A Dynamic Hypergraph-Based Encoder–Decoder Risk Model for Longitudinal Predictions of Knee Osteoarthritis Progression
Abstract
1. Introduction
1.1. Koa Incidence and Progression
- KOA Incidence: The main body of research in KOA identification and longitudinal prediction operates under the following directive: Given the patients’ data at an initial baseline time point, identify the presence and/or severity of the condition at a specific time point in the future. In [5], the authors develop a CNN architecture utilizing the Convolutional Block Attention Module (CBAM) [6] for automatic KL grade classification. Guan et al. [7] develop a series of deep learning models to identify the onset of the disease during a follow-up period of 48 months. A similar approach is adopted by the authors in [8], where maps are employed to diagnose radiographic KOA, while in [7], a series of deep learning models are developed and evaluated for the prediction of medial joint space loss at 48 months after the baseline visit. A Convolutional Variational Autoencoder (CVAE) was employed in [9] for the early detection of KOA incidence, within a short period of 24 months. An Adversarial Evolving Neural Network (AENN) was proposed by the authors in [10] for longitudinal grading of KOA severity. The authors in [11] develop a risk prediction Tool for Osteoarthritis Risk Prediction (TOARP) with the goal of producing estimates of KOA incidence over an 8-year period. The work in [12] evaluates a series of radiographic features to determine KOA incidence in patients with recently developed knee pain, within a time-frame of 5 years. Finally, in [13], a risk prediction model combining clinical, genetic, and biochemical risk factors is proposed to yield KL score predictions at regular points in the future.
- KOA progression: With regards to the problem of progression modeling, the main task lies in identifying the progression patterns of clinical biomarkers, such as the JSW, JSN, and KL grade, as the condition progresses through time. The authors in [14] develop a novel biomarker, which is then subsequently used as input to a series of standard machine learning models in order to identify KOA progression. A Siamese neural network is proposed in [15], using a diverse set of radiological features for the detection of KOA progression. In [16], the authors adopt a standard LASSO regression model combined with a clustering approach, utilizing X-ray readings with pain scores, to model the longitudinal progression of KOA. Finally, in [17], an end-to-end approach within the transformer-based framework is developed to predict the progression of KOA utilizing X-ray data. In [18], the authors predict the 3-year progression of JSN by performing fractal texture analysis (FTA) of the medial tibial plateau. Adopting a similar framework, the works in [19,20] predict KOA progression at and months ahead of baseline, respectively, by measuring the change in JSW and Joint-Space Alignment (JSA), respectively. The FTA method is employed as well in [21,22], whereby features characterizing the Trabecular Bone Texture (TBT) were utilized to predict JSN progression at 48 months ahead of baseline. Finally, in [23], a deep learning method utilizing both MRI and clinical data was employed to determine KOA progression by identifying changes in JSN over a period of 12 months.
1.2. Proposed Methodology
- Historical/prediction stage configuration: Most works in KOA incidence and progression forecasting treat the problems in a static way. Typically, they consider the baseline knee data and develop various risk models to make predictions at specific follow-up times. In this work, the prediction task is tackled in a dynamic manner, following the sequence-to-sequence learning approach. Concretely, we seek to produce a sequence of incidence/progression predictions of a knee at a prediction horizon by exploiting the dynamic evolution of the knee sequence during a previous historic period. To this end, the time domain is distinguished into a historic stage of P time steps and a prediction stage comprising the following T time steps.
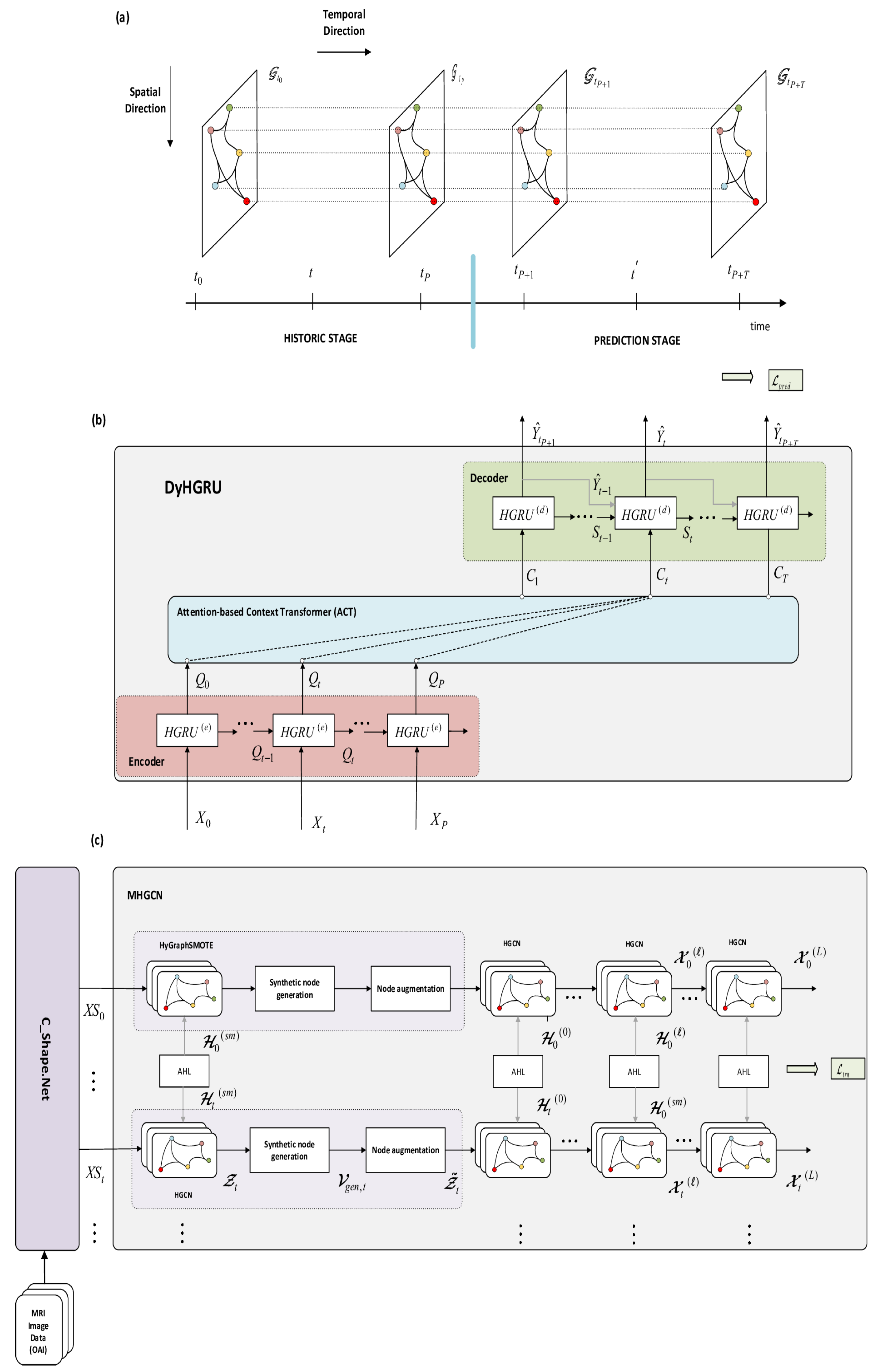
- Encoder–decoder architecture: The DyHGRU network adopts an encoder–decoder structure combined with the Attention-based Context Transformer (ACT). Both encoder and the decoder are composed of hypergraph-gated recurrent (HGRU) units, used to dynamically process the knee sequences along the historic and prediction stages. At the historic stage, the encoder receives the sequence of original knee data and creates a sequence of hidden states. Next, the ACT module filters this sequence, attending selectively to specific historic time steps to generate a sequence of context vectors that excites the decoder. Then, the decoder creates its own sequence of hidden states, along with the sequence of progression assessments, at the prediction stage.
- Spatio-temporal setting: The knee data are represented here in the spatio-temporal graph space. At each time step, the knees are regarded as a hypergraph, where nodes correspond to knees at this specific time. Along the temporal direction, knees are organized as a collection of interconnected hypergraphs containing the node sequences over time. These sequences are processed dynamically by the encoder. In the spatial direction, we apply the MHGCN network, which conducts multi-view HGCN convolutions on knee hypergraphs, at the different slices of the historical stage. This stage explores the pairwise relationships among the knees to obtain more comprehensive node representations. Knee relationships are created by constructing four different kinds of hyperedges (views), namely the shape descriptors and the three demographic factors of age, BMI, and gender.
- Imaging biomarkers: Previous works in KOA analysis utilize a variety of imaging biomarkers as inputs to the risk models, such as the trabecular bone texture (TBT), hip -angle, knee alignment, medial and lateral osteophyte scores, and mainly, features detected by traditional CNNs [24]. In this work, we opt to utilize the 3D global shape descriptors extracted by the recently proposed C_Shape.Net [25] using 3D MR images. This is a deep hypergraph convolutional network, designed to model the structural properties of the 3D knee cartilage volume. C_Shape.Net operates on a hypergraph of volumetric nodes, which are formed from triangular surface meshes of the cartilage. Nodes are equipped with a rich set of local features, such as spatial and geometric features of the faces, as well as volumetric measures, including thickness and volume values. In that respect, 3D shape descriptors are tightly connected to the joint space width (JSW) at a particular time step. When considering JSW at the different time steps of a knee sequence, we can also detect the trends of joint space narrowing (JSN) over time, which provides important evidence for the assessment of KOA progression. At the input, we also use the injury history sequence, which may notably affect the progress of KOA disease.
- Dataset balancing: Commonly, the number of knee progressors (minority class) is considerably smaller than non-progressors (majority class), rendering the knee dataset strongly imbalanced. This problem poses significant difficulties in both the learning process and the accuracy of the obtained results. Researchers in KOA progression usually face two alternatives: either retain the original dataset of imbalanced class distribution, which may undermine the robustness of predictions, since the classifier is biased towards the over-represented class of non-progressors, or cast a more balanced dataset of limited knees by disregarding the major portion of non-progressing knees. In this latter case, we are confronted with the data overfitting problem when deep risk models are to be trained. To tackle the dataset balancing problem in a systematic way, we propose the HyGraphSMOTE approach, which generates new synthetic knee progressors and progressing knee sequences via interpolation on existing knees of the minority class. This process is incorporated at the initial stage of the different convolutional branches pertaining to the MHGCN network.
- A novel dynamic hypergraph-based risk model (DyHRM) aiming to generate longitudinal predictions of KOA incidence and progression. The suggested model integrates two main parts, the DyHGRU and the MHGCN networks.
- The DyHGRU network with an encoder–decoder structure comprising hypergraph-gated recurrent units. It adopts the sequence-to-sequence learning, whereby historic sequences of knee shape data are transformed into label sequences of progression at the prediction stage.
- The HyGraphSMOTE method on hypergraphs, aiming to balance knee progressors against non-progressors in the dataset. The method applies oversampling of the minority node class, synthesizing new samples via interpolation on existing node pairs.
- The MHGCN network, which integrates data balancing and multi-view HGCN convolutions. Its scope is to acquire more representative node features by exploiting the hyperedge structures from both shape descriptors and various demographic factors.
- The adaptive hypergraph learning (AHL), used to automatically define the hyperedge structure during training. This mechanism is employed as an edge generator in the HyGraphSMOTE algorithm, as well as at each layer of the HGCN convolutions in MHGCN.
- The performance of the proposed methodology is evaluated and compared on both the KOA incidence and progression tasks, using different evaluation criteria to assess knee progress. We also conducted comprehensive ablation studies to investigate the impact of several factors in our approach, such as the effect of HyGraphSMOTE, the role of the ACT module, and HGRU units in DyHGRU.
2. Related Work
2.1. Graph and Hypergraph Convolutional Networks
2.2. Encoder–Decoder Framework
2.3. Class Imbalance Problem
3. Hypergraph Convolutional Networks
3.1. Hypergraph Spectral Convolutions
3.2. Adaptive Hypergraph Learning
4. Materials
5. Proposed KOA Predictor Network
5.1. Data Representation and Notations
5.2. Imbalance of Knee Progressors
6. Multi-View Hypergraph Convolutional Network
6.1. Adaptive Hypergraph Generation
6.2. HyGraphSMOTE Balancing Method
- Data Embedding: In this stage, the input data are transformed into an embedding feature space . Instead of applying minority oversampling over the original raw data, the new space provides more discriminating node representations, especially for the progressor ones. Since HyGraphSMOTE is designed on the hypergraph framework, the embedded features are acquired via a HGCN convolution:, where denotes the embedding of node at time t. Furthermore, and contain the learnable hyperedge weights and the filter parameters, respectively. denotes the incidence matrix at the SMOTE phase, as described in Equation (10). This matrix plays the role of a hyperedge generator, incorporating the multiple view hyperedge connections between the nodes. Here, is adaptively learned from node features using the attention-based AHL mechanism in Section 3.2: .
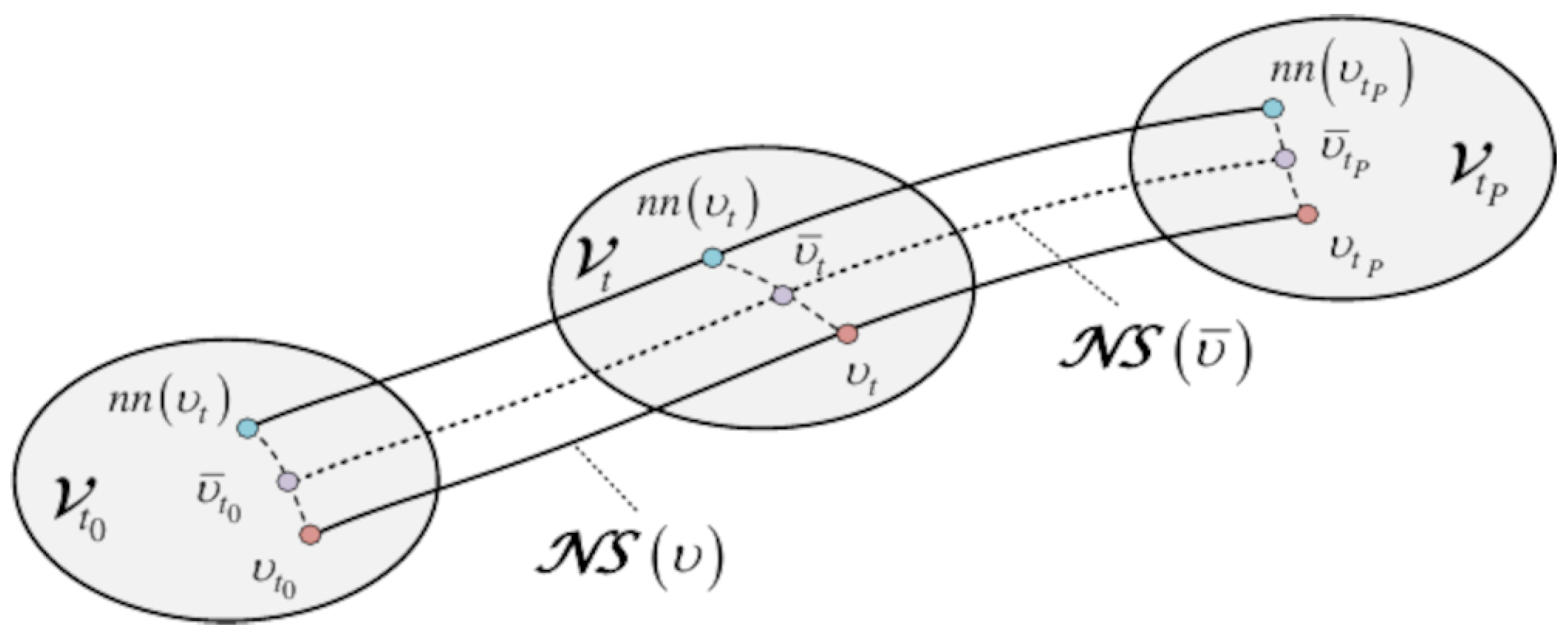
- Synthetic Node Generation: Let be a minority node at time t with an embedding feature . The first step is to find its nearest neighbor node in the embedding space, defined as follows:Minority oversampling generates two types of synthetic nodes by interpolating between the pairs . In the first main case, we consider that is also a minority node. The new node is created as follows:where is a uniformly distributed random variable in the range . Since both and are close neighbors of the minority class, node is crisply assigned to the same class. In the second case, the synthetic nodes are generated from mixed node pairs , considering that the nearest neighbor now belongs to the majority class. In that case, interpolation is applied on both the embedding and the label space, simultaneously. The feature and the label of the new mixed nodes are determined as follows:where , are the embedding and the label of the newly generated mixed node . Contrary to the previous case, the mixed nodes receive soft (fuzzy) label vectors, taking values between the minority and the majority ones. is a random variable in the range , where b is set to a value of in the experiments. This choice of b suggests that both the embedding and the label of node are placed by Equations (14) and (15) closer to the minority node rather than its nearest neighbor majority node . The incorporation of the mixed nodes tends to make the boundaries between classes smoother, thus facilitating better performance in the classification task. In the experiments, we apply a proportion of – of synthetic nodes, focusing primarily in generating new nodes from strictly minority node pairs and a smaller percentage from mixed pairs. The synthetic node generation on a minority class is carried out according to an oversampling rate , where the parameter controls the balance between the minority (progressor) and the majority (non-progressor) classes.The next step is to formulate the balanced graph at each time t via data augmentation. augments the existing node set with the set of synthetic nodes . The augmented contains a total of nodes. Similarly, concatenates the embeddings of the respective node sets. Finally, represents the hyperedge connections between existing nodes, as well as pairwise links between the synthetic nodes and the existing ones. This matrix is adaptively constructed again as
- Generation of Synthetic Sequences: The last stage of data balancing is to synthesize new progressive node sequences by repeatedly applying node oversampling over the different time steps. The sequence oversampling is visualized in Figure 3 and proceeds along the following steps:
- (a)
- At the historic stage, define the set of progressor nodes of the original dataset at the terminal step and restore their respective node sequences . The terminal nodes are distributed over the different KL classes of an inclusion set .
- (b)
- For each , find the nearest neighbor nodes from Equation (12). As an outcome, we obtain the nearest neighbor sequence of .
- (c)
- Generate a synthetic node sequence using either Equation (12) or Equations (14) and (15) to compute the features and labels of the pertaining nodes. Temporally, the new sequence evolves in the vicinity of ; hence, the corresponding nodes receive the same or similar labels as in the original sequence.
- (d)
- At the prediction stage, we assume that the label sequence follows a similar track as ; thus, we set for .
6.3. HGCN Convolutions
7. Dynamic Hypergraph Gated Recurrent Unit
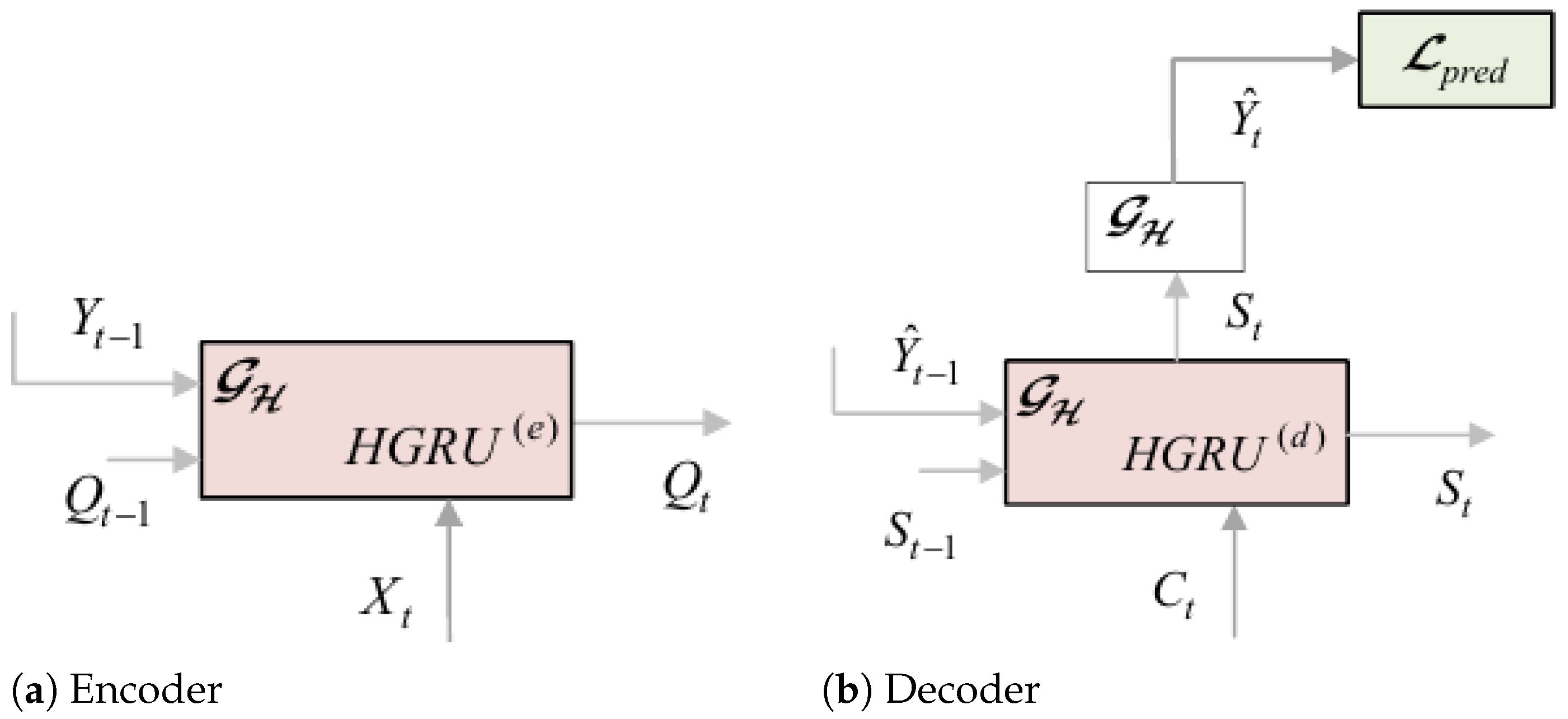
- Encoder: The encoder receives a sequence of knee data to generate a sequence of hidden states. The HGRU(e) model at time t is outlined in Figure 4a and is functionally described by , where , and are the external input, the d-dimensional hidden state of HGRU(e), and the ground true labels of nodes. We use the GRU as a building module, a variant of recurrent neural networks, which is suitable for effectively handling time-series data. Moreover, inspired by [59], we integrate the temporal capabilities of GRUs with the spatial HGCN convolutions to create the proposed HGRU(e) module with spatio-temporal processing capacities. This is achieved by replacing the linear transformations in GRUs with hypergraph aggregations. The computations involved in HGRU(e) are described as follows:The operator denotes concatenation of the input entries, and ⊙ represents the Hadamard product operator. and are the reset gate used to discard redundant previous information and the update gate that controls the output. The set of tunable parameters includes the following: and are the incidence matrix and the hyperedge weights involved in the HGCN convolutions in HGRU(e), while is an embedding matrix used for dimensionality matching of the labels . Furthermore, the sets , and are the filter parameters and bias terms pertaining to the , and , respectively. The set of learnable parameters discussed above are shared across all HGRU(e) units of the encoder for .
- Decoder: The decoder shares a similar structure as the encoder, comprising a total of THGRU(d) units (Figure 4b) for . It takes as input the context sequence and produces the sequence of label estimates at the prediction stage. Functionally, the decoder units are described by and , where are the context input and the output of the HGRU(d), and denotes the label estimates of nodes at time t. The respective convolutions of the HGRU(d) units are given as follows:where are the candidate hidden activations, and , are the reset gate and update gate, respectively. The first four equations (Equations (21)–(24) represent the internal convolution dynamics of the HGRU(d), while the last equation Equation (25)) is a convolution aiming to yield the label estimates of nodes from hidden states of the decoder at follow-up time t. The terminology and the usage of the various parameters in the decoder are similar to the ones in the encoder part.
- Attention-Based Context Transformer: The ACT module acts as an interface between the encoder and the decoder parts. Concretely, it transforms the sequence of hidden states acquired by the encoder into a sequence of context input vectors for the decoder. The context vectors at future times are determined by selectively applying multi-head attention to relevant time steps of the hidden sequence . Let denote the attention score between the weights and for a node . The superscript indicates the k-th attention head, . The attention scores are adaptively derived as follows:where the index indicates the v-th row of the respective matrices. The context value is obtained as a weighted sum from all historical time steps over the different attention heads:The calculations in Equations (26) and (27) are conducted in parallel, and the learnable parameters are shared across all nodes and time steps. According to Equation (27), the context vectors aim to identify the informative trends of the encoder evolution. Particularly, they seek to detect aggravation of JSN, which is a valuable indication to assess KOA progression. In that respect, the context sequence allows the decoder to acquire the correct track of and until the target follow-up time t is reached, where the progression level should be eventually evaluated.
- Network Training: Training is performed here following the semi-supervised learning (SSL) framework. In this setting, the input data comprises a labeled part of nodes and an unlabeled part of nodes, with . The former part contains nodes with historical shape sequences having labeled sequences of KOA grades in the prediction stage. The nodes in the latter part form the testing dataset; hence, their output label sequences are unknown. Under SSL, we exploit the shape content of both the training and testing data, which can yield better results.Learning of the DyHRM predictor is carried out in two phases. In phase 1, we conduct a node classification task to pretrain the branches of the MHGCN network at the historical time steps. The cross-entropy loss is used to match the estimates to the true labels in the KL classes :Pretraining aims to initialize the features of the real labeled and unlabeled nodes, and especially the synthetically ones generated by HyGraphSMOTE. Next, phase 2 performs end-to-end training of the entire network in Figure 2, including the DyHGRU, MHGCNs, and the C_Shape.net. The goal is mainly to optimize the future estimates given by the decoder using a regularized objective described as follows:where is a loss function defined on the progression classes :
8. Experimental Results
8.1. Configuration of Progression Tasks
8.2. Dataset Generation
8.3. Implementation Details
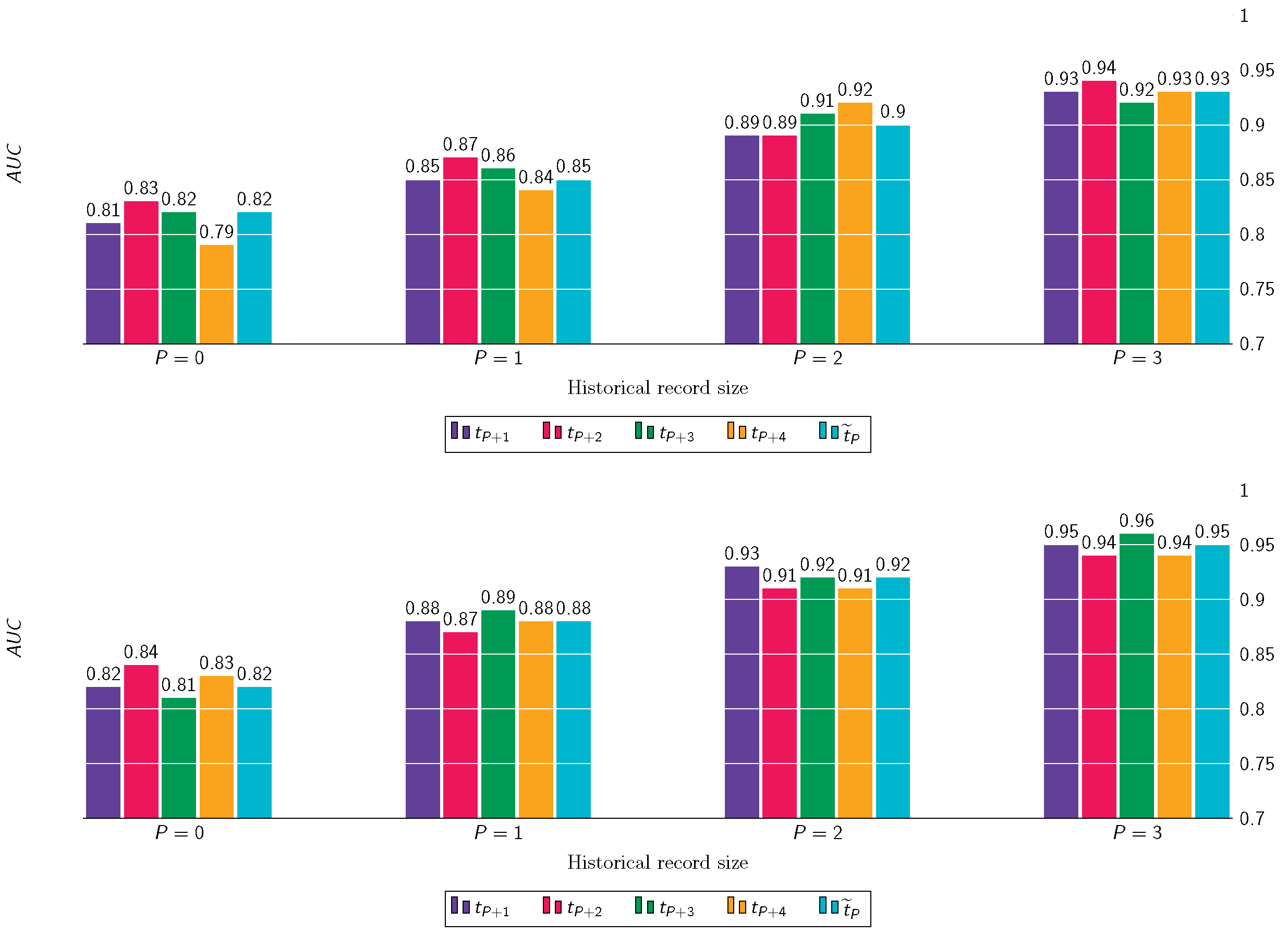
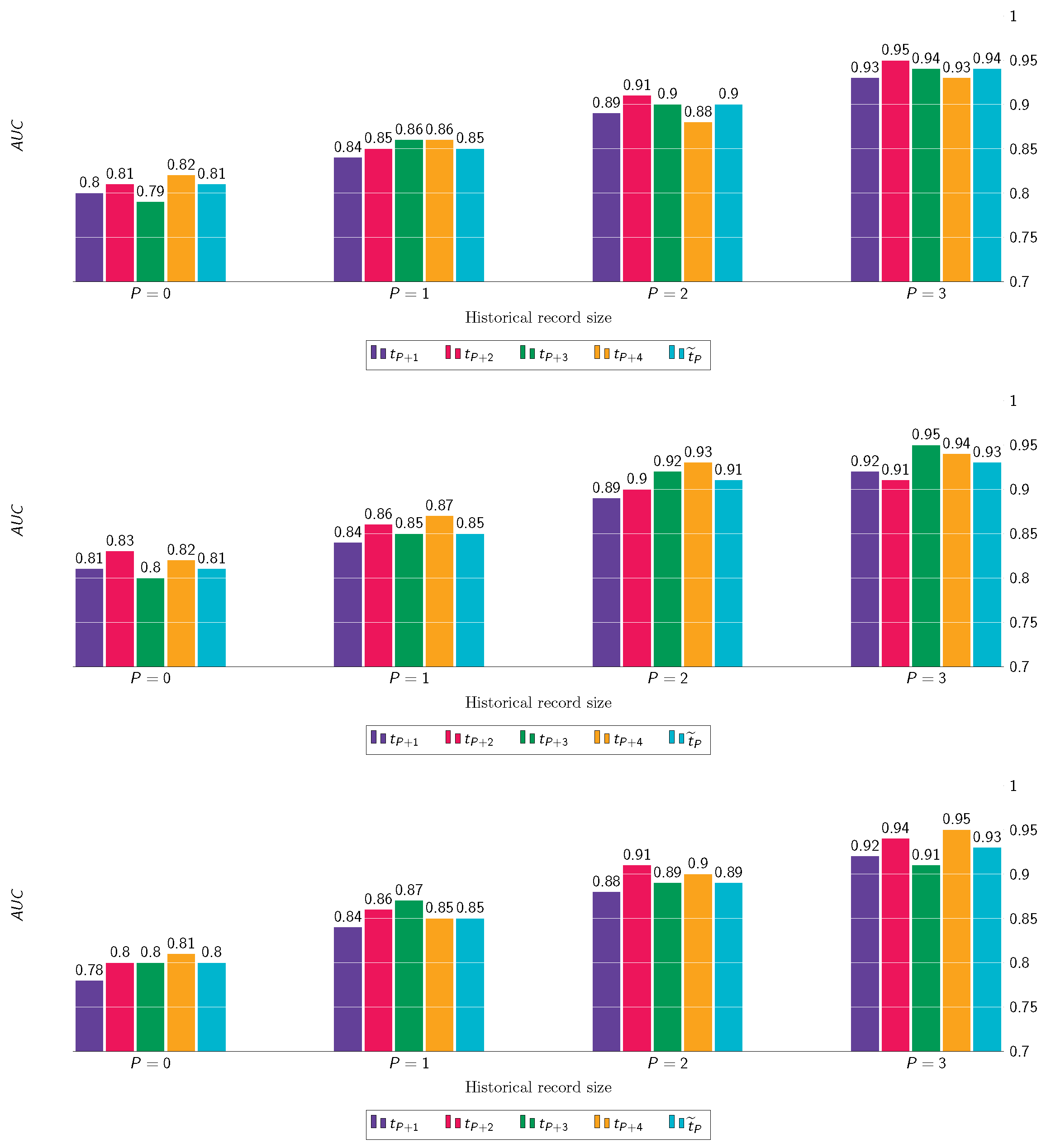
8.4. Longitudinal Predictions of KOA Incidence
8.5. Longitudinal Predictions of KOA Progression
8.6. Long-Term Predictions of KOA Incidence and Progression
8.7. Demographic-Based Accuracies
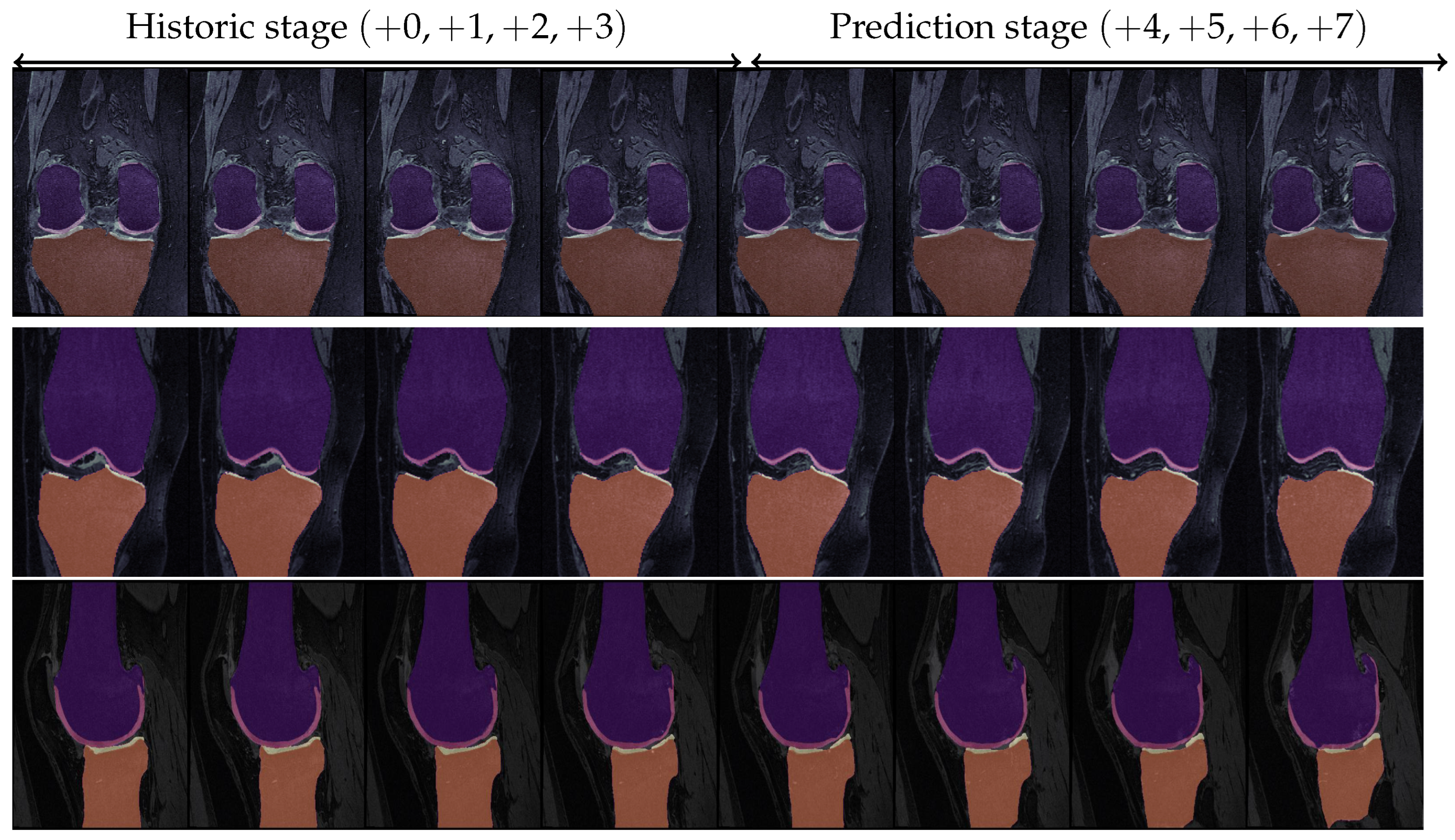
8.8. Test Case Demonstration
8.9. Ablation Studies
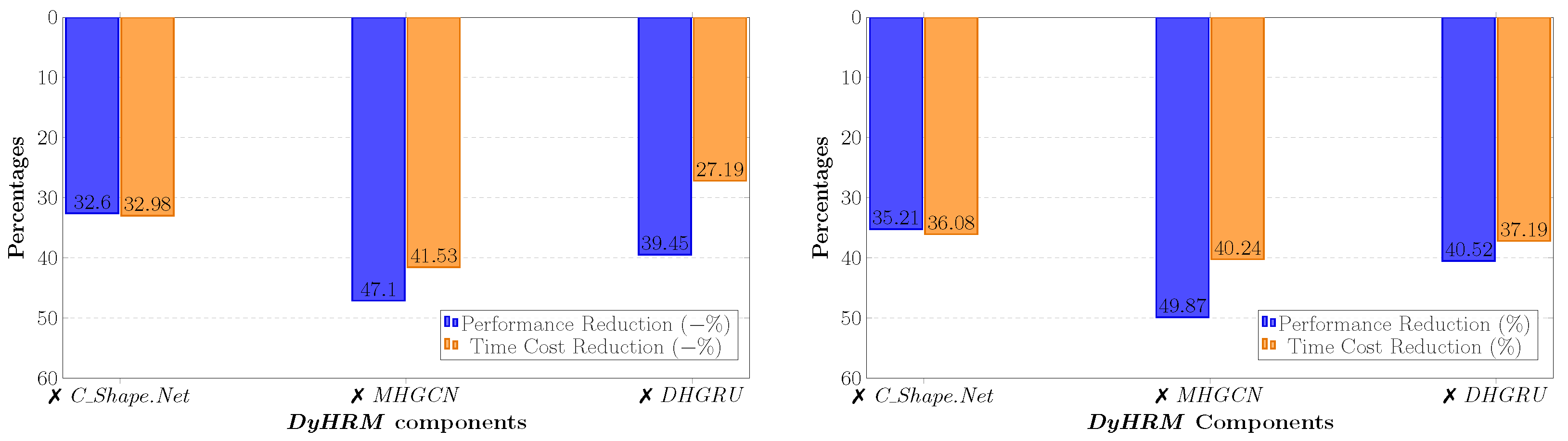
8.10. Parameter Importance and Computational Complexity
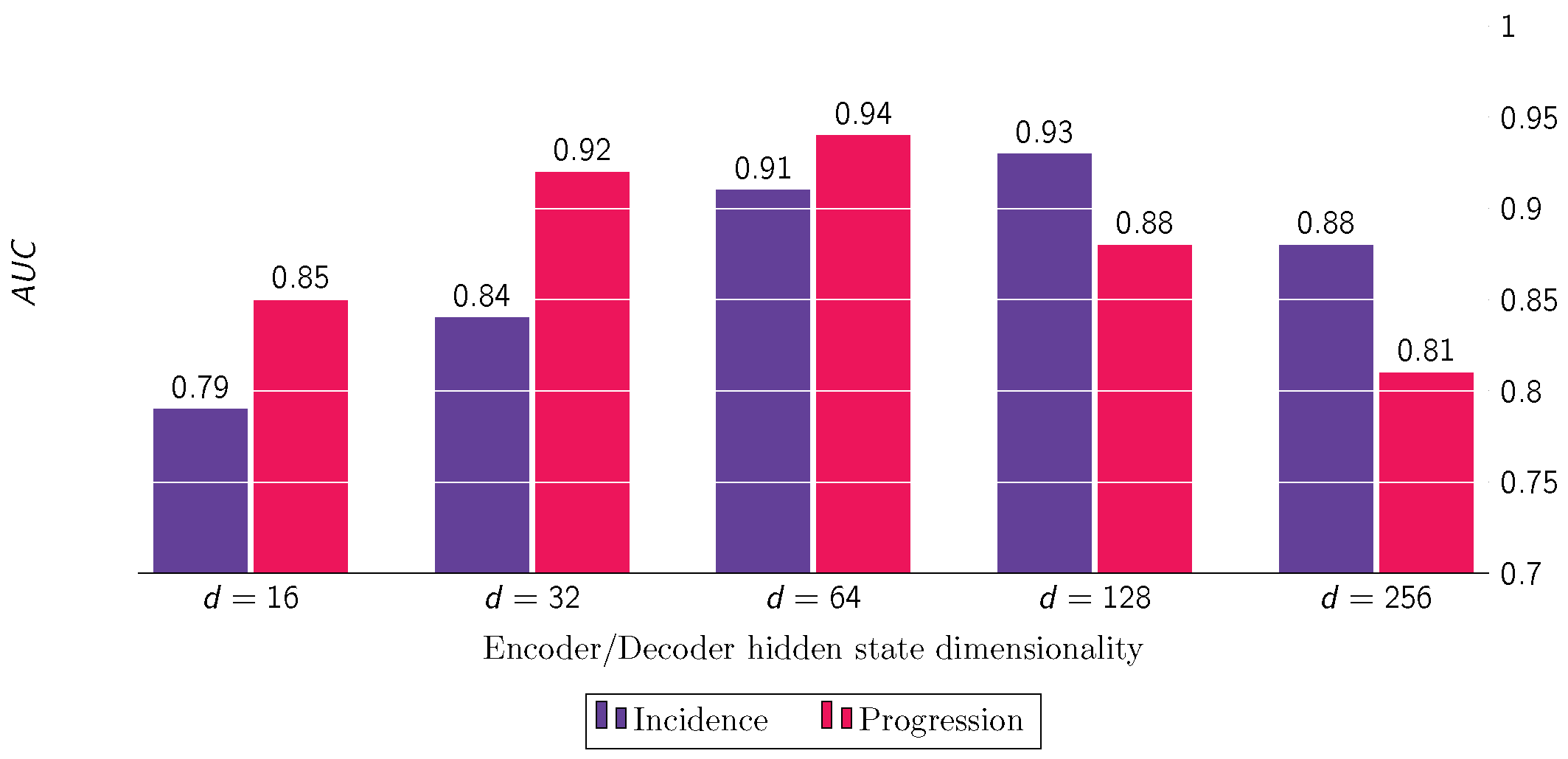
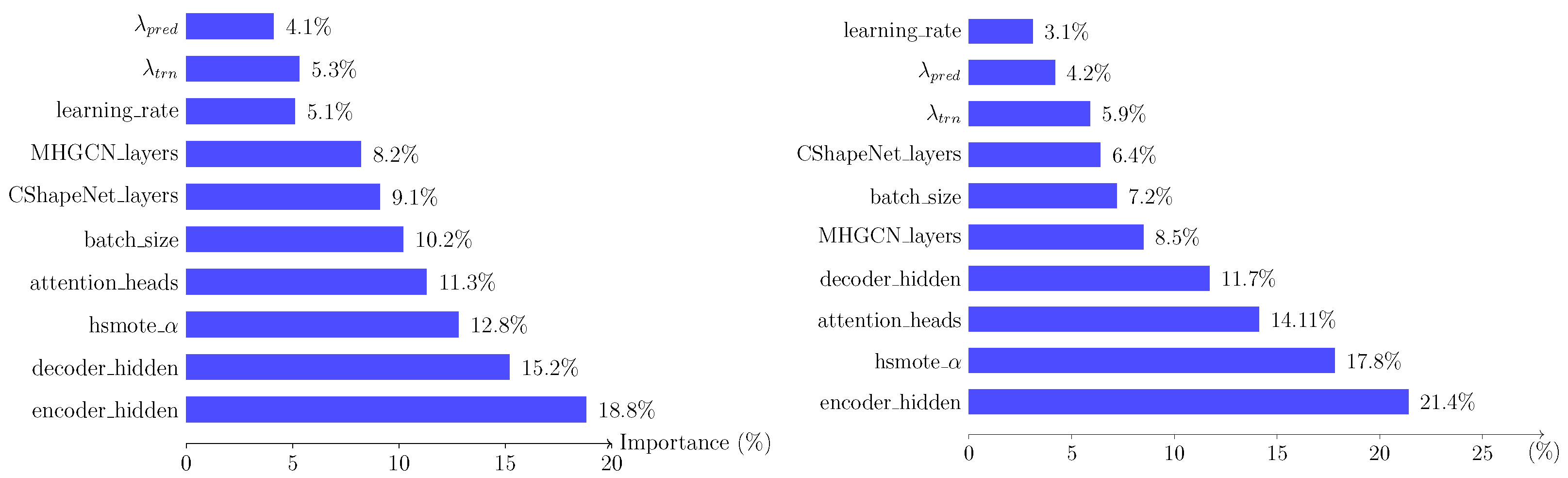
- The size of the hidden dimensions for the encoder and decoder parts of the HGRU units consistently score high in the ranking list, indicating the central role of this component in the overall model’s performance for both incidence and progression prediction tasks.
- The high score in both tasks of the parameter, controlling the balance between the minority and majority classes in the step, showcases the importance of implementing comprehensive balancing in datasets where a certain percentage of classes dominates the rest.
- The number of attention heads in the various components receives ranks in the medium to high tiers for both tasks. That is an expected result, especially given the previous observation of the high ranking of the encoder/decoder components. The attention mechanism is a crucial component of the transformer architecture, bridging the encoder and decoder units. In addition, the attention mechanism is extensively utilized across all the remaining major modules of the proposed model (hyperedge generation), further cementing its influence across the entire network.
- The number of layers for the C_Shape.Net and MHGCN components receive medium scores of relative importance. While the ablation studies performed in Section 8.9 suggest that the exclusion of these modules severely diminishes the models’ performance, the number of layers for both these part seems to not be as central, as long as these modules exist in the main model’s body.
- Finally, the components with the consistently lower scoring for both tasks seem to be the learning rate and the regularization terms for the two constituents of the loss function in Equation (29).
8.11. Comparative Analysis
- Temporal Fusion Transformer (TFT https://github.com/mattsherar/Temporal_Fusion_Transform (accessed on 6 August 2025)) [74]: The TFT features a complex architecture that utilizes variable selection networks (VSNs), static enrichment networks (SENs), and a temporal processing component comprising LSTM layers supplied with an attention mechanism to produce multi-horizon predictions. It is a multi-modular architecture with many innovative parts, but it can suffer from its extremely high parameter count, making it difficult to train on sequences of low to moderate length, leading to overfitting.
- Temporal Convolutional Attention Neural Network (TCAN https://github.com/YangLIN1997/TCAN-IJCNN2021/tree/main/model (accessed on 6 August 2025)) [75]: The model presented in this work features a sparse attention mechanism, specifically proposed to bypass the need for designing deep architectures in order to fully capture the spatio-temporal trends in data. The dilated temporal convolution operation allows the model to efficiently capture long-term dependencies while keeping the computational demands constrained. The main drawback of the work, however, is that the model cannot adequately represent spatial relationships among the data, limiting its appliance on datasets with a prominent spatio-temporal aspect.
- Adaptive Graph Convolutional Recurrent Network (AGCRNN https://github.com/LeiBAI/AGCRN (accessed on 6 August 2025)) [76]: In this study, the authors propose a modified graph convolutional operation where each graph convolutional layer learns its own embedding matrix. The model, at each layer, automatically learns the graph structure from the node embeddings, where the embedding parameters are specific to each node, thus bypassing the need for different embeddings with respect to the graph structure and the layer’s parameters. A major limitation of this work, however, is that it can only capture pairwise relationships between the nodes, thus not being generalizable to hypergraph data.
9. Conclusions
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
Appendix A
| Algorithm A1: HyGraphSMOTE Algorithm |
 |
References
- Wang, Z.; Xiao, Z.; Sun, C.; Xu, G.; He, J. Global, regional and national burden of osteoarthritis in 1990–2021: A systematic analysis of the global burden of disease study 2021. BMC Musculoskelet. Disord. 2024, 25, 1021. [Google Scholar] [CrossRef]
- Bastick, A.N.; Runhaar, J.; Belo, J.N.; Bierma-Zeinstra, S.M. Prognostic factors for progression of clinical osteoarthritis of the knee: A systematic review of observational studies. Arthritis. Res. Ther. 2015, 17, 152. [Google Scholar] [CrossRef]
- Mazzuca, S.A.; Brandt, K.D.; Katz, B.P.; Lane, K.A.; Buckwalter, K.A. Comparison of quantitative and semiquantitative indicators of joint space narrowing in subjects with knee osteoarthritis. Ann. Rheum. Dis. 2006, 65, 64–68. [Google Scholar] [CrossRef]
- Zhao, H.; Ou, L.; Zhang, Z.; Zhang, L.; Liu, K.; Kuang, J. The value of deep learning-based X-ray techniques in detecting and classifying K-L grades of knee osteoarthritis: A systematic review and meta-analysis. Eur. Radiol. 2024, 35, 327–340. [Google Scholar] [CrossRef]
- Zhang, B.; Tan, J.; Cho, K.; Chang, G.; Deniz, C.M. Attention-based CNN for KL Grade Classification: Data from the Osteoarthritis Initiative. In Proceedings of the 2020 IEEE 17th International Symposium on Biomedical Imaging (ISBI), Iowa City, IA, USA, 3–7 April 2020; pp. 731–735. [Google Scholar] [CrossRef]
- Woo, S.; Park, J.; Lee, J.; Kweon, I.S. CBAM: Convolutional Block Attention Module. arXiv 2018, arXiv:1807.06521. [Google Scholar] [CrossRef]
- Guan, B.; Liu, F.; Haj-Mirzaian, A.; Demehri, S.; Samsonov, A.; Neogi, T.; Guermazi, A.; Kijowski, R. Deep learning risk assessment models for predicting progression of radiographic medial joint space loss over a 48-MONTH follow-up period. Osteoarthr. Cartil. 2020, 28, 428–437. [Google Scholar] [CrossRef]
- Pedoia, V.; Lee, J.; Norman, B.; Link, T.; Majumdar, S. Diagnosing osteoarthritis from T2 maps using deep learning: An analysis of the entire Osteoarthritis Initiative baseline cohort. Osteoarthr. Cartil. 2019, 27, 1002–1010. [Google Scholar] [CrossRef] [PubMed]
- Alexopoulos, A.; Hirvasniemi, J.; Tumer, N. Early detection of knee osteoarthritis using deep learning on knee magnetic resonance images. Osteoarthr. Imaging 2023, 3, 100112. [Google Scholar] [CrossRef]
- Hu, K.; Wu, W.; Li, W.; Simic, M.; Zomaya, A.; Wang, Z. Adversarial Evolving Neural Network for Longitudinal Knee Osteoarthritis Prediction. IEEE Trans. Med. Imaging 2022, 41, 3207–3217. [Google Scholar] [CrossRef]
- Joseph, G.B.; McCulloch, C.E.; Nevitt, M.C.; Neumann, J.; Gersing, A.S.; Kretzschmar, M.; Schwaiger, B.J.; Lynch, J.A.; Heilmeier, U.; Lane, N.E.; et al. Tool for Osteoarthritis Risk Prediction (TOARP) over 8 years using Baseline Clinical Data, X-ray, and MR imaging - Data from the Osteoarthritis Initiative. J. Magn. Reson. Imaging 2018, 47, 1517–1526. [Google Scholar] [CrossRef]
- Kinds, M.B.; Marijnissen, A.C.A.; Vincken, K.L.; Viergever, M.A.; Drossaers-Bakker, K.W.; Bijlsma, J.W.J.; Bierma-Zeinstra, S.M.A.; Welsing, P.M.J.; Lafeber, F.P.J.G. Evaluation of separate quantitative radiographic features adds to the prediction of incident radiographic osteoarthritis in individuals with recent onset of knee pain: 5-year follow-up in the CHECK cohort. Osteoarthr. Cartil. 2012, 20, 548–556. [Google Scholar] [CrossRef][Green Version]
- Kerkhof, H.J.M.; Bierma-Zeinstra, S.M.A.; Arden, N.K.; Metrustry, S.; Castano-Betancourt, M.; Hart, D.J.; Hofman, A.; Rivadeneira, F.; Oei, E.H.G.; Spector, T.D.; et al. Prediction model for knee osteoarthritis incidence, including clinical, genetic and biochemical risk factors. Ann. Rheum. Dis. 2013, 73, 2116–2121. [Google Scholar] [CrossRef]
- Du, Y.; Almajalid, R.; Shan, J.; Zhang, M. A Novel Method to Predict Knee Osteoarthritis Progression on MRI Using Machine Learning Methods. IEEE Trans. Nanobiosci. 2018, 17, 228–236. [Google Scholar] [CrossRef]
- Almhdie-Imjabbar, A.; Nguyen, K.L.; Toumi, H.; Jennane, R.; Lespessailles, E. Prediction of knee osteoarthritis progression using radiological descriptors obtained from bone texture analysis and Siamese neural networks: Data from OAI and MOST cohorts. Arthritis Res. Ther. 2022, 24, 66. [Google Scholar] [CrossRef]
- Halilaj, E.; Le, Y.; Hicks, J.; Hastie, T.; Delp, S. Modeling and predicting osteoarthritis progression: Data from the osteoarthritis initiative. Osteoarthr. Cartil. 2018, 26, 1643–1650. [Google Scholar] [CrossRef]
- Panfilov, E.; Saarakkala, S.; Nieminen, M.T.; Tiulpin, A. End-To-End Prediction of Knee Osteoarthritis Progression With Multi-Modal Transformers. arXiv 2023, arXiv:2307.00873. [Google Scholar] [CrossRef]
- Kraus, V.B.; Feng, S.; Wang, S.; White, S.; Ainslie, M.; Brett, A.; Holmes, A.; Charles, H.C. Trabecular morphometry by fractal signature analysis is a novel marker of osteoarthritis progression. Arthritis Reuhmatol. 2009, 60, 3711–3722. [Google Scholar] [CrossRef]
- Kraus, V.B.; Feng, S.; Wang, S.; White, S.; Ainslie, M.; Graver, M.P.H.L.; Brett, A.; Eckstein, F.; Hunter, D.J.; Lane, N.E.; et al. Subchondral Bone Trabecular Integrity Predicts and Changes Concurrently With Radiographic and Magnetic Resonance Imaging–Determined Knee Osteoarthritis Progression. Arthritis Rheumatol. 2013, 65, 1812–1821. [Google Scholar] [CrossRef]
- Kraus, V.B.; Collins, J.E.; Charles, H.C.; Pieper, C.F.; Whitley, L.; Losina, E.; Nevitt, M.; Hoffmann, S.; Roemer, F.; Guermazi, A.; et al. Predictive Validity of Radiographic Trabecular Bone Texture in Knee Osteoarthritis. Arthritis Rheumatol. 2018, 70, 80–87. [Google Scholar] [CrossRef] [PubMed]
- Janvier, T.; Jennane, R.; Toumi, H.; Lespessailles, E. Subchondral tibial bone texture predicts the incidence of radiographic knee osteoarthritis: Data from the Osteoarthritis Initiative. Osteoarthr. Cartil. J. 2017, 25, 2047–2054. [Google Scholar] [CrossRef] [PubMed]
- Woloszynski, T.; Podsiadlo, P.; Stachowiak, G.W.; Kurzynski, M.; Lohm, L.S.; Englund, M. Prediction of progression of radiographic knee osteoarthritis using tibial trabecular bone texture. Arthritis Rheumatol. 2012, 64, 688–695. [Google Scholar] [CrossRef]
- Schiratti, J.B.; Dubois, R.; Herent, P.; Cahané, D.; Dachary, J.; Clozel, T.; Wainrib, G.; Keime-Guibert, F.; Lalande, A.; Pueyo, M.; et al. A deep learning method for predicting knee osteoarthritis radiographic progression from MRI. Arthritis Res. Ther. 2021, 23, 262. [Google Scholar] [CrossRef]
- Almhdie-Imjabbar, A.; Toumi, H.; Lespessailles, E. Radiographic Biomarkers for Knee Osteoarthritis: A Narrative Review. Life 2023, 13, 237. [Google Scholar] [CrossRef]
- Theocharis, J.B.; Chadoulos, C.G.; Symeonidis, A.L. A Novel Approach Based on Hypergraph Convolutional Neural Networks for Cartilage Shape Description and Longitudinal Prediction of Knee Osteoarthritis Progression. Mach. Learn. Knowl. Extr. 2025, 7, 40. [Google Scholar] [CrossRef]
- Wu, Z.; Pan, S.; Chen, F.; Long, G.; Zhang, C.; Yu, P.S. A Comprehensive Survey on Graph Neural Networks. IEEE Trans. Neural Netw. Learn. Syst. 2021, 32, 4–24. [Google Scholar] [CrossRef] [PubMed]
- Defferrard, M.; Bresson, X.; Vandergheynst, P. Convolutional Neural Networks on Graphs with Fast Localized Spectral Filtering. arXiv 2017, arXiv:1606.09375. [Google Scholar] [CrossRef]
- Zeng, H.; Zhou, H.; Srivastava, A.; Kannan, R.; Prasanna, V. GraphSAINT: Graph Sampling Based Inductive Learning Method. arXiv 2020, arXiv:1907.04931. [Google Scholar] [CrossRef]
- Veličković, P.; Cucurull, G.; Casanova, A.; Romero, A.; Liò, P.; Bengio, Y. Graph Attention Networks. arXiv 2018, arXiv:1710.10903. [Google Scholar] [CrossRef] [PubMed]
- Kipf, T.N.; Welling, M. Semi-Supervised Classification with Graph Convolutional Networks. arXiv 2017, arXiv:1609.02907. [Google Scholar] [CrossRef]
- Gilmer, J.; Schoenholz, S.S.; Riley, P.F.; Vinyals, O.; Dahl, G.E. Neural Message Passing for Quantum Chemistry. In Proceedings of the Proceedings—International Conference on Machine Learning, Sydney, Australia, 6–11 August 2017. [Google Scholar]
- Hamilton, W.; Ying, Z.; Leskovec, J. Inductive Representation Learning on Large Graphs. arXiv 2017, arXiv:1706.02216. [Google Scholar] [CrossRef]
- Zhang, S.; Tong, H.; Xu, J.; Maciejewski, R. Graph convolutional networks: A comprehensive review. Comput. Soc. Netw. 2019, 6, 11. [Google Scholar] [CrossRef]
- Ying, R.; He, R.; Chen, K.; Eksombatchai, P.; Hamilton, W.L.; Leskovec, J. Graph convolutional neural networks for web-scale recommender systems. In Proceedings of the ACM SIGKDD International Conference on Knowledge Discovery and Data Mining, London, UK, 19–23 August 2018; pp. 974–983. [Google Scholar] [CrossRef]
- Song, L.; Zhang, Y.; Wang, Z.; Gildea, D. A Graph-to-Sequence Model for AMR-to-Text Generation. In Proceedings of the 56th Annual Meeting of the Association for Computational Linguistics, Melbourne, Australia, 15–20 July 2018; pp. 1616–1626. [Google Scholar]
- Ma, Z.; Jiang, Z.; Zhang, H. Hyperspectral Image Classification Using Feature Fusion Hypergraph Convolution Neural Network. IEEE Trans. Geosci. Remote Sens. 2022, 60, 1–14. [Google Scholar] [CrossRef]
- Chadoulos, C.; Tsaopoulos, D.; Symeonidis, A.; Moustakidis, S.; Theocharis, J. Dense Multi-Scale Graph Convolutional Network for Knee Joint Cartilage Segmentation. Bioengineering 2024, 11, 278. [Google Scholar] [CrossRef] [PubMed]
- Feng, Y.; You, H.; Zhang, Z.; Ji, R.; Gao, Y. Hypergraph Neural Networks. AAAI 2019, 33, 3558–3565. [Google Scholar] [CrossRef]
- Bai, J.; Gong, B.; Zhao, Y.; Lei, F.; Yan, C.; Gao, Y. Multi-Scale Representation Learning on Hypergraph for 3D Shape Retrieval and Recognition. IEEE Trans. Image Process. 2021, 30, 5327–5338. [Google Scholar] [CrossRef]
- Bai, S.; Zhang, F.; Torr, P.H. Hypergraph convolution and hypergraph attention. Pattern Recognit. 2021, 110, 107637. [Google Scholar] [CrossRef]
- Chai, S.; Jain, R.K.; Mo, S.; Liu, J.; Yang, Y.; Li, Y.; Tateyama, T.; Lin, L.; Chen, Y.W. A Novel Adaptive Hypergraph Neural Network for Enhancing Medical Image Segmentation. In Proceedings of the Medical Image Computing and Computer Assisted Intervention–MICCAI 2024; Linguraru, M.G., Dou, Q., Feragen, A., Giannarou, S., Glocker, B., Lekadir, K., Schnabel, J.A., Eds.; Springer Nature: Cham, Switzerland, 2024; pp. 23–33. [Google Scholar]
- Jing, W.; Wang, J.; Di, D.; Li, D.; Song, Y.; Fan, L. Multi-modal hypergraph contrastive learning for medical image segmentation. Pattern Recognit. 2025, 165, 111544. [Google Scholar] [CrossRef]
- Antelmi, A.; Cordasco, G.; Polato, M.; Scarano, V.; Spagnuolo, C.; Yang, D. A Survey on Hypergraph Representation Learning. ACM Comput. Surv. 2023, 56, 1–38. [Google Scholar] [CrossRef]
- Sutskever, I.; Vinyals, O.; Le, Q.V. Sequence to Sequence Learning with Neural Networks. arXiv 2014, arXiv:1409.3215. [Google Scholar] [CrossRef]
- Staudemeyer, R.C.; Morris, E.R. Understanding LSTM – a tutorial into Long Short-Term Memory Recurrent Neural Networks. arXiv 2019, arXiv:1909.09586. [Google Scholar]
- Dey, R.; Salem, F.M. Gate-Variants of Gated Recurrent Unit (GRU) Neural Networks. arXiv 2017, arXiv:1701.05923. [Google Scholar] [CrossRef]
- Niu, Z.; Yu, Z.; Tang, W.; Wu, Q.; Reformat, M. Wind power forecasting using attention-based gated recurrent unit network. Energy 2020, 196, 117081. [Google Scholar] [CrossRef]
- Kao, I.F.; Zhou, Y.; Chang, L.C.; Chang, F.J. Exploring a Long Short-Term Memory based Encoder-Decoder framework for multi-step-ahead flood forecasting. J. Hydrol. 2020, 583, 124631. [Google Scholar] [CrossRef]
- Li, Q.; Li, Z.; Shangguan, W.; Wang, X.; Li, L.; Yu, F. Improving soil moisture prediction using a novel encoder-decoder model with residual learning. Comput. Electron. Agric. 2022, 195, 106816. [Google Scholar] [CrossRef]
- Shamshad, F.; Khan, S.; Zamir, S.W.; Khan, M.H.; Hayat, M.; Khan, F.S.; Fu, H. Transformers in medical imaging: A survey. Med Image Anal. 2023, 88, 102802. [Google Scholar] [CrossRef] [PubMed]
- Yan, Z.; Zhai, D.H.; Xia, Y. DMS-GCN: Dynamic Mutiscale Spatiotemporal Graph Convolutional Networks for Human Motion Prediction. arXiv 2021, arXiv:2112.10365. [Google Scholar] [CrossRef]
- Ghosh, P.; Yao, Y.; Davis, L.S.; Divakaran, A. Stacked Spatio-Temporal Graph Convolutional Networks for Action Segmentation. arXiv 2019, arXiv:1811.10575. [Google Scholar] [CrossRef]
- Wang, F.; Du, X.; Zhang, W.; Nie, L.; Wang, H.; Zhou, S.; Ma, J. Remote Sensing LiDAR and Hyperspectral Classification with Multi-Scale Graph Encoder–Decoder Network. Remote Sens. 2024, 16, 3912. [Google Scholar] [CrossRef]
- Cheng, Y.; Zhu, W.; Li, D.; Wang, L. Multi-label classification of arrhythmia using dynamic graph convolutional network based on encoder-decoder framework. Biomed. Signal Process. Control 2024, 95, 106348. [Google Scholar] [CrossRef]
- Wang, X.; Si, H.; Zhang, F.; Zhou, X.; Sun, D.; Lyu, W.; Yang, Q.; Tang, J. HGTS-Former: Hierarchical HyperGraph Transformer for Multivariate Time Series Analysis. arXiv 2025, arXiv:2508.02411. [Google Scholar]
- Zhang, W.; Qiu, H. HCLGT-DRP: Hypergraph contrastive learning and graph transformer for drug response prediction. Expert Syst. Appl. 2026, 297, 129320. [Google Scholar] [CrossRef]
- Wu, J.; Gao, Z.; Jing, G.; Li, Q.; Zhang, Y. HyperMM: Satellite Image Sequence Prediction via Hypergraph-Enhanced Motion Matrix. IEEE Trans. Geosci. Remote Sens. 2025. early access. [Google Scholar] [CrossRef]
- Tian, J.; Lu, P.; Sha, H. HCNS:A deep learning model for identifying essential proteins based on hypergraph convolution and sequence features. Anal. Biochem. 2025, 707, 115949. [Google Scholar] [CrossRef]
- Huang, X.; Ye, Y.; Ding, W.; Yang, X.; Xiong, L. Multi-mode dynamic residual graph convolution network for traffic flow prediction. Inf. Sci. 2022, 609, 548–564. [Google Scholar] [CrossRef]
- Qin, Y.; Fang, Y.; Luo, H.; Zhao, F.; Wang, C. DMGCRN: Dynamic Multi-Graph Convolution Recurrent Network for Traffic Forecasting. arXiv 2021, arXiv:2112.02264. [Google Scholar] [CrossRef]
- Chawla, N.V.; Bowyer, K.W.; Hall, L.O.; Kegelmeyer, W.P. SMOTE: Synthetic Minority Over-sampling Technique. J. Artif. Intell. Res. 2002, 16, 321–357. [Google Scholar] [CrossRef]
- Chen, C.; Shen, W.; Yang, C.; Fan, W.; Liu, X.; Li, Y. A New Safe-Level Enabled Borderline-SMOTE for Condition Recognition of Imbalanced Dataset. IEEE Trans. Instrum. Meas. 2023, 72, 1–10. [Google Scholar] [CrossRef]
- Mathew, J.; Luo, M.; Pang, C.K.; Chan, H.L. Kernel-based SMOTE for SVM classification of imbalanced datasets. In Proceedings of the IECON 2015—41st Annual Conference of the IEEE Industrial Electronics Society, Yokohama, Japan, 9–12 November 2015; pp. 1127–1132. [Google Scholar] [CrossRef]
- Zhao, L.; Shang, Z.; Qin, A.; Zhang, T.; Zhao, L.; Wei, Y.; Tang, Y.Y. A cost-sensitive meta-learning classifier: SPFCNN-Miner. Future Gener. Comput. Syst. 2019, 100, 1031–1043. [Google Scholar] [CrossRef]
- Chawla, N.; Lazarevic, A.; Hall, L.; Bowyer, K. SMOTEBoost: Improving Prediction of the Minority Class in Boosting. In Proceedings of the European Conference on Principles of Data Mining and Knowledge Discovery; Springer: Berlin/Heidelberg, Germany, 2003; Volume 838, pp. 107–119. [Google Scholar] [CrossRef]
- Zhao, T.; Zhang, X.; Wang, S. GraphSMOTE: Imbalanced Node Classification on Graphs with Graph Neural Networks. In Proceedings of the 14th ACM International Conference on Web Search and Data Mining, Virtual, 8–12 March 2021; ACM: New York, NY, USA, 2021; pp. 833–841. [Google Scholar] [CrossRef]
- Chen, D.; Lin, Y.; Zhao, G.; Ren, X.; Li, P.; Zhou, J.; Sun, X. Topology-imbalance learning for semi-supervised node classification. In Proceedings of the 35th International Conference on Neural Information Processing Systems, Red Hook, NY, USA, 6–14 December 2021. NIPS ’21. [Google Scholar]
- Zhou, M.; Gong, Z. GraphSR: A Data Augmentation Algorithm for Imbalanced Node Classification. arXiv 2023, arXiv:2302.12814. [Google Scholar] [CrossRef]
- Li, W.Z.; Wang, C.D.; Xiong, H.; Lai, J.H. GraphSHA: Synthesizing Harder Samples for Class-Imbalanced Node Classification. arXiv 2023, arXiv:2306.09612. [Google Scholar] [CrossRef]
- Wang, J.; Yang, J.; Lidun. Wacml: Based on graph neural network for imbalanced node classification algorithm. Multimed. Syst. 2024, 30, 258. [Google Scholar] [CrossRef]
- Peterfy, C.; Schneider, E.; Nevitt, M. The osteoarthritis initiative: Report on the design rationale for the magnetic resonance imaging protocol for the knee. Osteoarthr. Cartil. 2008, 16, 1433–1441. [Google Scholar] [CrossRef]
- Ambellan, F.; Tack, A.; Ehlke, M.; Zachow, S. Automated segmentation of knee bone and cartilage combining statistical shape knowledge and convolutional neural networks: Data from the Osteoarthritis Initiative. Med. Image Anal. 2019, 52, 109–118. [Google Scholar] [CrossRef] [PubMed]
- Frazier, P.I. A Tutorial on Bayesian Optimization. arXiv 2018, arXiv:1807.02811. [Google Scholar] [CrossRef]
- Lim, B.; Arik, S.O.; Loeff, N.; Pfister, T. Temporal Fusion Transformers for interpretable multi-horizon time series forecasting. Int. J. Forecast. 2021, 37, 1748–1764. [Google Scholar] [CrossRef]
- Lin, Y.; Koprinska, I.; Rana, M. Temporal Convolutional Attention Neural Networks for Time Series Forecasting. In Proceedings of the 2021 International Joint Conference on Neural Networks (IJCNN), Virtual, 18–22 July 2021; pp. 1–8. [Google Scholar] [CrossRef]
- Zhang, H.; Zou, Y.; Yang, X.; Yang, H. A temporal fusion transformer for short-term freeway traffic speed multistep prediction. Neurocomputing 2022, 500, 329–340. [Google Scholar] [CrossRef]
- Wesseling, J.; Boers, M.; Viergever, M.; Hilberdink, W.; Lafeber, F.; Dekker, J.; Bijlsma, J. Cohort profile: Cohort Hip and Cohort Knee (CHECK) study. Int. J. Epidemiol. 2014, 45, 36–44. [Google Scholar] [CrossRef]


| Inclusion Set | Evaluation Criteria | ||||
|---|---|---|---|---|---|
| Incidence | - | ||||
| Progression | |||||
| 0 | 714 | 509 | 448 | 329 | 479 | 302 | 0 | 0 | 0 | ||
| 1 | 1441 | 1017 | 729 | 918 | 1002 | 514 | 0 | 0 | 0 | ||
| 2 | 1102 | 779 | 524 | 678 | 881 | 451 | 1921 | 1127 | 958 | ||
| 3 | 0 | 0 | 0 | 513 | 762 | 434 | 1640 | 874 | 743 | ||
| 7263 | 7263 | 7263 | |||||||||
| Hyperparameter | Search Space | Incidence | Progression |
|---|---|---|---|
| 1 cm | |||
| 4 | 3 | ||
| 4 | 2 | ||
| 256 | 64 | ||
| 8 | 4 | ||
| 256 | 128 | ||
| 256 | 64 |
| Method | Dataset | Source | Inclusion Set | Definition | Pred. Target | Results |
|---|---|---|---|---|---|---|
| Guan et al. [7] | OAI | X-ray + {Dem., Injury hist., Tbf. angle} | Baseline | at | ||
| Pedoia et al. [8] | OAI | MRI | ||||
| Alexopoulos et al. [9] | OAI | MRI | at | |||
| Du et al. [14] | OAI | MRI | - | - | at | |
| Janvier et al. [21] | OAI | X-ray | at | |||
| Joseph et al. [11] | OAI | MRI | at | 1 cm7 | ||
| Kerkhof et al. [13] | Rotterdam H. | Imaging variables | - | |||
| Kinds et al. [12] | CHECK | X-ray | at | 1cm9 | ||
| Woloszynski et al. [22] | LU | MRI | at | |||
| Proposed | OAI | MRI + {Dem., Injury hist.} | at | |||
| at |
| Method | Dataset | Source | Inclusion Set | Definition | Pred. Target | Results |
|---|---|---|---|---|---|---|
| Imjabbar et al. [15] | OAI | X-ray | KL at | |||
| Imjabbar et al. [15] | MOST | X-ray | KL at | |||
| Guan et al. [7] | OAI | X-ray | KL at | |||
| Janvier et al. [21] | OAI | X-ray | KL at | |||
| Woloszynski et al. [22] | LUH | X-ray | KL at | |||
| Kraus et al. [19] | Pfizer | X-ray | KL at | |||
| Kraus et al. [20] | FNIH | X-ray | KL at | |||
| Kraus et al. [18] | POP | X-ray | KL at | |||
| Halilaj et al. [16] | OAI | X-ray | - | - | at | |
| Schiratti et al. [23] | OAI | MRI | 1 cm ≤ ≤ | at | ||
| Panfilov et al. [17] | OAI | MRI, X-ray | - | - | at | |
| Proposed | OAI | MRI + {Dem., Injury hist.} | at | at | ||
| at | ||||||
| Gender | Age | BMI | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Male | Female | Normal | Pre-Obese | Obese | |||||||||
| Inc. | |||||||||||||
| Progr. | |||||||||||||
| Knee #1 (Top Row) | Knee #2 (Medium Row) | Knee #3 (Bottom Row) | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Actual | ||||||||||||||
| Predicted | ||||||||||||||
| Full model configuration | ||||
| w/o HyGraphSMOTE | ||||
| w/o Demographics [all] | ||||
| w/o Demographics [Age] | ||||
| w/o Demographics [BMI] | ||||
| w/o Demographics [Gender] | ||||
| w/o Injury | ||||
| w/o SSL | ||||
| w/o Pretraining | ||||
| w/o ACT |
| Full model configuration | ||||
| w/o HyGraphSMOTE | ||||
| w/o Demographics [all] | ||||
| w/o Demographics [Age] | ||||
| w/o Demographics [BMI] | ||||
| w/o Demographics [Gender] | ||||
| w/o Injury | ||||
| w/o SSL | ||||
| w/o Pretraining | ||||
| w/o ACT |
| Component | Parameters (Inc.) | Percentage (%) | Parameters (Progr.) | Percentage (%) |
|---|---|---|---|---|
| C_Shape.Net | 1,072,421 | 26.67% | 943,972 | 52.39% |
| MHGCN | 1,755,940 | 43.68% | 468,900 | 26.02% |
| Encoder | 185,856 | 4.62% | 74,496 | 4.13% |
| Decoder | 739,461 | 18.37% | 246,658 | 13.70% |
| ACT | 266,240 | 6.66% | 67,584 | 3.75% |
| Total |
| Incidence | Progression | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| DyHRM | |||||||||
| TFT | |||||||||
| TCAN | |||||||||
| AGCRNN | |||||||||
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Theocharis, J.B.; Chadoulos, C.G.; Symeonidis, A.L. A Dynamic Hypergraph-Based Encoder–Decoder Risk Model for Longitudinal Predictions of Knee Osteoarthritis Progression. Mach. Learn. Knowl. Extr. 2025, 7, 94. https://doi.org/10.3390/make7030094
Theocharis JB, Chadoulos CG, Symeonidis AL. A Dynamic Hypergraph-Based Encoder–Decoder Risk Model for Longitudinal Predictions of Knee Osteoarthritis Progression. Machine Learning and Knowledge Extraction. 2025; 7(3):94. https://doi.org/10.3390/make7030094
Chicago/Turabian StyleTheocharis, John B., Christos G. Chadoulos, and Andreas L. Symeonidis. 2025. "A Dynamic Hypergraph-Based Encoder–Decoder Risk Model for Longitudinal Predictions of Knee Osteoarthritis Progression" Machine Learning and Knowledge Extraction 7, no. 3: 94. https://doi.org/10.3390/make7030094
APA StyleTheocharis, J. B., Chadoulos, C. G., & Symeonidis, A. L. (2025). A Dynamic Hypergraph-Based Encoder–Decoder Risk Model for Longitudinal Predictions of Knee Osteoarthritis Progression. Machine Learning and Knowledge Extraction, 7(3), 94. https://doi.org/10.3390/make7030094









