A Machine Learning-Based Model to Predict In-Hospital Mortality of Lung Cancer Patients: A Population-Based Study of 523,959 Cases
Abstract
Highlights
- Our model including six epidemiological components was successfully validated on both internal and external validation.
- Risk stratification by the model showed significantly different survival patterns even after discharge.
- Three well-developed interfaces are friendly to both physicians and patients for prognosis-related conversations.
- Our model with easily accessible variables showed its robustness in inferring its predictive value with respect to in-hospital mortality of lung cancer patients.
- The model is highly applicable in follow-up.
- Its applications are useful to clinical in the assistance of strategic planning and the improvement of end-of-life care.
Abstract
1. Introduction
2. Materials and Methods
2.1. Data Processing
2.2. Defining In-Hospital Mortality
2.3. Model Training
2.4. Model Evaluation
2.5. Analysis Platform
3. Results
3.1. Patient Characteristics
3.2. Model Training
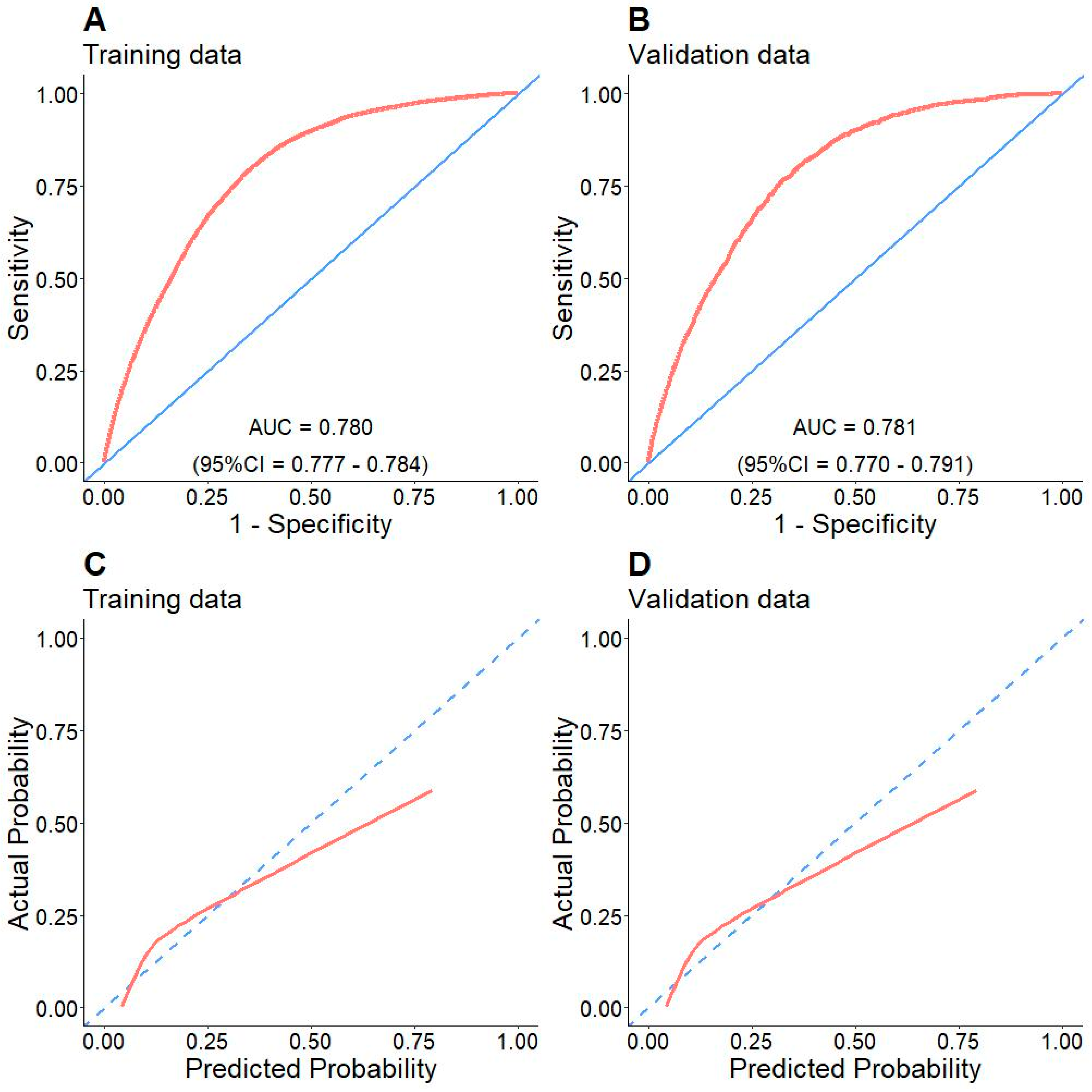
3.3. Model Evaluation
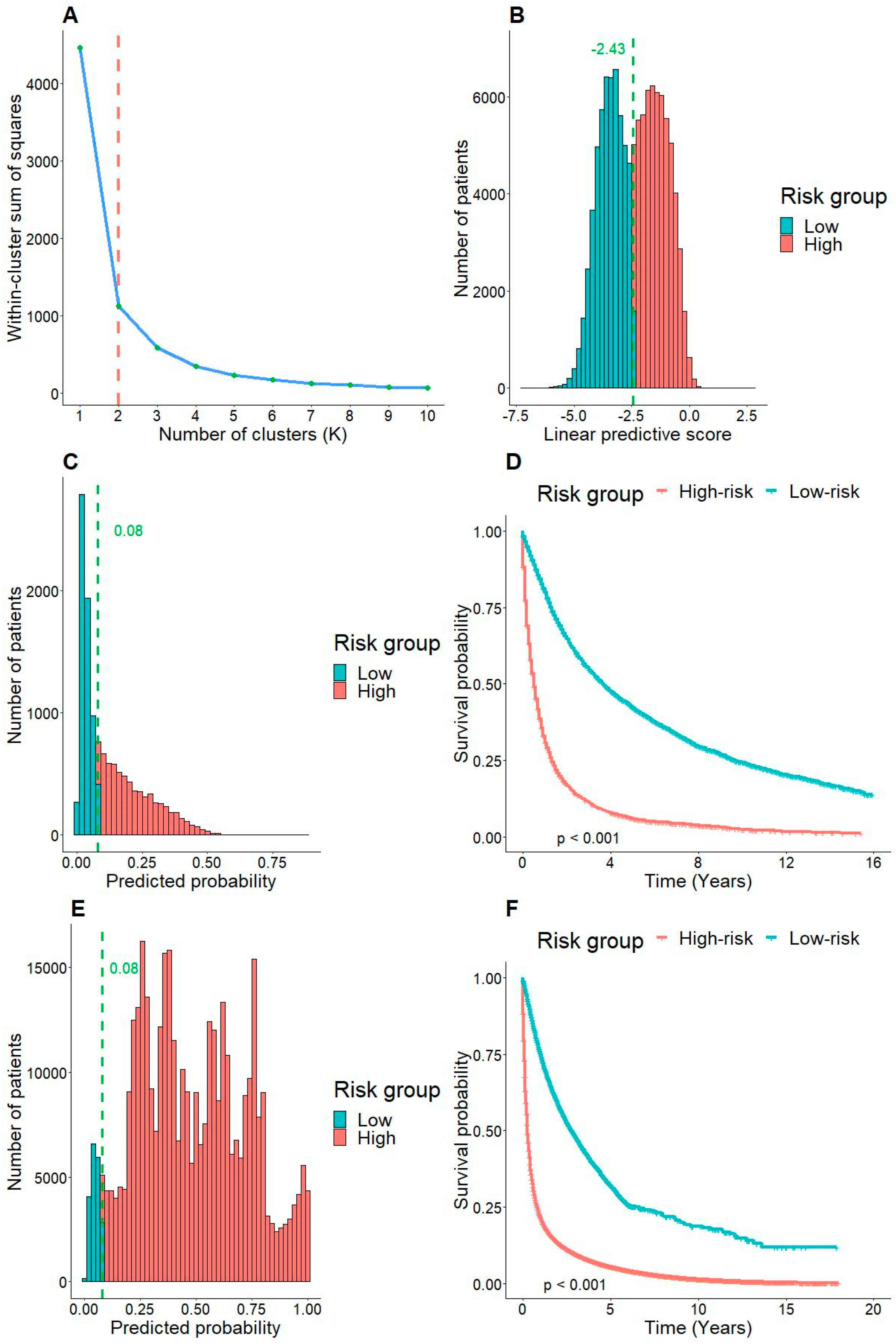
3.4. Risk Stratification by the Model
3.5. Applications of the Model
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Siegel, R.L.; Miller, K.D.; Fuchs, H.E.; Jemal, A. Cancer statistics, 2022. CA A Cancer J. Clin. 2022, 72, 7–33. [Google Scholar] [CrossRef] [PubMed]
- Barta, J.A.; Powell, C.A.; Wisnivesky, J.P. Global Epidemiology of Lung Cancer. Ann. Glob. Health 2019, 85, 8. [Google Scholar] [CrossRef]
- Sung, H.; Ferlay, J.; Siegel, R.L.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA A Cancer J. Clin. 2021, 71, 209–249. [Google Scholar] [CrossRef]
- Alexander, M.; Kim, S.Y.; Cheng, H. Update 2020: Management of Non-Small Cell Lung Cancer. Lung 2020, 198, 897–907. [Google Scholar] [CrossRef] [PubMed]
- Thai, A.A.; Solomon, B.J.; Sequist, L.V.; Gainor, J.F.; Heist, R.S. Lung cancer. Lancet 2021, 398, 535–554. [Google Scholar] [CrossRef]
- Long, K.; Suresh, K. Pulmonary toxicity of systemic lung cancer therapy. Respirology 2020, 25, 72–79. [Google Scholar]
- Howlader, N.; Forjaz, G.; Mooradian, M.J.; Meza, F.; Kong, C.Y.; Cronin, K.A.; Mariotto, A.B.; Lowy, D.R.; Feuer, E.J. The Effect of Advances in Lung-Cancer Treatment on Population Mortality. N. Engl. J. Med. 2020, 383, 640–649. [Google Scholar] [CrossRef] [PubMed]
- Zugazagoitia, J.; Paz-Ares, L. Extensive-Stage Small-Cell Lung Cancer: First-Line and Second-Line Treatment Options. J. Clin. Oncol. 2022, 40, 671–680. [Google Scholar] [CrossRef]
- Horn, L.; Mansfield, A.S.; Szczęsna, A.; Havel, L.; Krzakowski, M.; Hochmair, M.J.; Huemer, F.; Losonczy, G.; Johnson, M.L.; Nishio, M.; et al. First-Line Atezolizumab plus Chemotherapy in Extensive-Stage Small-Cell Lung Cancer. N. Engl. J. Med. 2018, 379, 2220–2229. [Google Scholar] [PubMed]
- Paz-Ares, L.; Dvorkin, M.; Chen, Y.; Reinmuth, N.; Hotta, K.; Trukhin, D.; Statsenko, G.; Hochmair, M.J.; Özgüroğlu, M.; Ji, J.H.; et al. Durvalumab plus platinum-etoposide versus platinum-etoposide in first-line treatment of extensive-stage small-cell lung cancer (CASPIAN): A randomised, controlled, open-label, phase 3 trial. Lancet 2019, 394, 1929–1939. [Google Scholar] [CrossRef]
- Criss, S.D.; Cao, P.; Bastani, M.; Haaf, K.T.; Chen, Y.; Sheehan, D.F.; Erik FBlom, E.F.; Toumazis, I.; Jeon, J.; de Koning, H.J.; et al. Cost-Effectiveness Analysis of Lung Cancer Screening in the United States: A Comparative Modeling Study. Ann. Intern. Med. 2019, 171, 796–804. [Google Scholar] [CrossRef] [PubMed]
- Davidoff, A.J.; Canavan, M.E.; Prsic, E.; Saphire, M.; Wang, S.Y.; Presley, C.J. End-of-life care trajectories among older adults with lung cancer. J. Geriatr. Oncol. 2022, 14, 101381. [Google Scholar] [CrossRef] [PubMed]
- Temel, J.S.; Petrillo, L.A.; Greer, G.A. Patient-Centered Palliative Care for Patients with Advanced Lung Cancer. J. Clin. Oncol. 2022, 40, 626–634. [Google Scholar] [CrossRef]
- Stapelfeld, C.; Dammann, C.; Maser, E. Sex-specificity in lung cancer risk. Int. J. Cancer 2019, 146, 2376–2382. [Google Scholar] [CrossRef]
- Tolwin, Y.; Gillis, R.; Peled, N. Gender and lung cancer-SEER-based analysis. Ann. Epidemiol. 2020, 46, 14–19. [Google Scholar] [CrossRef] [PubMed]
- Komici, K.; Bencivenga, L.; Navani, N.; D’Agnano, V.; Guerra, G.; Bianco, A.; Rengo, G.; Perrotta, F. Frailty in Patients with Lung Cancer: A Systematic Review and Meta-Analysis. Chest 2022, 162, 485–497. [Google Scholar] [CrossRef]
- Shen, H.; Deng, G.; Chen, Q.; Qian, J. The incidence, risk factors and predictive nomograms for early death of lung cancer with synchronous brain metastasis: A retrospective study in the SEER database. BMC Cancer 2021, 21, 825. [Google Scholar] [CrossRef]
- Skaug, K.; Eide, G.E.; Gulsvik, A. Hospitalisation days in patients with lung cancer in a general population. Respir. Med. 2009, 103, 1941–1948. [Google Scholar] [CrossRef][Green Version]
- Xia, Y.; Ma, H.; Buckeridge, D.L.; Brisson, M.; Sander, B.; Chan, A.; Verma, A.; Ganser, I.; Kronfli, N.; Mishra, S.; et al. Mortality trends and length of stays among hospitalized patients with COVID-19 in Ontario and Québec (Canada): A population-based cohort study of the first three epidemic waves. Int. J. Infect. Dis. 2022, 121, 1–10. [Google Scholar] [CrossRef]
- Nazábal, A.; Olmos, P.M.; Ghahramani, Z.; Valera, I. Handling incomplete heterogeneous data using VAEs. Pattern Recognit. 2020, 107, 107501. [Google Scholar] [CrossRef]
- Le, M.K.; Oishi, N.; Vuong, H.G.; Kondo, T. Survival analyses of soft tissue pleomorphic sarcomas and a proposed leiomyosarcoma-specific dynamic nomogram: A large population-based study. Pathol.-Res. Pract. 2022, 237, 153999. [Google Scholar] [CrossRef] [PubMed]
- Balch, C.M.; Morrow, M.; Fritz, A.G.; Page, D.L.; Greene, F.L.; Haller, D.G.; Fleming, I.D. AJCC Cancer Staging Manual, 6th ed.; Springer: New York, NY, USA, 2013. [Google Scholar]
- Maharaj, R.; Raffaele, I.; Wendon, J. Rapid response systems: A systematic review and meta-analysis. Crit. Care 2015, 19, 254. [Google Scholar] [CrossRef] [PubMed]
- Majeed, J.; Chawla, S.; Bondar, E.; Chimonas, S.; Martin, S.C.; O’Sullivan, M.; Jones, D. Rapid Response Team Activations in Oncologic Ambulatory Sites: Characteristics, Interventions, and Outcomes. JCO Oncol. Pract. 2022, 18, e1961–e1970. [Google Scholar] [CrossRef] [PubMed]
- Stanzione, A.; Verde, F.; Romeo, V.; Boccadifuoco, F.; Mainenti, P.P.; Maurea, S. Radiomics and machine learning applications in rectal cancer: Current update and future perspectives. World J. Gastroenterol. 2021, 27, 5306–5321. [Google Scholar] [CrossRef]



| Variable | Train (n = 115,145) | Validation (n = 13,017) | Test (n = 395,797) | p-Value |
|---|---|---|---|---|
| Age (years) | 71 (2–85) | 71 (19–85) | 69 (0–90.1) | <0.001 |
| Gender | <0.001 | |||
| Women | 56,592 (49.1%) | 6411 (49.3%) | 169,427 (42.8%) | |
| Men | 58,553 (50.9%) | 6606 (50.7%) | 226,370 (57.2%) | |
| Race | <0.001 | |||
| AIAN 1 | 657 (0.6%) | 72 (0.6%) | 1757 (0.4%) | |
| API 2 | 11,176 (9.7%) | 1262 (9.7%) | 28,408 (7.2%) | |
| Black | 9882 (8.6%) | 1107 (8.5%) | 28,956 (7.3%) | |
| White | 93,237 (81.0%) | 10,557 (85.0%) | 336,046 (84.9%) | |
| Not reported | 193 (0.2%) | 19 (0.1%) | 630 (0.2%) | |
| Tumor size (cm) | 3.4 (0–60.0) | 3.5 (0.1–4.9) | 3.4 (0–60.0) | 0.494 |
| AJCC stage | <0.001 | |||
| Stage I | 35,057 (30.4%) | 3968 (30.5%) | 31,265 (24.0%) | |
| Stage II | 9976 (8.7%) | 1123 (8.6%) | 9246 (7.1%) | |
| Stage III | 25,315 (22.0%) | 2876 (22.1%) | 27,869 (21.4%) | |
| Stage IV | 44,797 (38.9%) | 5050 (38.0%) | 62,021 (47.6%) | |
| T stage | <0.001 | |||
| T1 | 35,394 (30.7%) | 3957 (30.4%) | 31,465 (26.2%) | |
| T2 | 35,094 (30.5%) | 4061 (31.2%) | 34,654 (28.8%) | |
| T3 | 16,047 (13.9%) | 1774 (13.6%) | 16,704 (13.9%) | |
| T4 | 28,610 (24.8%) | 3225 (24.8%) | 37,345 (31.1%) | |
| N stage | <0.001 | |||
| N0 | 54,856 (47.6%) | 6181 (47.5%) | 54,623 (43.7%) | |
| N1 | 10,863 (9.4%) | 1209 (9.3%) | 11,669 (9.3%) | |
| N2 | 36,268 (31.5%) | 4167 (32.0%) | 42,436 (34.0%) | |
| N3 | 13,158 (11.4%) | 1460 (11.2%) | 16,188 (13.0%) | |
| M stage | <0.001 | |||
| M0 | 70,348 (61.1%) | 7967 (61.2%) | 75,528 (55.0%) | |
| M1 | 44,797 (38.9%) | 5050 (38.8%) | 61,740 (45.0%) | |
| Surgery | <0.001 | |||
| No | 80,031 (69.5%) | 9076 (69.7%) | 169,300 (42.8%) | |
| Yes | 35,114 (30.5%) | 3941 (30.3%) | 226,497 (57.2%) | |
| In-hospital mortality | <0.001 | |||
| No | 100,360 (87.2%) | 11,302 (86.8%) | 311,582 (80.6%) | |
| Yes | 14,785 (12.8%) | 1715 (13.2%) | 74,905 (19.4%) | |
| Survival time (month) | 12 (0–191) | 12 (0–191) | 8 (0–539) | 0.494 |
| Variable | Univariate | Multivariate | ||||
|---|---|---|---|---|---|---|
| OR 1 | 95%CI 2 | p-Value | OR | 95%CI | p-Value | |
| Age (years) | 1.04 | 1.04–1.04 | <0.001 | 1.05 | 1.05–1.05 | <0.001 |
| Gender | ||||||
| Female | 1.00 | |||||
| Male | 1.30 | 1.26–1.35 | <0.001 | 1.20 | 1.16–1.25 | <0.001 |
| Race | ||||||
| AIAN * | 1.00 | |||||
| Asian | 1.12 | 0.87–1.45 | 0.390 | |||
| Black | 1.28 | 0.99–1.66 | 0.056 | |||
| White | 0.62 | 0.33–1.14 | 0.122 | |||
| Not reported | 1.27 | 0.99–1.63 | 0.059 | |||
| Tumor size (cm) | 1.16 | 1.15–1.16 | <0.001 | 1.07 | 1.06–1.07 | <0.001 |
| T stage | ||||||
| T1 | 1.00 | 1.00 | ||||
| T2 | 2.15 | 2.04–2.28 | <0.001 | 1.18 | 1.10–1.25 | <0.001 |
| T3 | 3.39 | 3.19–3.61 | <0.001 | 1.28 | 1.19–1.37 | <0.001 |
| T4 | 5.09 | 4.83–5.37 | <0.001 | 1.64 | 1.54–1.76 | <0.001 |
| N stage | ||||||
| N0 | 1.00 | 1.00 | ||||
| N1 | 1.70 | 1.59–1.81 | <0.001 | 1.00 | 0.93–1.08 | 0.950 |
| N2 | 3.00 | 2.88–3.12 | <0.001 | 1.23 | 1.17–1.30 | <0.001 |
| N3 | 2.87 | 2.72–3.03 | <0.001 | 1.09 | 1.03–1.16 | 0.005 |
| M stage | ||||||
| M0 | 1.00 | n/a | n/a | |||
| M1 | 5.58 | 5.36–5.80 | <0.001 | n/a | n/a | |
| AJCC stages | ||||||
| I | 1.00 | 1.00 | ||||
| II | 1.99 | 1.78–2.23 | <0.001 | 1.59 | 1.41–1.80 | <0.001 |
| III | 4.07 | 3.77–4.40 | <0.001 | 2.48 | 2.26–2.72 | <0.001 |
| IV | 12.25 | 11.43–13.14 | <0.001 | 8.20 | 7.58–8.97 | <0.001 |
| Variable | Score |
|---|---|
| Age (years) | 2.5 scores per 10 years |
| Gender | |
| Women | 0 |
| Men | 1 |
| Tumor size (cm) | 3 scores per 10 cm |
| AJCC stage | |
| Stage I | 0 |
| Stage II | 2.5 |
| Stage III | 4.5 |
| Stage IV | 10.5 |
| T stage | |
| T1 | 0 |
| T2 | 0.7 |
| T3 | 1.2 |
| T4 | 2.5 |
| N stage | |
| N0-N1 | 0 |
| N2-N3 | 1 |
| Risk group | Total score |
| Low-risk | <26 |
| High-risk | ≥26 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tran, Q.N.N.; Le, M.-K.; Kondo, T.; Moriguchi, T. A Machine Learning-Based Model to Predict In-Hospital Mortality of Lung Cancer Patients: A Population-Based Study of 523,959 Cases. Adv. Respir. Med. 2023, 91, 310-323. https://doi.org/10.3390/arm91040025
Tran QNN, Le M-K, Kondo T, Moriguchi T. A Machine Learning-Based Model to Predict In-Hospital Mortality of Lung Cancer Patients: A Population-Based Study of 523,959 Cases. Advances in Respiratory Medicine. 2023; 91(4):310-323. https://doi.org/10.3390/arm91040025
Chicago/Turabian StyleTran, Que N. N., Minh-Khang Le, Tetsuo Kondo, and Takeshi Moriguchi. 2023. "A Machine Learning-Based Model to Predict In-Hospital Mortality of Lung Cancer Patients: A Population-Based Study of 523,959 Cases" Advances in Respiratory Medicine 91, no. 4: 310-323. https://doi.org/10.3390/arm91040025
APA StyleTran, Q. N. N., Le, M.-K., Kondo, T., & Moriguchi, T. (2023). A Machine Learning-Based Model to Predict In-Hospital Mortality of Lung Cancer Patients: A Population-Based Study of 523,959 Cases. Advances in Respiratory Medicine, 91(4), 310-323. https://doi.org/10.3390/arm91040025






