Abstract
Fluorescence labeling of gold-aryl nanoparticles, AuNPs-COOH, was achieved by the covalent derivatization with dansyl chloride (DNS-Cl) reagent (5-naphthalene-1-sulfonyl chloride) for potential ssDNA recognition. The fluorescent gold nanoparticles of AuNPs-C6H4-4-COO-dansyl (AuNPs-DNS) of spherical shape and a size of 19.3 ± 8.3 nm were synthesized in a carbonate-bicarbonate buffer (pH = 10.6) at 37 °C. The fluorescence emission at 475 nm was acquired using fluorescence spectroscopy and investigated using time-resolved photoluminescence. The conjugation of ssDNA to AuNPs-DNS using the freeze-thaw and salt-aging methods was confirmed by fluorescence emission quenching, gel electrophoresis separation, and lifetime decrease. Conjugated ssDNA to AuNPs-DNS using the freeze-thaw method was more efficient than the salt-aging method. The purity of ssDNA upon conjugation was measured with optical density, and the obtained A260/A280 ratio was in the range of 1.7–2.0. This research can be applied to other nucleotide recognition and theranostics.
1. Introduction
The significance of the surface derivatization of metal nanoparticles using covalent routes has been increasing rapidly because of the crucial role of the obtained nanostructures in biomedical applications [1,2,3,4,5,6]. AuNP surface modifications are generally achieved using thiols, amines, phosphines, silica, and biomolecules [7]. These processes follow either covalent-based modifications or noncovalent interactions. Organothiols, disulfides, cysteine-based, and Au-S-bonded gold nanoparticles are most extensively investigated due to their chemically inert outer surface hydrocarbon tails. Several reports have shown the surface derivatization of these thiol ligands through terminal carboxylate and amine functional moieties [7,8]. However, disadvantages in gold-thiolate interfaces originating from their decreased longevity under harsh conditions restrict their role in a wide range of applications [9,10], hence, the construction of robust gold-carbon interfaces has attracted great interest [11]. Aryl-functionalized gold nanosurfaces originating from diazoniums provide immense stability due to covalent Au-C bonds in various solvents and under harsh conditions. Nevertheless, their fluorescence applications are not well explored due to their innate inability to generate fluorescence. Dansyl chloride (DNS-Cl) (DNS-Cl = 5-naphthalene-1-sulfonyl chloride) is a widely used sulfonyl compound as a fluorescent probe in immunofluorescence assays to modify amino acids and proteins via coupling to amine and carboxyl functional groups. Dansylation of carboxyl, amine, and hydroxyl functional groups has shown green fluorescence emission upon coupling [12,13,14,15,16]. The derivatization using DNS-Cl has been established to determine minute amounts of peptides and proteins, understand the functionalized interface layers of polymers, and detect oxygen-containing functional groups [14,16]. Recently, derivatization has been extended to nanocomposites and nanomaterials. The dansyl-labeled AgNPs@SiO2 nanocomposites showed enhanced fluorescence upon derivatization using the amine functional group of aminopropyltrimethoxysilane [17]. The fluorescence of DNS glutathione-capped AuNPs was used for the detection of bovine serum albumin protein [18]. Another dansyl-derivatized AgNP containing 2-aminothiophenyl functional groups was synthesized [19].
Recently, aryldiazonium gold(III) salts reduced gold-aryl NPs have been widely recognized [1,2]. Noncovalent surface modification of gold-aryl NPs is described for the immobilization of proteins, drugs, and DNA [20,21,22]. Recognition of DNA via diazonium routes has been described in a few reports on electrochemical grafted salts on various electrodes. Generally, DNA is functionalized with a diazonium salt, electrochemically grafted, and finally recognized by electrochemistry or fluorescence [23,24]. However, there is no single report on the recognition of DNA over fluorescent gold-aryl NPs. Earlier described DNA recognition over gold-aryl NPs using a polyelectrolyte method [21]. Polydiallyldimethylammonium chloride (PDADMAC) is a biocompatible cationic polyelectrolyte that is used in combination with AuNPs to enhance their cellular uptake and biocompatibility [21]. DNA was stabilized and protected against nuclease degradation over gold-aryl NPs in the presence of the positive polyelectrolyte PDADMAC [21].
An example of the bio/nano interface is the interaction between single-stranded DNA and gold nanoparticles (AuNPs). Aptamer-gold nanoparticle conjugates are effectively utilized as a calorimetric detection tool for several target molecules [25,26,27]. The physical features of AuNPs have been linked to the molecular recognition and programmability of DNA. Hence, nanomedicine [28] and nanotechnology [29] have all seen significant advancements, and supporting this information, there are two recent methods developed in the recognition of DNA over AuNPs. In the first method, using freezing-thaw, a reagent-less conjugation of ssDNA with AuNPs consumes 2–60 min depending on freezing speeds [30,31]. The freeze-thaw method is time-efficient and simple. In the second method, using salt-aging, DNA was mixed with AuNPs. This involves a particular concentration of salt, such as sodium chloride, which is mixed with AuNPs and incubated overnight with continuous agitation to increase the number of recognized DNA strands [32,33,34]. Concerning previous studies, the conjugates displayed different colors after freeze-thawing, in which 13 nm citrate-capped AuNPs (cit-AuNPs) were red, and as expected, the red color turned blue upon freezing and remained blue after thawing [30], and both the methods still retained their stability [29,32].
Gold nanoparticles exhibit strong surface plasmon resonance when illuminated with visible light. The origin of this absorption can be simply described as a collective resonant oscillation of the free electrons in the conduction band of the gold surface [35]. Plasmons in gold nanoparticles with a size much smaller than the incident light wavelength are nonpropagating-excitations and termed localized surface plasmons (LSPs), and photons with a frequency in resonance with the oscillation can excite the LSPs. The resultant oscillation of such plasmons with incident light is responsible for the absorption band [36]. Meanwhile, the photoluminescent properties of nanoscale gold clusters are well known and critically important in many applications. The interband electronic transitions from s to p and radiative recombination to d band holes of the photo-excited nanoclusters are responsible for PL characteristics [37]. However, the quantum confinement effects on localized surface states govern gold nanoparticles’ luminescence nature. Although gold nanoparticles have been extensively used in colorimetric applications, their fluorescence applications are not well explored due to their innate inability to generate fluorescence. In the present work, we utilized dansyl chloride to surface coordinate with the carboxylate functional group on post-synthetic AuNPs-COOH, thus, generating fluorescent gold-aryl nanoparticles (AuNPs-dansyl). This work emphasizes the derivatization of gold-aryl NPs functionalized with carboxylic functional groups (AuNPs-COOH) using DNS-Cl in basic media to form AuNPs-C6H4-4-COO-dansyl. Dansyl chloride reacted with the carboxyl functional groups to generate intense green fluorescence under UV light. This surface reaction extends the possibility of a new realm of post-synthetic modification of gold-aryl nanoparticles. Essentially, modifying the surface of AuNPs with a tunable photophysical probe provides attractive features. The synthesized fluorescent AuNPs-DNS were further utilized for the evaluation of ssDNA conjugation using two methods: (1) freeze-thaw and (2) salt-aging. Moreover, this surface reaction extends the possibility of a new post-synthetic modification of gold-aryl NPs.
2. Experimental Methods
2.1. Chemicals
Sodium nitrite, hydrochloric acid (36.5–38.0%), 4-aminobenzoic acid, dansyl chloride, NaCl, agarose, glycerol, EDTA disodium salt, Tris, and glacial acetic acid were purchased from Sigma-Aldrich. HAuCl4 was synthesized in our lab. Sodium borohydride was purchased from Fisher Chemicals. Sodium carbonate and sodium bicarbonate were purchased from SDS Fine. The ssDNA sequences of P1-ACGATCAGATACCGTCGTAATCTT and P2-GAACCCAAAGACTTTGATTTCTCAT were purchased from Alpha DNA, Montreal, QC, Canada.
2.2. Instruments
The synthesized AuNPs-DNS were characterized with a high-resolution transmission electron microscope (HR-TEM, Field Emission JEM-2100F, JEOL, Akishima, Japan). For the analyses, the nanoparticles in the water/diethyl ether mixture were immobilized on a TEM grid of FORMVAR FCF 400 Cu (Electron Microscopy Sciences, Hatfield, PA, USA). The size of nanoparticles was measured using the ImageJ program. UV-visible absorption spectra were measured using a scanning spectrophotometer (Spectro UV-2510TS, Labomed Inc., Los Angeles, CA, USA) in the 200–800 nm range with 2 nm resolution.
Fluorescence emission spectra were recorded on Shimadzu RF-6000 Spectrophotometer. Lifetime data were obtained in the time-domain using the time-correlated single-photon counting (TCSPC) method using a picosecond pulsed diode laser (Edinburgh instruments) of 406.2 nm and a pulse width of 53.2 ps. Fluorescence lifetime decays were analyzed using a 2-exponential model, which is a good fit to the fluorescence decay curve using the least-squares method of fit analysis with a χ2 value of ~1.0. Fluorescence lifetime decays were analyzed using F980 software. Attenuated total reflectance Fourier transform infrared (ATR-FTIR) spectra were recorded on a Bruker (Platinum ATR) Tensor II FT-IR spectrophotometer. X-ray powder diffraction (XRD) data were collected using Bruker D8 ADVANCE diffractometer with Cu-Kα X-ray source at λ = 1.5406 Å operating at 40 kV tube voltage and 40 mA current.
A Renishaw Ramascope was used for Raman measurements of solid samples. All Raman measurements were collected at laser excitation wavelengths of 785 nm in the range of 200–3000 cm−1. ζ-potential analyses of the nanoparticles in solution were carried out using Anton Paar Litesizer 500 and analyzed using KalliopeTM software. Nanodrop Thermo Scientific 2000, was used to quantify the AuNPs-DNS coupled DNA, which was then separated by BioRad DNA Gel Electrophoreses and captured with Gel Doc EZ System, Biorad Laboratories.
2.3. Synthesis
2.3.1. Synthesis of AuNPs-COOH
Water-soluble aryldiazonium gold(III) salt [HOOC-4-C6H4N≡N]AuCl4 was synthesized following our published procedure [38]. The gold-aryl NPs were synthesized by reducing the salt using 0.1 M NaBH4. Briefly, 0.48 g of the salt was dissolved in 50 mL of DI water, and 0.1 M NaBH4 was added dropwise to the solution with stirring (>300 rpm). The resulting purple NPs were purified using a 20-cm long cellulose membrane dialysis tubing with a 25 mm width in 200 mL of DI water overnight at room temperature and stored in the fridge for further use.
2.3.2. Synthesis of AuNPs-DNS
Briefly, 5.0 mL of AuNPs-COOH colloidal solution, in a glass vial, was added to an equal volume of 1.0 M Na2CO3-NaHCO3 buffer (pH 10.6). The solution was stirred for a few minutes, and then a 1/4 volume of DNS-Cl (5 mg/mL) in acetone was added and mixed well by vigorous shaking. Finally, the mixture was placed in the dark at 37 °C for 1.5 h. The AuNPs-DNS formed a beige color with intense green fluorescence under UV light and were then stored in the fridge for further use.
2.4. Methods of ssDNA Conjugation with AuNPs-DNS
2.4.1. Freeze-Thaw Method
The ssDNA of 100 μM and AuNPs-DNS were mixed and frozen at −20 °C for 2 h followed by −80 °C for 1 h. The freezing conditions were effective within 1 h. Subsequently, the experiment was continued at −80 °C since −20 °C was not suitable for the conjugation of DNA with AuNPs-DNS.
2.4.2. Salt-Aging Method
In the salt-aging method, AuNPs-DNS were mixed with 300 mM NaCl and 100 μm of each DNA strand, followed by continuous agitation for 14–16 h or overnight incubation.
After conjugation, using both methods, the conjugates were separated from the unconjugated AuNPs-DNS by centrifugation at 14,000 RPM for 15 min. The pellets were washed 3 times using 5 mM phosphate buffer (pH 7) and resuspended with the same buffer for further analysis. Nanodrop (260/280 nm absorbance) was used to verify the purity and concentration of AuNPs-DNS/ssDNA conjugates. The conjugates were combined with an equal volume of glycerol and were run for 30 min at 100 volts using 1.5% agarose gel. Bands were visualized using the Biorad Laboratories Gel Doc EZ.
3. Results and Discussion
3.1. Synthesis and Characterization
AuNPs-DNS was synthesized by the covalent derivatization of AuNPs-COOH using DNS-Cl under a basic pH of 10.6 at 37 °C, Figure 1A. Dansylation requires basic media and the pH of Na2CO3-NaHCO3 buffer provides the required pH for the reaction to proceed to completion.
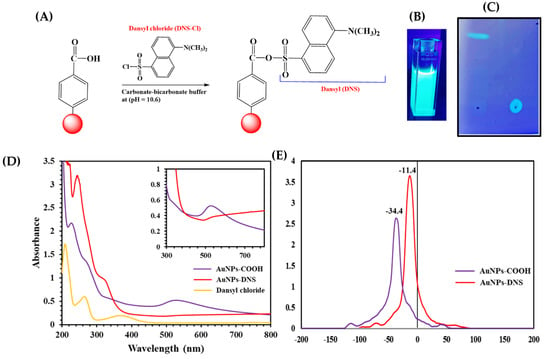
Figure 1.
(A) Schematic representation showing facile surface derivatization of AuNPs-COOH using DNS-Cl. (B) Cuvette of AuNPs-DNS under UV light, (C,D) UV-visible spectra showing AuNPs-COOH, DNS-Cl, and AuNPs-DNS. (E) Thin-layer chromatography (TLC) for dansyl chloride and AuNPs-DNS UV light irradiation.
Literature examples described the dansylation of aromatic carboxylic acid groups to form esters and primary amines to form amides, which were completed in Na2CO3-NaHCO3 at pH 11 to form fluorescent derivatives under UV light [12,13,14,15,16]. The coupling of SO2Cl with -COOH functional groups resulted in the ester formation of -COO-SO2-. The literature presented two routes for the coupling of DNS with the organic shell instilled on the nanoparticles. First, in the synthesis of Ag@SiO2 NPs/DNS, the organic silicon shell attached to Ag@SiO2-linker-NH2 was derivatized with DNS [17]. In the second route for the synthesis of AuNPs-glutathione-DNS, sodium borohydride was added to form the AuNPs core in the presence of the already synthesized glutathione-DNS organic shell. The pH was kept at <1 to form a superlattice [18]. In our synthesis route, AuNPs modified with the aryl-COOH shell were synthesized first, followed by DNS coupling with the carboxylic functional groups to generate fluorescent AuNPs-DNS, Figure 1B and Figure S1. Our route avoided the possible hydrolysis of the ester linkage if sodium borohydride was used in the reduction of the aryldiazonium gold(III) precursor.
Thin-layer chromatography (TLC) was used to confirm the complete conversion of carboxylic acid to the ester upon dansylation. Initial assessment of successful dansylation of AuNPs was detected using a TLC silica sheet, which showed a bright green fluorescence under short and long UV light, Figure 1C. Previous reports on dansylation indicated successful product separation using TLC [12,13]. Glutathione capped AuNPs showed terminal amino acid conjugation using TLC [39]. Our samples were spotted on TLC plates and were resolved using CH3OH: CHCl3 (1:2) solvent mixtures. Separation using TLC revealed that DNS-Cl traveled far up the plate while AuNPs-DNS was immobile.
UV-visible spectra showed plasmon peaks of the parent AuNPs-COOH and AuNPs-DNS at 530 nm and a broad peak starting at 545 nm, Figure 1D. Gold plasmon peak broadening upon derivatization indicates increased particle size. The proposed reaction corroborates this observation as the covalent bonding increased particle size. In addition, dansyl chloride (in ethanol) spectra resulted in three maximum peaks of absorbance at 375, 267, and 210 nm, and the AuNPs-DNS (in water) showed the maxima at 329 and 246 nm. The shift in values is due to solvent media affecting electronic absorption spectra [40]. The surface charges of the nanoparticles were probed using zeta-potential (ζ-potential). The surface charges of the nanoparticles were probed using zeta-potential (ζ-potential). Generally, for AuNPs-COOH, the ζ-potential ranges from −25 to −35 mV, which is indicative of a stable colloidal suspension due to electrostatic repulsion. Here, we observed the neutralization of charge from −34.4 mV for AuNPs-COOH to −11.4 mV for AuNPs-dansyl. Possibly, this induces the aggregation of the particles and indicates the bulk arylation on the gold surface, Figure 1E.
ATR-FTIR measurements of AuNPs-COOH and AuNPs-DNS indicated the presence of a strong carboxyl C=O band at 1638 and 1647 cm−1, whereas the C=O band at 1697 cm−1 was observed only in AuNPs-DNS. Dansyl chloride vibrational bands, corresponding to ring deformation at 1371 cm−1, υ CC + β CHring at 1236 cm−1, and r CH3 + υsym SO2 + υ CC at 1093 cm−1, were observed in AuNPs-DNS spectra, Figure S2 [40,41]. Raman spectra acquired using 785 nm laser excitation confirmed the dansylation of AuNPs-COOH. The bands at 600, 1076, and 1610 cm−1 were assigned to ring deformation in the aryl shell of AuNPs-COOH. A characteristic band at 1610 cm−1 assigned to the C=O functional group in AuNPs-COOH was observed as a shoulder with a υ C-C band at 1567 cm−1 in AuNPs-DNS. Dansyl chloride spectra showed υ CC + β CHring band at 1409 cm−1, ring deformation band at 1355 cm−1, and γ CCC band at 580 cm−1, which were also observed in AuNPs-DNS, Figure 2A [40,41]. Similar stretching vibration assignments based on FTIR and Raman measurements were assigned to 4-iodobenzene sulfonyl chloride using experimental and theoretical analysis [42].
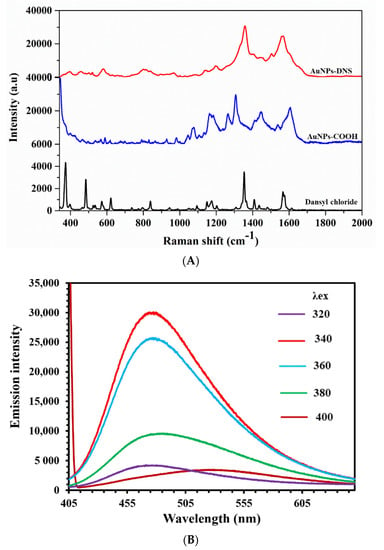
Figure 2.
(A) Raman spectra at λex = 785 nm for dansyl chloride, AuNPs-COOH, and AuNPs-DNS. (B) Fluorescence emission spectra of AuNPs-DNS at different excitation wavelengths.
Excitation of AuNPs-DNS at 340 nm displayed a fluorescence emission peak at 475 nm, Figure 2B. The intense green fluorescence of AuNPs-DNS distinguishes it from the DNS-Cl ligand, Figure S3. The obtained lifetime values for DNS-Cl were as short as τ1 of 0.12 ns and τ2 of 0.54 ns, whereas longer lifetime values of τ1 of 0.89 ns and τ2 of 10.37 ns for AuNPs-DNS were observed, Figure S4 and Table S1. The τ2 value of AuNPs-DNS significantly increased due to dansylation.
TEM images show the morphology of the synthesized AuNPs-DNS, Figure 3. They are generally spherical, with a size of 19.3 ± 8.3 nm. The measured lattice spacing of 0.20 nm, which corresponds to the (200) plane of metallic gold, indicates that aryldiazonium gold(III) salt was completely reduced by NaBH4 to AuNPs, and also, dansylation did not alter the spherical morphology of the parent AuNPs. In our previous studies, spherical AuNPs were also synthesized using aryldiazonium gold(III) salts [38]. The P-XRD pattern showed characteristic diffraction peaks for face-centered cubic (fcc) AuNPs reflections (PDF Card #4-784) at 2θ = 38.1° and 44.1°, Figure S5.
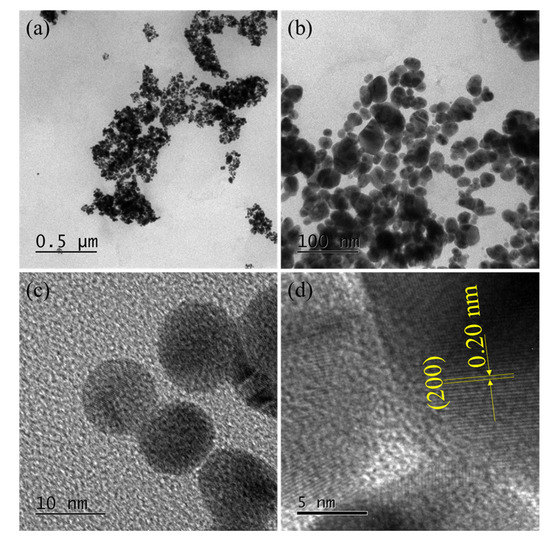
Figure 3.
TEM images of AuNPs-DNS. (a,b) AuNPs with low magnification and (c,d) AuNPs with high magnification.
3.2. Freeze-Thaw and Salt-Aging Method for ssDNA Conjugation
The conjugation of ssDNA with the fluorescent AuNPs-DNS using freeze-thaw and salt-aging methods is described. In both methods, the fluorescence emission from AuNPs-DNS was weakened after ssDNA conjugation, as revealed by the concentration and gel electrophoresis of the conjugates. The freeze-thaw method displayed weaker fluorescence emission than the salt-aging method, as shown in Figure 4A,B.
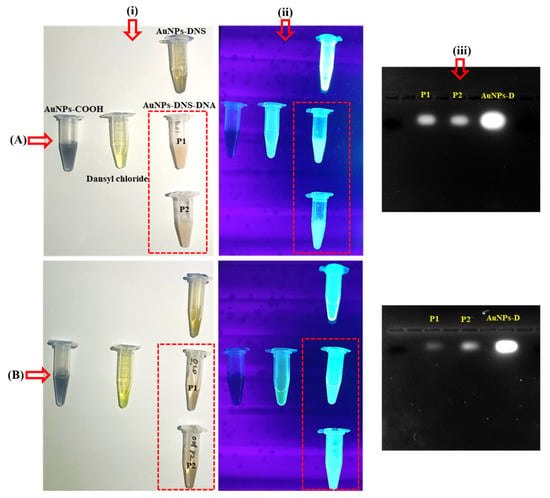
Figure 4.
Images of samples after ssDNA conjugation with AuNPs-DNS. (A) Freeze-thaw method and (B) salt-aging method (i) daylight images, (ii) UV light images, and (iii) separation of conjugated DNA by gel electrophoresis.
Comparing the results of ssDNA quantification using nanodrop, which describes the concentration of conjugated DNA for each oligo per microliter, the study found that P1 was higher than P2, Figure 5A. The purity of ssDNA upon conjugation was measured by optical density, and the obtained A260/A280 ratio was in the range of 1.7–2.0. Overall, we noticed an increase in the concentration in the freeze-thaw method over the salt-aging method, and DNA purity was nearly 1.8, Figure 5B. This clearly explains the good quality ssDNA in the freeze-thaw method, however, it is lower in the salt-aging method”. Additionally, the fluorescence of the samples in daylight and under UV light indicates that the intensity is much lower in the freeze-thaw method than in the salt-aging method.
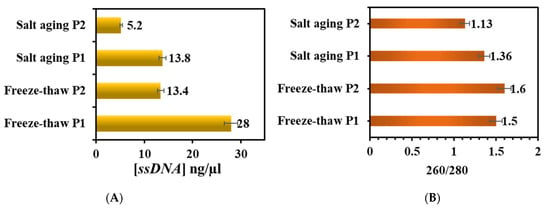
Figure 5.
Optical density measurements showing ssDNA (A) concentration and (B) purity values upon immobilization on AuNPs-dansyl.
Finally, to validate the ssDNA recognition, the samples were run on an agarose gel, which revealed that the band intensities were lower in both processes compared with AuNPs-DNS, but higher in the salt-aging method. P2 showed lower intensity, which supports the 260/280 ratio, Figure 4(iii).
Literature work showed that chloride anions were adsorbed on the surface of AuNPs and decreased DNA conjugation. This competition explains the poor DNA conjugation in the presence of NaCl [43]. Dansyl chloride is a powerful fluorescent agent that can be used to identify the amino terminals of peptide chains. Dansyl chloride can specifically react with the chain’s N-terminal alpha-amino group to produce dansyl-peptide, and the dansyl-amino acid produced by hydrolyzing the dansyl-peptide also has powerful fluorescent properties. Having these properties, dansyl chloride has been used in the application of sequencing for a long time due to its susceptibility to hydrolysis by water and hydroxyl ions [14]. In addition, the salt-aging method is less reliable since it requires a long time and validation, and the results depend on the oligos properties [34,44,45] to achieve an efficient AuNPs-DNS-ssDNA-conjugation without aggregation. A procedure was developed which involved continuous additions and progressive increases in salt concentrations over two days [44]. For example, multilayer-coated GNPs were prepared by sequential modification of GNPs with PEI, a cationic polymer, and finally with antisense-oligonucleotides to target the mecA gene and to suppress methicillin resistance of methicillin-resistant S. aureus [46]. To further support how important the GNPs’ properties with DNA conjugation are, a study was conducted on porous-GNPS and the steric repulsion between 20 layers around the particles, keeping the porous GNPs relatively stable. Furthermore, when compared to GNPs and Tween GNPs, porous GNPs showed a considerable increase in DNA loading [47]. DNA hybridized to GNPs has undergone kinetic and thermodynamic experiments, which revealed that ssDNA first adheres to the GNPs before gradually diffusing over their surface. A DNA hairpin’s secondary structure prevents GNPs and DNA from interacting with each other, immediately enhancing the stability of the attached DNA. This characteristic can be applied to the targeted delivery of specific cancer cells, provided AuNPs are attached with respective nucleic acid sequences. For example, a study was conducted on prostate cancer by attaching GNPs to anti-PSMA antibodies by using an oligonucleotide that was complementary to the sequence of the anti-PSMA (prostate-specific membrane antigen) [48]. Another innovative study conducted by Chang et al., 2006, used DNA functionalized GNPs to create a chip-based DNA biobarcode sensor to find target DNA sequences. In the case of biosensors or biobarcoding, the target DNA can be assessed using complementary DNA attached to GNPs. DNA is assessed using complementary DNA attached to GNPs and subsequent detection of the amplified DNA instead of the original target DNA [49].
3.3. Assessment of Fluorescence from AuNPs-DNS/ssDNA Conjugates
Fluorescence emission intensities were compared for the two ssDNA primers conjugated with AuNPs-DNS. In the presence of both P1 and P2 sequences, the fluorescence emission intensity of ssDNA sequences was significantly quenched and redshifted, Figure 6A,B. AuNPs-DNS showed an excitation wavelength at 340 nm, corresponding to well-resolved fluorescence emission at 475 nm. In addition, ssDNA conjugation resulted in an excitation shift to 320 nm and a fluorescence emission maximum at 515 nm. Previous reports demonstrated the use of fluorescence emission quenching in assessing ssDNA conjugation as a specific hybridization of ssDNA-modified AuNPs with complementary oligonucleotides [50]. Fluorescence emission quenching and shifting perhaps reflected the efficient interactions of ssDNA with AuNPs-DNS.
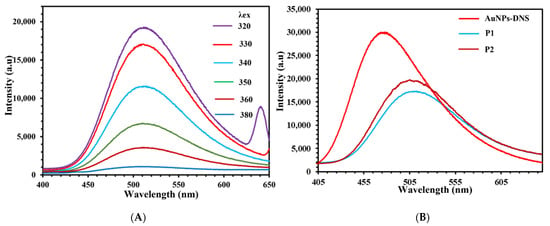
Figure 6.
Fluorescence emission spectra of (A) AuNPs-DNS + ssDNA (P1) at different excitation wavelengths, (B) peak shifts for AuNPs-DNS upon P1 and P2 immobilization.
Fluorescence lifetimes of AuNPs-DNS/ssDNA bioconjugates were measured to probe the ssDNA recognition. The observed biexponential decay suggests that the addition of both P1 and P2 ssDNA sequences remarkably reduced the lifetime of the AuNPs-DNS fluorophores, Figure S6. The excited-state lifetime of AuNPs-DNS decreased from 10.37 to 2.7 ns for the salt-aging method and 0.6 ns for the freeze-thaw method, Table S2. The decreased lifetime of one of the decay components supports the observed quenching [51,52]. There are different quenching mechanisms proposed for nanoparticle-quencher interactions, such as non-radiative recombination, energy and electron transfer, and surface adsorption. Factors including spectral overlap, the orientation of the transition dipoles, and donor-acceptor distance matter in the rate of energy transfer between donor-acceptors [53]. Time-resolved fluorescence lifetime measurements were used to validate the quenching mechanisms of ssDNA interactions with AuNPs-DNS. After the addition of ssDNA, the excited-state lifetime of AuNPs-DNS showed a dynamic quenching since the excited-state quenching and fluorescence lifetime values were changed remarkably [52,53,54,55].
4. Conclusions
Fluorescent gold nanoparticles of AuNPs-DNS were synthesized and characterized. The dansylation of gold-aryl nanoparticles in a basic medium resulted in fluorescent gold nanoparticles. We successfully conjugated ssDNA with AuNPs-DNS with two methods, viz., freeze-thaw and salt-aging. The freeze-thaw method was more efficient than the salt-aging method for ssDNA conjugation to AuNPs-DNS. Fluorescence intensity was quenched by ssDNA conjugation. Fluorescent lifetime and optical density measurements showed a decreased exciton lifetime with good purity of ssDNA after conjugation. This method can be applied to DNA biosensing and related bioanalytical applications.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/colloids6030042/s1, Figure S1: Visible light and UV irradiated photographs; Figure S2: ATR-FTIR spectra; Figure S3: Fluorescent emission spectra; Figure S4: Time resolved photoluminescence measurements; Table S1: Time-resolved fluorescent lifetime measurements; Figure S5. P-XRD spectra; Figure S6: Time resolved photoluminescence measurements for ssDNA recognition; Table S2: Time-resolved fluorescent lifetime measurements
Author Contributions
Conceptualization, J.B.M.P. and G.A.K.; methodology, J.B.M.P. and G.A.K.; Validation, R.O.A.O., C.H. and S.K. formal analysis, R.O.A.O., C.H. and A.A.M.; investigation, J.B.M.P. and G.A.K.; resources, R.O.A.O., C.H. and A.A.M.; writing—original draft preparation, J.B.M.P. and G.A.K.; writing—review and editing, A.A.M.; supervision, R.O.A.O., C.H., A.A.M.; funding acquisition, C.H. and A.A.M. All authors have read and agreed to the published version of the manuscript.
Funding
AAM acknowledges the University of Sharjah support of competitive grants, 160-2142-029-P and 150-2142-017-P. CH acknowledges the support of the National Research Foundation of Korea (NRF), grant funded by the Korean government (MSIT) (2021R1A4A1032746 and 2021R1A2C1093183).
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Not applicable.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Ahmad, A.A.L.; Parambath, J.B.M.; Postnikov, P.S.; Guselnikova, O.; Chehimi, M.M.; Bruce, M.R.M.; Bruce, A.E.; Mohamed, A.A. Conceptual Developments of Aryldiazonium Salts as Modifiers for Gold Colloids and Surfaces. Langmuir 2021, 37, 8897–8907. [Google Scholar] [CrossRef] [PubMed]
- Li, D.; Luo, Y.; Onidas, D.; He, L.; Jin, M.; Gazeau, F.; Pinson, J.; Mangeney, C. Surface functionalization of nanomaterials by aryl diazonium salts for biomedical sciences. Adv. Colloid Interface Sci. 2021, 294, 102479. [Google Scholar] [CrossRef] [PubMed]
- Hetemi, D.; Noël, V.; Pinson, J. Grafting of Diazonium Salts on Surfaces: Application to Biosensors. Biosensors 2020, 10, 4. [Google Scholar] [CrossRef] [PubMed]
- Hameed, M.K.; Parambath, J.B.; Gul, M.T.; Khan, A.A.; Park, Y.; Han, C.; Mohamed, A.A. Arylated gold nanostars aided SERS study of breast cancer cells. Appl. Surf. Sci. 2022, 583, 152504. [Google Scholar] [CrossRef]
- Panicker, S.; Ahmady, I.; Han, C.; Chehimi, M.; Mohamed, A. On demand release of ionic silver from gold-silver alloy nanoparticles: Fundamental antibacterial mechanisms study. Mater. Today Chem. 2020, 16, 100237. [Google Scholar] [CrossRef]
- Sandomierski, M.; Voelkel, A. Diazonium Modification of Inorganic and Organic Fillers for the Design of Robust Composites: A Review. J. Inorg. Organomet. Polym. Mater. 2020, 31, 1–21. [Google Scholar] [CrossRef]
- Alex, S.; Tiwari, A. Functionalized Gold Nanoparticles: Synthesis, Properties and Applications—A Review. J. Nanosci. Nanotechnol. 2015, 15, 1869–1894. [Google Scholar] [CrossRef]
- Templeton, A.C.; Hostetler, M.J.; Kraft, A.C.T.; Murray, R.W. Reactivity of Monolayer-Protected Gold Cluster Molecules: Steric Effects. J. Am. Chem. Soc. 1998, 120, 1906–1911. [Google Scholar] [CrossRef]
- Shewchuk, D.M.; McDermott, M.T. Comparison of Diazonium Salt Derived and Thiol Derived Nitrobenzene Layers on Gold. Langmuir 2009, 25, 4556–4563. [Google Scholar] [CrossRef]
- Civit, L.; Fragoso, A.; O’Sullivan, C.K. Thermal stability of diazonium derived and thiol-derived layers on gold for application in genosensors. Electrochem. Commun. 2010, 12, 1045–1048. [Google Scholar] [CrossRef]
- Laurentius, L.; Stoyanov, S.R.; Gusarov, S.; Kovalenko, A.; Du, R.; Lopinski, G.P.; McDermott, M.T. Diazonium-Derived Aryl Films on Gold Nanoparticles: Evidence for a Carbon–Gold Covalent Bond. ACS Nano 2011, 5, 4219–4227. [Google Scholar] [CrossRef] [PubMed]
- Bartzatt, R. Dansylation of aromatic, aliphatic, and medicinal carboxylic acid compounds in 1 M Na2CO3 buffer. Anal. Chim. Acta 2003, 488, 203–209. [Google Scholar] [CrossRef]
- Bartzatt, R. Dansylation of hydroxyl and carboxylic acid functional groups. J. Biochem. Biophys. Methods 2001, 47, 189–195. [Google Scholar] [CrossRef]
- Gray, W.R. [8] End-group analysis using dansyl chloride. Methods Enzymol. 1972, 25, 121–138. [Google Scholar] [CrossRef] [PubMed]
- Holmes-Farley, S.R.; Whitesides, G.M. Fluorescence properties of dansyl groups covalently bonded to the surface of oxidatively functionalized low-density polyethylene film. Langmuir 1986, 2, 266–281. [Google Scholar] [CrossRef]
- Murgia, S.; Falchi, A.M.; Meli, V.; Schillén, K.; Lippolis, V.; Monduzzi, M.; Rosa, A.; Schmidt, J.; Talmon, Y.; Bizzarri, R.; et al. Cubosome formulations stabilized by a dansyl-conjugated block copolymer for possible nanomedicine applications. Colloids Surf. B: Biointerfaces 2015, 129, 87–94. [Google Scholar] [CrossRef]
- Szczepańska, E.; Synak, A.; Bojarski, P.; Niedziałkowski, P.; Wcisło, A.; Ossowski, T.; Grobelna, B. Dansyl-Labelled Ag@SiO2 Core-Shell Nanostructures—Synthesis, Characterization, and Metal-Enhanced Fluorescence. Materials 2020, 13, 5168. [Google Scholar] [CrossRef]
- Shibu, E.S.; Muhammed, M.A.H.; Kimura, K.; Pradeep, T. Fluorescent superlattices of gold nanoparticles: A new class of functional materials. Nano Res. 2009, 2, 220–234. [Google Scholar] [CrossRef]
- Tharmaraj, V.; Pitchumani, K. A highly selective ratiometric fluorescent chemosensor for Cu(ii) based on dansyl-functionalized thiol stabilized silver nanoparticles. J. Mater. Chem. B 2013, 1, 1962–1967. [Google Scholar] [CrossRef]
- Hameed, M.; Panicker, S.; Abdallah, S.H.; Khan, A.A.; Han, C.; Chehimi, M.M.; Mohamed, A.A. Protein-Coated Aryl Modified Gold Nanoparticles for Cellular Uptake Study by Osteosarcoma Cancer Cells. Langmuir 2020, 36, 11765–11775. [Google Scholar] [CrossRef]
- Panicker, S.; Ahmady, I.M.; Almehdi, A.M.; Workie, B.; Sahle-Demessie, E.; Han, C.; Chehimi, M.M.; Mohamed, A.A. Gold-Aryl nanoparticles coated with polyelectrolytes for adsorption and protection of DNA against nuclease degradation. Appl. Organomet. Chem. 2019, 33, e4803. [Google Scholar] [CrossRef]
- Bosak, A.; Saraf, N.; Willenberg, A.; Kwan, M.W.C.; Alto, B.W.; Jackson, G.W.; Batchelor, R.H.; Nguyen-Huu, T.D.; Sankarapani, V.; Parks, G.D.; et al. Aptamer–gold nanoparticle conjugates for the colorimetric detection of arboviruses and vector mosquito species. RSC Adv. 2019, 9, 23752–23763. [Google Scholar] [CrossRef] [PubMed]
- Saraf, N.; Bosak, A.; Willenberg, A.; Das, S.; Willenberg, B.J.; Seal, S. Colorimetric detection of epinephrine using an optimized paper-based aptasensor. RSC Adv. 2017, 7, 49133–49143. [Google Scholar] [CrossRef]
- Saraf, N.; Villegas, M.; Willenberg, B.J.; Seal, S. Multiplex Viral Detection Platform Based on a Aptamers-Integrated Microfluidic Channel. ACS Omega 2019, 4, 2234–2240. [Google Scholar] [CrossRef] [PubMed]
- Ahmady, I.M.; Hameed, M.K.; Almehdi, A.M.; Arooj, M.; Workie, B.; Sahle-Demessie, E.; Han, C.; Mohamed, A.A. Green and cytocompatible carboxyl modified gold–lysozyme nanoantibacterial for combating multidrug-resistant superbugs. Biomater. Sci. 2019, 7, 5016–5026. [Google Scholar] [CrossRef] [PubMed]
- Chung, D.-J.; Oh, S.-H.; Komathi, S.; Gopalan, A.I.; Lee, K.P.; Choi, S.-H. One-step modification of various electrode surfaces using diazonium salt compounds and the application of this technology to electrochemical DNA (E-DNA) sensors. Electrochim. Acta 2012, 76, 394–403. [Google Scholar] [CrossRef]
- Yang, L.; Xu, Y.; Wang, X.; Zhu, J.; Zhang, R.; He, P.; Fang, Y. The application of β-cyclodextrin derivative functionalized aligned carbon nanotubes for electrochemically DNA sensing via host–guest recognition. Anal. Chim. Acta 2011, 689, 39–46. [Google Scholar] [CrossRef]
- Giljohann, D.A.; Seferos, D.S.; Daniel, W.L.; Massich, M.D.; Patel, P.C.; Mirkin, C.A. Gold Nanoparticles for Biology and Medicine*. In Spherical Nucleic Acids; Jenny Stanford Publishing: Singapore, 2020; pp. 55–90. [Google Scholar] [CrossRef]
- Cutler, J.I.; Auyeung, E.; Mirkin, C.A. Spherical Nucleic Acids. J. Am. Chem. Soc. 2012, 134, 1376–1391. [Google Scholar] [CrossRef]
- Liu, B.; Liu, J. Freezing Directed Construction of Bio/Nano Interfaces: Reagentless Conjugation, Denser Spherical Nucleic Acids, and Better Nanoflares. J. Am. Chem. Soc. 2017, 139, 9471–9474. [Google Scholar] [CrossRef]
- Liu, B.; Liu, J. Freezing-Driven DNA Adsorption on Gold Nanoparticles: Tolerating Extremely Low Salt Concentration but Requiring High DNA Concentration. Langmuir 2019, 35, 6476–6482. [Google Scholar] [CrossRef]
- Storhoff, J.J.; Elghanian, R.; Mucic, R.C.; Mirkin, C.A.; Letsinger, R.L. One-Pot Colorimetric Differentiation of Polynucleotides with Single Base Imperfections Using Gold Nanoparticle Probes. J. Am. Chem. Soc. 1998, 120, 1959–1964. [Google Scholar] [CrossRef]
- Hurst, S.J.; Lytton-Jean, A.K.R.; Mirkin, C.A. Maximizing DNA Loading on a Range of Gold Nanoparticle Sizes. Anal. Chem. 2006, 78, 8313–8318. [Google Scholar] [CrossRef] [PubMed]
- Demers, L.M.; Mirkin, C.A.; Mucic, R.C.; Reynolds, R.A.; Letsinger, R.L.; Elghanian, R.; Viswanadham, G. A Fluorescence-Based Method for Determining the Surface Coverage and Hybridization Efficiency of Thiol-Capped Oligonucleotides Bound to Gold Thin Films and Nanoparticles. Anal. Chem. 2000, 72, 5535–5541. [Google Scholar] [CrossRef] [PubMed]
- Link, S.; El-Sayed, M.A. Optical Properties and Ultrafast Dynamics of Metallic Nanocrystals. Annu. Rev. Phys. Chem. 2003, 54, 331–366. [Google Scholar] [CrossRef]
- Amendola, V.; Pilot, R.; Frasconi, M.; Marago, O.M.; Iatì, M.A. Surface plasmon resonance in gold nanoparticles: A review. J. Phys. Condens. Matter 2017, 29, 203002. [Google Scholar] [CrossRef] [PubMed]
- Alluhaybi, H.; Ghoshal, S.; Alsobhi, B.; Shamsuri, W.W. Visible photoluminescence from gold nanoparticles: A basic insight. Optik 2019, 192, 162936. [Google Scholar] [CrossRef]
- Ahmad, A.A.L.; Panicker, S.; Chehimi, M.M.; Monge, M.; Lopez-De-Luzuriaga, J.M.; Mohamed, A.A.; Bruce, A.E.; Bruce, M.R.M. Synthesis of water-soluble gold–aryl nanoparticles with distinct catalytic performance in the reduction of the environmental pollutant 4-nitrophenol. Catal. Sci. Technol. 2019, 9, 6059–6071. [Google Scholar] [CrossRef]
- Harper, B.; Sinche, F.; Wu, R.H.; Gowrishankar, M.; Marquart, G.; Mackiewicz, M.; Harper, S.L. The Impact of Surface Ligands and Synthesis Method on the Toxicity of Glutathione-Coated Gold Nanoparticles. Nanomaterials 2014, 4, 355–371. [Google Scholar] [CrossRef]
- Karabacak, M.; Cinar, M.; Kurt, M.; Poiyamozhi, A.; Sundaraganesan, N. The spectroscopic (FT-IR, FT-Raman, UV and NMR) first order hyperpolarizability and HOMO–LUMO analysis of dansyl chloride. Spectrochim. Acta Part A Mol. Biomol. Spectrosc. 2014, 117, 234–244. [Google Scholar] [CrossRef]
- Socrates, G. Infrared Characteristics Group Frequencies; John Wiley and Sons: New York, NY, USA, 1980. [Google Scholar]
- Arivazhagan, M.; Prabhakaran, S.; Gayathri, R. Molecular structure, vibrational spectroscopic, first hyperpolarizability, NBO and HOMO, LUMO studies of P-Iodobenzene sulfonyl chloride. Spectrochim. Acta Part A Mol. Biomol. Spectrosc. 2011, 82, 332–339. [Google Scholar] [CrossRef]
- Bell, S.E.J.; Sirimuthu, N.M.S. Surface-Enhanced Raman Spectroscopy (SERS) for Sub-Micromolar Detection of DNA/RNA Mononucleotides. J. Am. Chem. Soc. 2006, 128, 15580–15581. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Servos, M.R.; Liu, J. Instantaneous and Quantitative Functionalization of Gold Nanoparticles with Thiolated DNA Using a pH-Assisted and Surfactant-Free Route. J. Am. Chem. Soc. 2012, 134, 7266–7269. [Google Scholar] [CrossRef] [PubMed]
- Xu, Q.; Lou, X.; Wang, L.; Ding, X.; Yu, H.; Xiao, Y. Rapid, Surfactant-Free, and Quantitative Functionalization of Gold Nanoparticles with Thiolated DNA under Physiological pH and Its Application in Molecular Beacon-Based Biosensor. ACS Appl. Mater. Interfaces 2016, 8, 27298–27304. [Google Scholar] [CrossRef]
- Beha, M.J.; Ryu, J.S.; Kim, Y.S.; Chung, H.J. Delivery of antisense oligonucleotides using multi-layer coated gold nanoparticles to methicillin-resistant S. aureus for combinatorial treatment. Mater. Sci. Eng. C 2021, 126, 112167. [Google Scholar] [CrossRef] [PubMed]
- Hakimian, F.; Ghourchian, H. Simple and rapid method for synthesis of porous gold nanoparticles and its application in improving DNA loading capacity. Mater. Sci. Eng. C 2019, 103, 109795. [Google Scholar] [CrossRef]
- Javier, D.J.; Nitin, N.; Levy, M.; Ellington, A.; Richards-Kortum, R. Aptamer-Targeted Gold Nanoparticles as Molecular-Specific Contrast Agents for Reflectance Imaging. Bioconj. Chem. 2008, 19, 1309–1312. [Google Scholar] [CrossRef]
- Chang, T.-L.; Tsai, C.-Y.; Sun, C.-C.; Uppala, R.; Chen, C.-C.; Lin, C.-H.; Chen, P.-H. Electrical detection of DNA using gold and magnetic nanoparticles and bio bar-code DNA between nanogap electrodes. Microelectron. Eng. 2006, 83, 1630–1633. [Google Scholar] [CrossRef]
- Lapiene, V.; Kukolka, F.; Kiko, K.; Arndt, A.; Niemeyer, C.M. Conjugation of Fluorescent Proteins with DNA Oligonucleotides. Bioconj. Chem. 2010, 21, 921–927. [Google Scholar] [CrossRef]
- Nazarenko, I.; Pires, R.; Lowe, B.; Obaidy, M.; Rashtchian, A. Effect of primary and secondary structure of oligodeoxyribonucleotides on the fluorescent properties of conjugated dyes. Nucleic Acids Res. 2002, 30, 2089–2195. [Google Scholar] [CrossRef]
- Parambath, J.B.M.; Kanan, S.M.; Mohamed, A.A. Tryptophan capped gold-aryl nanoparticles for energy transfer study with SARS-CoV-2 spike proteins. Soft Mater. 2022, 16, 1–9. [Google Scholar] [CrossRef]
- Chakraborty, S.; Mukherjee, S. Effects of protecting groups on luminescent metal nanoclusters: Spectroscopic signatures and applications. Chem. Commun. 2021, 58, 29–47. [Google Scholar] [CrossRef] [PubMed]
- Swierczewska, M.; Lee, S.; Chen, X. The design and application of fluorophore–gold nanoparticle activatable probes. Phys. Chem. Chem. Phys. 2011, 13, 9929–9941. [Google Scholar] [CrossRef] [PubMed]
- Dulkeith, E.; Morteani, A.C.; Niedereichholz, T.; Klar, T.A.; Feldmann, J.; Levi, S.A.; van Veggel, F.C.J.M.; Reinhoudt, D.N.; Möller, M.; Gittins, D.I. Fluorescence Quenching of Dye Molecules near Gold Nanoparticles: Radiative and Nonradiative Effects. Phys. Rev. Lett. 2002, 89, 203002. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).