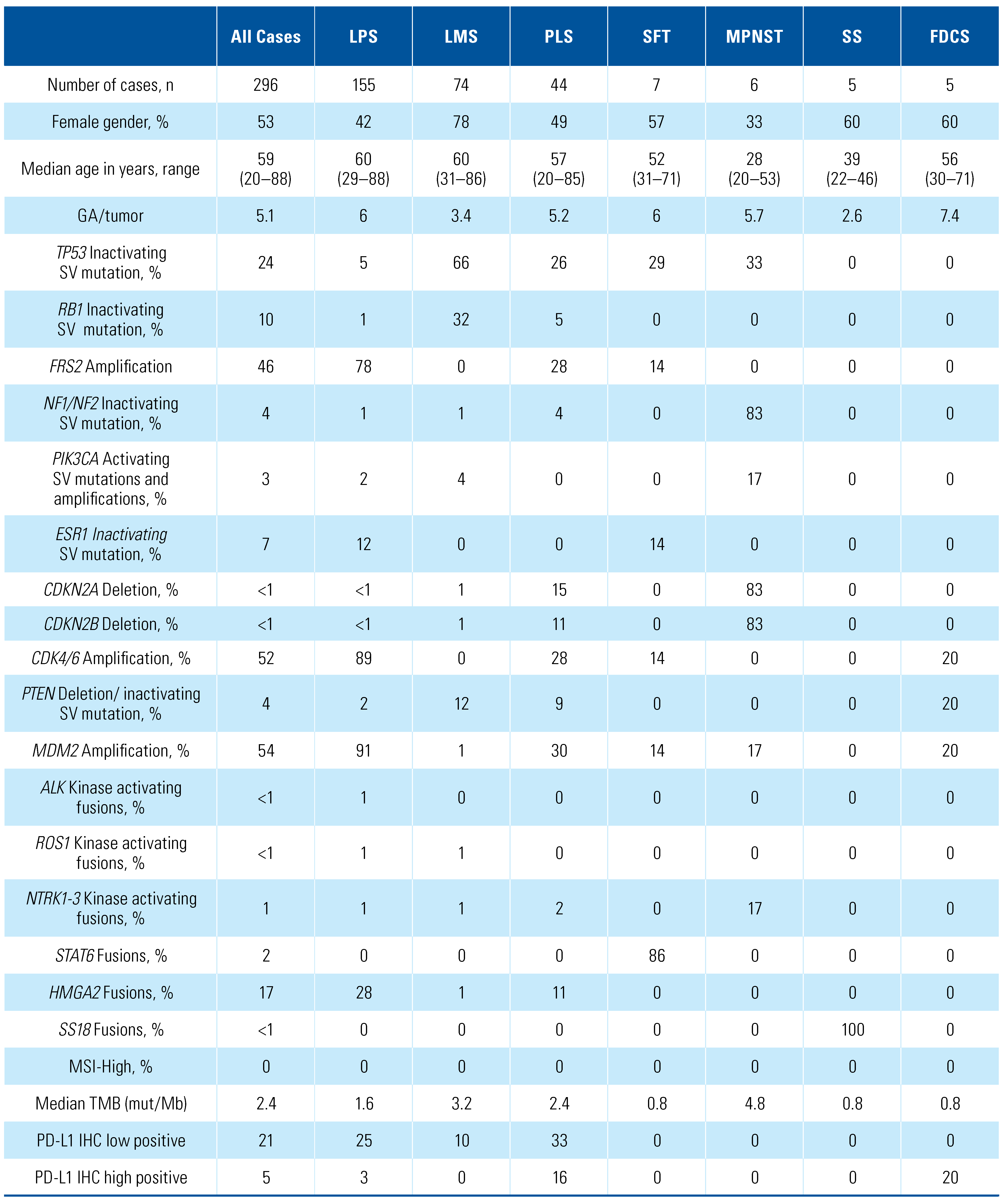
3. Results
4. Discussion
5. Conclusions
Supplementary Materials
Conflicts of Interest
Abbreviations
| CGP | comprehensive genomic profiling |
| FDCS | follicular dendritic cell sarcoma |
| GA | genomic alteration |
| ICI | immune checkpoint inhibitor |
| LMS | leiomyosarcoma |
| LPS | liposarcoma |
| MPNST | malignant peripheral nerve sheath tumors |
| MSI | microsatellite instability |
| OS | overall survival |
| PFS | progression-free survival |
| PLS | pleomorphic sarcoma |
| PRS | primary retroperitoneal sarcoma |
| SFT | solitary fibrous tumors |
| SS | synovial sarcomas |
| TMB | tumor mutational burden |
| TKI | tyrosine kinase inhibitor |
References
- WHO Classification of Tumours Editorial Board. WHO Classification of Tumours of Soft Tissue and Bone, 5th ed.; IARC Press: Lyon, France, 2020. [Google Scholar]
- Gatta, G.; Capocaccia, R.; Botta, L.; Mallone, S.; De Angelis, R.; Ardanaz, E.; et al. Burden and centralised treatment in Europe of rare tumours: results of RARECAREnet—a population-based study. Lancet Oncol. 2017, 18, 1022–1039. [Google Scholar] [CrossRef] [PubMed]
- Gronchi, A.; Miceli, R.; Shurell, E.; Eilber, F.C.; Eilber, F.R.; Anaya, D.A.; et al. Outcome prediction in primary resected retroperitoneal soft tissue sarcoma: histology-specific overall survival and disease- free survival nomograms built on major sarcoma center data sets. J Clin Oncol. 2013, 31, 1649–1655. [Google Scholar] [CrossRef] [PubMed]
- Toulmonde, M.; Bonvalot, S.; Méeus, P.; Stoeckle, E.; Riou, O.; Isambert, N.; et al. Retroperitoneal sarcomas: patterns of care at diagnosis, prognostic factors and focus on main histological subtypes: a multicenter analysis of the French Sarcoma Group. Ann Oncol. 2014, 25, 735–742. [Google Scholar] [CrossRef] [PubMed]
- Taylor, B.S.; Barretina, J.; Maki, R.G.; Antonescu, C.R.; Singer, S.; Ladanyi, M. Advances in sarcoma genomics and new therapeutic targets. Nat Rev Cancer. 2011, 11, 541–557. [Google Scholar] [CrossRef] [PubMed]
- Pederzoli, F.; Bandini, M.; Marandino, L.; Ali, S.M.; Madison, R.; Chung, J.; et al. Targetable gene fusions and aberrations in genitourinary oncology. Nat Rev Urol. 2020, 17, 613–625. [Google Scholar] [CrossRef] [PubMed]
- Yakirevich, E.; Madison, R.; Fridman, E.; Mangray, S.; Carneiro, B.A.; Lu, S.; et al. Comprehensive genomic profiling of adult renal sarcomas provides insight into disease biology and opportunities for targeted therapies. Eur Urol Oncol. 2021, 4, 282–288. [Google Scholar] [CrossRef] [PubMed]
- Frampton, G.M.; Fichtenholtz, A.; Otto, G.A.; Wang, K.; Downing, S.R.; He, J.; et al. Development and validation of a clinical cancer genomic profiling test based on massively parallel DNA sequencing. Nat Biotechnol. 2013, 31, 1023–1031. [Google Scholar] [CrossRef] [PubMed]
- He, J.; Abdel-Wahab, O.; Nahas, M.K.; Wang, K.; Rampal, R.K.; Intlekofer, A.M.; et al. Integrated genomic DNA/RNA profiling of hematologic malignancies in the clinical setting. Blood. 2016, 127, 3004–3014. [Google Scholar] [CrossRef] [PubMed]
- Gao, J.; Aksoy, B.A.; Dogrusoz, U.; Dresdner, G.; Gross, B.; Sumer, S.O.; et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci Signal. 2013, 6, pl1. [Google Scholar] [CrossRef]
- Cerami, E.; Gao, J.; Dogrusoz, U.; Gross, B.E.; Sumer, S.O.; Aksoy, B.A.; et al. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2012, 2, 401–404. [Google Scholar] [CrossRef]
- Chalmers, Z.R.; Connelly, C.F.; Fabrizio, D.; Gay, L.; Ali, S.M.; Ennis, R.; et al. Analysis of 100,000 human cancer genomes reveals the landscape of tumor mutational burden. Genome Med. 2017, 9, 34. [Google Scholar] [CrossRef]
- Trabucco, S.E.; Gowen, K.; Maund, S.L.; Sanford, E.; Fabrizio, D.A.; Hall, M.J.; et al. A novel next-generation sequencing approach to detecting microsatellite instability and pan-tumor characterization of 1000 microsatellite instability-high cases in 67,000 patient samples. J Mol Diagn. 2019, 21, 1053–1066. [Google Scholar] [CrossRef]
- Nazzani, S.; Preisser, F.; Bandini, M.; Marchioni, M.; Tian, Z.; Soulières, D.; et al. Surgically treated retroperitoneal sarcoma: a population- based competing risks analysis. Eur Urol Oncol. 2018, 1, 346–351. [Google Scholar] [CrossRef] [PubMed]
- Bonvalot, S.; Gronchi, A.; Le Péchoux, C.; Swallow, C.J.; Strauss, D.; Meeus, P.; et al. Preoperative radiotherapy plus surgery versus surgery alone for patients with primary retroperitoneal sarcoma (EORTC-62092: STRASS): a multicentre, open-label, randomised, phase 3 trial. Lancet Oncol. 2020, 21, 1366–1377. [Google Scholar] [CrossRef] [PubMed]
- van der Graaf, W.T.A.; Blay, J.-Y.; Chawla, S.P.; Kim D-W, Bui-Nguyen, B.; Casali, P.G.; et al. Pazopanib for metastatic soft-tissue sarcoma (PALETTE): a randomised, double-blind, placebo-controlled phase 3 trial. Lancet. 2012, 379, 1879–1886. [Google Scholar] [CrossRef]
- Schöffski, P.; Chawla, S.; Maki, R.G.; Italiano, A.; Gelderblom, H.; Choy, E.; et al. Eribulin versus dacarbazine in previously treated patients with advanced liposarcoma or leiomyosarcoma: a randomised, open-label, multicentre, phase 3 trial. Lancet. 2016, 387, 1629–1637. [Google Scholar] [CrossRef] [PubMed]
- Lucchesi, C.; Khalifa, E.; Laizet, Y.; Soubeyran, I.; Mathoulin-Pelissier, S.; Chomienne, C.; et al. Targetable alterations in adult patients with soft-tissue sarcomas: insights for personalized therapy. JAMA Oncol. 2018, 4, 1398–1404. [Google Scholar] [CrossRef] [PubMed]
- Abeshouse, A.; Adebamowo, C.; Adebamowo, S.N.; Akbani, R.; Akeredolu, T.; Ally, A.; et al. Comprehensive and integrated genomic characterization of adult soft tissue sarcomas. Cell 2017, 171, 950–965. e28. [Google Scholar] [CrossRef]
- Arnaud-Coffin, P.; Brahmi, M.; Vanacker, H.; Eberst, L.; Tredan, O.; Attignon, V.; et al. Therapeutic relevance of molecular screening program in patients with metastatic sarcoma: analysis from the ProfiLER 01 trial. Transl Oncol. 2020, 13, 100870. [Google Scholar] [CrossRef]
- Somaiah, N.; Beird, H.C.; Barbo, A.; Song, J.; Shaw, K.R.M.; Wang, W.-L.; et al. Targeted next generation sequencing of well-differentiated/ dedifferentiated liposarcoma reveals novel gene amplifications and mutations. Oncotarget. 2018, 9, 19891–19899. [Google Scholar] [CrossRef]
- Sriraman, A.; Dickmanns, A.; Najafova, Z.; Johnsen, S.A.; Dobbelstein, M. CDK4 inhibition diminishes p53 activation by MDM2 antagonists. Cell Death Dis. 2018, 9, 1–16. [Google Scholar] [CrossRef] [PubMed]
- Gluck, W.L.; Gounder, M.M.; Frank, R.; Eskens, F.; Blay, J.Y.; Cassier, P.A.; et al. Phase 1 study of the MDM2 inhibitor AMG 232 in patients with advanced P53 wild-type solid tumors or multiple myeloma. Invest New Drugs. 2020, 38, 831–843. [Google Scholar] [CrossRef] [PubMed]
- Dickson, M.A.; Schwartz, G.K.; Keohan, M.L.; D’Angelo, S.P.; Gounder, M.M.; Chi, P.; et al. Progression-free survival among patients with well- differentiated or dedifferentiated liposarcoma treated with CDK4 inhibitor palbociclib: a phase 2 clinical trial. JAMA Oncol. 2016, 2, 937–940. [Google Scholar] [CrossRef]
- Dickson, M.A.; Tap, W.D.; Keohan, M.L.; D’Angelo, S.P.; Gounder, M.M.; Antonescu, C.R.; et al. Phase II trial of the CDK4 inhibitor PD0332991 in patients with advanced CDK4-amplified well-differentiated or dedifferentiated liposarcoma. J Clin Oncol. 2013, 31, 2024–2028. [Google Scholar] [CrossRef] [PubMed]
- Infante, J.R.; Cassier, P.A.; Gerecitano, J.F.; Witteveen, P.O.; Chugh, R.; Ribrag, V.; et al. A Phase I study of the cyclin-dependent kinase 4/6 inhibitor ribociclib (LEE011) in patients with advanced solid tumors and lymphomas. Clin Cancer Res. 2016, 22, 5696–5705. [Google Scholar] [CrossRef] [PubMed]
- Chudasama, P.; Mughal, S.S.; Sanders, M.A.; Hübschmann, D.; Chung, I.; Deeg, K.I.; et al. Integrative genomic and transcriptomic analysis of leiomyosarcoma. Nat Commun. 2018, 9, 144. [Google Scholar] [CrossRef] [PubMed]
- George, S.; Miao, D.; Demetri, G.D.; Adeegbe, D.; Rodig, S.J.; Shukla, S.; et al. Loss of PTEN is associated with resistance to anti-pd-1 checkpoint blockade therapy in metastatic uterine leiomyosarcoma. Immunity. 2017, 46, 197–204. [Google Scholar] [CrossRef] [PubMed]
- Bui, N.Q.; Przybyl, J.; Trabucco, S.E.; Frampton, G.; Hastie, T.; van de Rijn, M.; et al. A clinico-genomic analysis of soft tissue sarcoma patients reveals CDKN2A deletion as a biomarker for poor prognosis. Clin Sarcoma Res. 2019, 9, 12. [Google Scholar] [CrossRef]
- Doebele, R.C.; Drilon, A.; Paz-Ares, L.; Siena, S.; Shaw, A.T.; Farago, A.F.; et al. Entrectinib in patients with advanced or metastatic NTRK fusion- positive solid tumours: integrated analysis of three phase 1–2 trials. Lancet Oncol. 2020, 21, 271–282. [Google Scholar] [CrossRef]
- Dodd, R.D.; Mito, J.K.; Eward, W.C.; Chitalia, R.; Sachdeva, M.; Ma, Y.; et al. NF1 deletion generates multiple subtypes of soft-tissue sarcoma that respond to MEK Inhibition. Mol Cancer Ther. 2013, 12, 1906–1917 doi org/101158/1535. [Google Scholar] [CrossRef]
- D’Angelo, S.P.; Shoushtari, A.N.; Agaram, N.P.; Kuk, D.; Qin, L.-X.; Carvajal, R.D.; et al. Prevalence of tumor-infiltrating lymphocytes and PD-L1 expression in the soft tissue sarcoma microenvironment. Hum Pathol. 2015, 46, 357–365. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.R.; Moon, Y.J.; Kwon, K.S.; Bae, J.S.; Wagle, S.; Kim, K.M.; et al. Tumor infiltrating PD1-positive lymphocytes and the expression of pd-l1 predict poor prognosis of soft tissue sarcomas. PLoS One. 2013, 8, e82870. [Google Scholar] [CrossRef] [PubMed]
- Tawbi, H.A.; Burgess, M.; Bolejack, V.; Van Tine, B.A.; Schuetze, S.M.; Hu, J.; et al. Pembrolizumab in advanced soft-tissue sarcoma and bone sarcoma (SARC028): a multicentre, two-cohort, single-arm, open- label, phase 2 trial. Lancet Oncol. 2017, 18, 1493–1501. [Google Scholar] [CrossRef]
- D’Angelo, S.P.; Mahoney, M.R.; Tine, B.A.V.; Atkins, J.; Milhem, M.M.; Jahagirdar, B.N.; et al. Nivolumab with or without ipilimumab treatment for metastatic sarcoma (Alliance A091401): two open- label, non-comparative, randomised, phase 2 trials. Lancet Oncol. 2018, 19, 416–426. [Google Scholar] [CrossRef]
 |
This is an open access article under the terms of a license that permits non-commercial use, provided the original work is properly cited. © 2021 The Authors. Société Internationale d'Urologie Journal, published by the Société Internationale d'Urologie, Canada.
Share and Cite
Necchi, A.; Basile, G.; Pederzoli, F.; Bandini, M.; Grivas, P.; Bratslavsky, G.; Spiess, P.E.; Killian, J.K.; Lin, D.I.; Williams, E.; et al. Primary Adult Retroperitoneal Sarcoma: A Comprehensive Genomic Profiling Study. Soc. Int. Urol. J. 2021, 2, 216-228. https://doi.org/10.48083/VOGF2319
Necchi A, Basile G, Pederzoli F, Bandini M, Grivas P, Bratslavsky G, Spiess PE, Killian JK, Lin DI, Williams E, et al. Primary Adult Retroperitoneal Sarcoma: A Comprehensive Genomic Profiling Study. Société Internationale d’Urologie Journal. 2021; 2(4):216-228. https://doi.org/10.48083/VOGF2319
Chicago/Turabian StyleNecchi, Andrea, Giuseppe Basile, Filippo Pederzoli, Marco Bandini, Petros Grivas, Gennady Bratslavsky, Philippe E. Spiess, J. Keith Killian, Douglas I. Lin, Erik Williams, and et al. 2021. "Primary Adult Retroperitoneal Sarcoma: A Comprehensive Genomic Profiling Study" Société Internationale d’Urologie Journal 2, no. 4: 216-228. https://doi.org/10.48083/VOGF2319
APA StyleNecchi, A., Basile, G., Pederzoli, F., Bandini, M., Grivas, P., Bratslavsky, G., Spiess, P. E., Killian, J. K., Lin, D. I., Williams, E., Ramkissoon, S., Severson, E. A., Alexander, B. M., Venstrom, J., Reddy, P., McGregor, K., Elvin, J. A., Schrock, A. B., Pavlick, D. C., ... Ross, J. S. (2021). Primary Adult Retroperitoneal Sarcoma: A Comprehensive Genomic Profiling Study. Société Internationale d’Urologie Journal, 2(4), 216-228. https://doi.org/10.48083/VOGF2319




