A Novel Prediction Model for Multimodal Medical Data Based on Graph Neural Networks
Abstract
1. Introduction
2. Materials and Methods
2.1. Graph Data Structure for Feature Expression
2.2. GDS Construction Based on Similarity Measurement
2.3. The Learning of GDS Using a Graph Neural Network
- Sampling: The model first samples the neighborhood node information of the original object node. The rule of sampling is as follows: firstly, set one is the original object node, and the remaining n nodes are the neighboring nodes. Set m as a fixed number of nodes for each sampling, and if n > = m, perform sampling without dropout; otherwise, perform sampling with dropout until m nodes are sampled. It is worth noting that GraphSage can achieve significant generalization of non-adjacent nodes without the involvement of the whole graph structure during the sampling process.
- Aggregation: For the sampled information, the next step is to aggregate it according to a specific rule. This process involves combining the feature vectors of a node and its neighbors, along with their respective weights, to capture the graph’s structure and generate new neighborhood embeddings. The aggregator function is described by Equation (3). In a GraphSAGE model, there are three primary types of aggregators: the mean aggregator, LSTM aggregator, and pooling aggregator. Additionally, convolution-based GCN operators can also be utilized as aggregators.where σ represents the non-linear activation function, represents the weight matrix of aggregators, represents vector concatenation, and represents the feature vector of the node.
- Prediction: The results will be input into the downstream machine learning classifiers for training, enabling node prediction.
2.4. Disease Prediction Model Based on GraphSAGE
| Algorithm 1: EPGC algorithm |
| Input: Multimodal data ; the depth of graph; weight matrix ; non-linear activation function ; aggregator. Adjacency function: . Output: , the result of the following prediction: 1: Extract features from non-numerical data in to obtain numerical data . 2: Normalize , . 3: Calculate the Pearson correlation coefficient between each sample in according to Equation (2). 4: Construct the graph . 5: Sample the node information of and generate feature vector . 6: Aggregate feature information, , and generate new feature vectors. 7: Normalize feature vectors, . 8: Generate new node embedding, . 9: Classify feature vectors and output the prediction result of diseases. |
3. Experimental Section
3.1. Experimental Data
3.2. Experimental Design
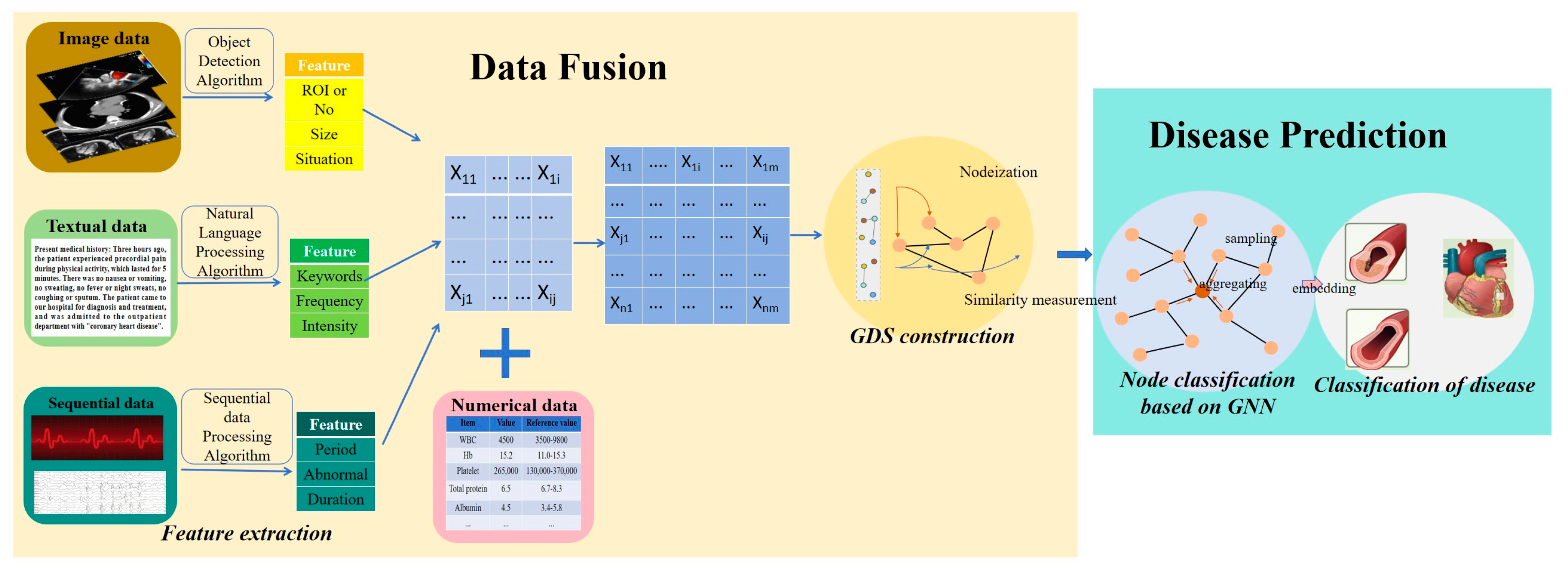
3.2.1. Data Fusion and Disease Prediction
- Data preprocessing: This mainly involves standardizing the format and dimensions of experimental data and converting the multimodal data into numerical data. We use AI-based feature extraction to convert these multimodal data into numerical data. This needs to be implemented based on AI algorithms, as shown in Figure 3. It mainly focuses on image data (CT, MRI, etc.), textual data (symptom description, etc.), and sequential data (electrocardiogram, electroencephalogram, etc.). The ECG data describe the features of a patient’s ECG-related bands and was preprocessed by the dataset provider. Additionally, some individual features in the original data include descriptive text data, such as the severity of heart valve disease, which is categorized as N, Mild, Moderate, and Severe to describe the patient’s condition. For consistency and to perform a numerical analysis, we convert these descriptive categories into numerical values: 0 for N, 1 for Mild, 2 for Moderate, and 3 for Severe. In addition, the multimodal medical data are characterized by dimensional inconsistency. To address this, we apply mean normalization to the original data. This step helps to mitigate the impact of varying dimensions on model learning and ensures that the classifier accuracy is not adversely affected by these differences.
- GDS construction: Each patient is represented as a node, and the similarity between patients is represented as an edge. The edges are determined using the Pearson correlation coefficient. According to the Pearson correlation coefficient, an edge is drawn between two nodes if their correlation coefficient is greater than or equal to 0.5, i.e., , indicating a strong similarity and thus a connection between them. Conversely, if the coefficient is 0.5 or less, i.e., , this indicates a weak similarity and no connection edge is drawn between the nodes.
- Disease prediction—designing the network learning models: The constructed GDS is undirected. We propose using a GraphSAGE network, which samples the node information with random walks on the graph and generates the node features. Given that the types and sizes of experimental data are relatively simple, we propose using a two-layer GNN network, utilizing a GCN aggregator and mean aggregator, respectively. This design approach enhances the extraction of features from the graph structure. The expression of the GCN aggregator is shown in Equation (4), where represents the feature vector of node, represents the non-linear activation function, represents the adjacency weight matrix of node, and is the normalized Laplacian matrix. The mean aggregator is shown in Equation (5), where represents the feature vector of node , and g represents the , which is the adjacent point of the node.
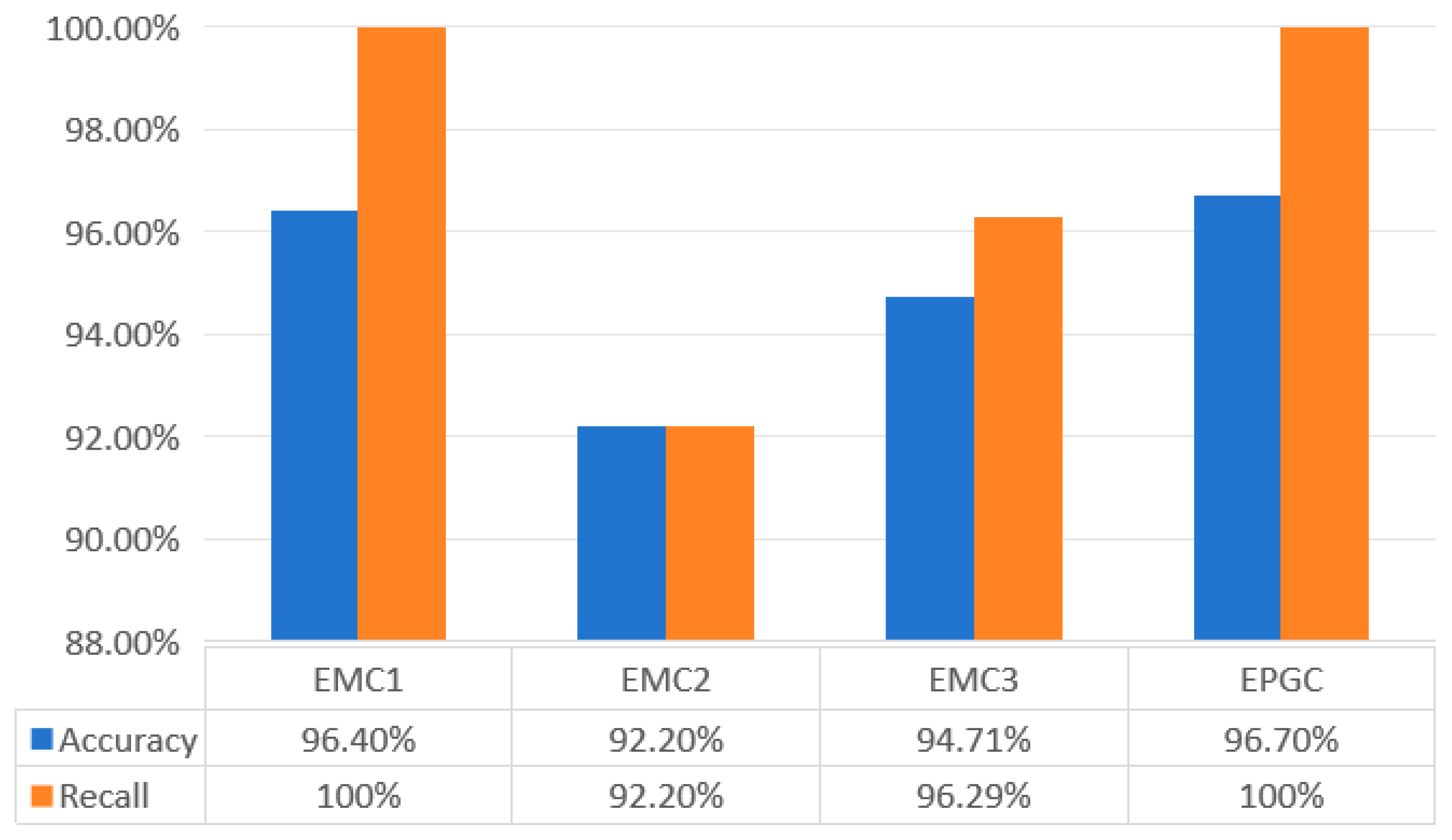
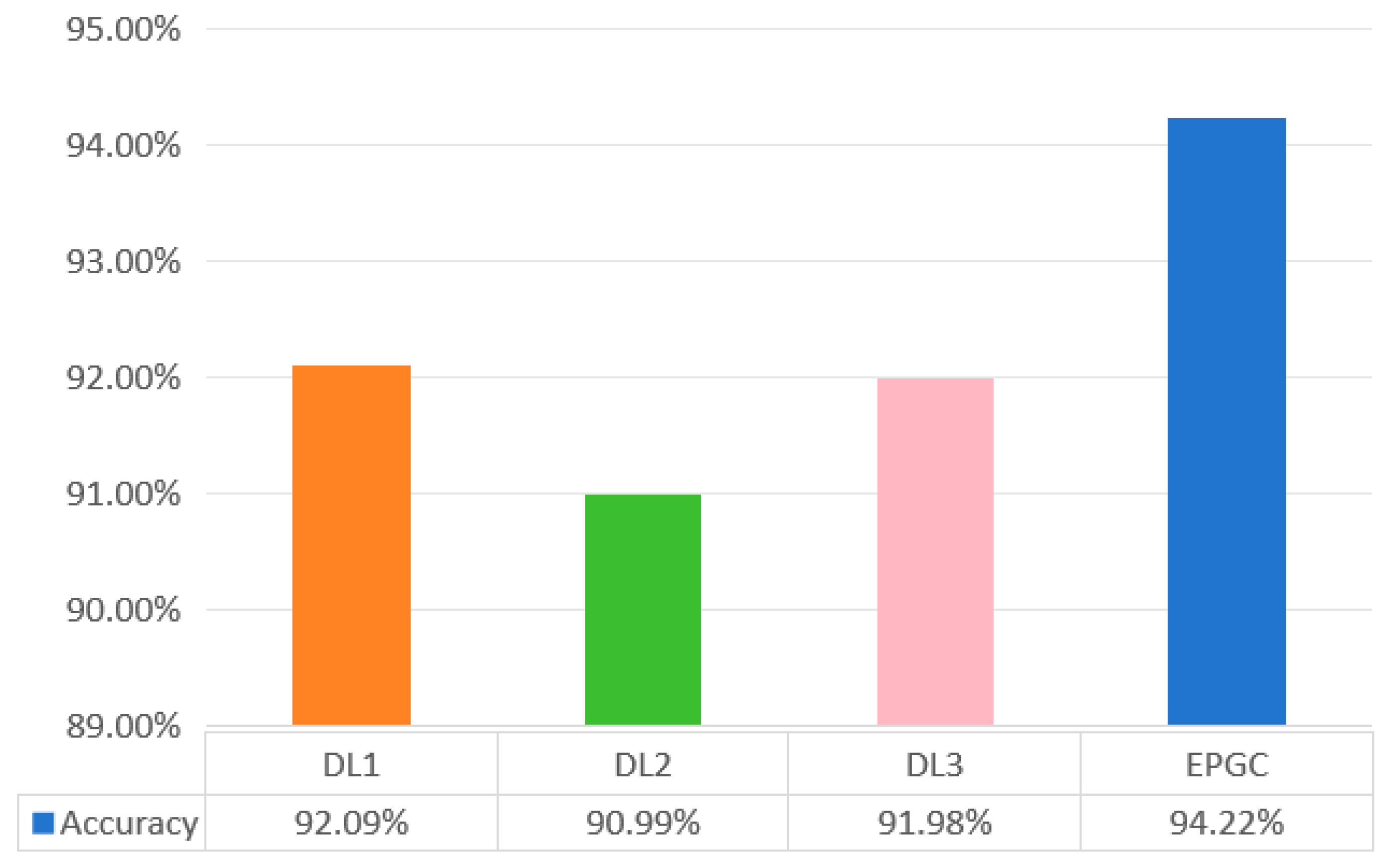
3.2.2. Control Group Experiment
3.2.3. Model Evaluation
4. Results
5. Discussion
6. Conclusions
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
Correction Statement
References
- Jiang, S.; Wang, T.; Zhang, K. Data-driven decision-making for precision diagnosis of digestive diseases. Biomed. Eng. Online 2023, 22, 87. [Google Scholar] [CrossRef] [PubMed]
- Khoa, L.D.V.; Yen, L.M.; Nhat, P.T.H.; Duong, H.T.H.; Thuy, D.B.; Zhu, T.; Greeff, H.; Clifton, D.; Thwaites, C.L. Vital sign monitoring using wearable devices in a Vietnamese intensive care unit. BMJ Innov. 2021, 7, s1–s5. [Google Scholar] [CrossRef]
- Diagnosis—Coronary Heart Disease. Available online: https://www.nhs.uk/conditions/coronary-heart-disease/diagnosis/ (accessed on 17 January 2024).
- Cai, Q.; Wang, H.; Li, Z.; Liu, X. A Survey on Multimodal Data-Driven Smart Healthcare Systems: Approaches and Applications. IEEE Access 2019, 7, 133583–133599. [Google Scholar] [CrossRef]
- Beam, A.L.; Kompa, B.; Schmaltz, A.; Fried, I.; Weber, G.; Palmer, N.; Shi, X.; Cai, T.; Kohane, I.S. Clinical Concept Embeddings Learned from Massive Sources of Multimodal Medical Data. Pac. Symp. Biocomput. 2020, 25, 295–306. [Google Scholar]
- Lipkova, J.; Chen, R.J.; Chen, B.; Lu, M.Y.; Barbieri, M.; Shao, D.; Vaidya, A.J.; Chen, C.; Zhuang, L.; Williamson, D.F.K.; et al. Artificial intelligence for multimodal data integration in oncology. Cancer Cell 2022, 40, 1095–1110. [Google Scholar] [CrossRef]
- Wei, T.; Tiwari, P.; Pandey, H.M.; Moreira, C.; Jaiswal, A.K. Multimodal medical image fusion algorithm in the era of big data. In Neural Computing & Applications; Springer: Berlin/Heidelberg, Germany, 2020. [Google Scholar] [CrossRef]
- Acosta, J.N.; Falcone, G.J.; Rajpurkar, P.; Topol, F.J. Multimodal biomedical AI. Nat. Med. 2022, 28, 773–1784. [Google Scholar] [CrossRef]
- Kline, A.; Wang, H.; Li, Y.; Dennis, S.; Hutch, M.; Xu, Z.; Wang, F.; Cheng, F.; Luo, Y. Multimodal machine learning in precision health: A scoping review. Npj Digit. Med. 2022, 5, 171. [Google Scholar] [CrossRef] [PubMed]
- Stahlschmidt, S.R.; Ulfenborg, B.; Synnergren, J. Multimodal deep learning for biomedical data fusion: A review. Brief. Bioinform. 2022, 23, bbab569. [Google Scholar] [CrossRef]
- Azam, M.A.; Khan, K.B.; Salahuddin, S.; Rehman, E.; Khan, S.A.; Khan, M.A.; Kadry, S.; Gandomi, A.H. A review on multimodal medical image fusion: Compendious analysis of medical modalities, multimodal databases, fusion techniques and quality metrics. Comput. Biol. Med. 2022, 144, 105253. [Google Scholar] [CrossRef] [PubMed]
- Guo, Z.; Li, X.; Huang, H.; Guo, N.; Li, Q. Deep Learning-Based Image Segmentation on Multimodal Medical Imaging. IEEE Trans. Radiat. Plasma Med. Sci. 2019, 3, 162–169. [Google Scholar] [CrossRef]
- Duan, J.; Xiong, J.; Li, Y.; Ding, W. Deep learning based multimodal biomedical data fusion: An overview and comparative review. Inf. Fusion 2024, 112, 102536. [Google Scholar] [CrossRef]
- Chen, H.; Gao, M.; Zhang, Y. Attention-Based Multi-NMF Deep Neural Network with Multimodality Data for Breast Cancer Prognosis Model. Biomed. Res. Int. 2019, 2019, 9523719. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.; Ju, J.; Guo, L.; Ji, B.; Shi, S.; Yang, Z.; Gao, S.; Yuan, X.; Tian, G.; Liang, Y.; et al. Prediction of HER2-positive breast cancer recurrence and metastasis risk from histopathological images and clinical information via multimodal deep learning. Comput. Struct. Biotechnol. J. 2021, 20, 333–342. [Google Scholar] [CrossRef]
- Alizadehsani, R.; Hosseini, M.J.; Khosravi, A.; Khozeimeh, F.; Roshanzamir, M.; Sarrafzadegan, M. Non-invasive detection of coronary artery disease in high-risk patients based on the stenosis prediction of separate coronary arteries. Comput. Methods Programs Biomed. 2018, 162, 119–127. [Google Scholar] [CrossRef]
- Golovenkin, S.E.; Dorrer, M.G.; Nikulina, S.Y.; Orlova, Y.V.; Pelipeckaya, E.V. Evaluation of the effectiveness of using artificial intelligence to predict the response of the human body to cardiovascular diseases. J. Phys. Conf. Ser. 2020, 1679, 042017. [Google Scholar] [CrossRef]
- Yavru, I.B.; Gunduz, S.Y. Predicting Myocardial Infarction Complications and Outcomes with Deep Learning. Eskişehir Tech. Univ. J. Sci. Technol. A-Appl. Sci. Eng. 2022, 23, 184–194. [Google Scholar] [CrossRef]
- Hu, S.L. Research on Medical Multi-Source Data Fusion Based on Big Data. Recent Adv. Comput. Sci. Commun. 2022, 15, 376–387. [Google Scholar] [CrossRef]
- Bronstein, M.M.; Bruna, J.; LeCun, Y.; Szlam, A.; Vandergheynst, P. Geometric Deep Learning: Going beyond Euclidean data. IEEE Signal Process. Mag. 2017, 34, 18–42. [Google Scholar] [CrossRef]
- Wang, L.; Zhang, Y.; Feng, J. On the Euclidean distance of images. IEEE Trans. Pattern Anal. Mach. Intell. 2005, 27, 1334–1339. [Google Scholar] [CrossRef]
- He, H.; Wu, D. Transfer Learning for Brain—Computer Interfaces: A Euclidean Space Data Alignment Approach. IEEE Trans. Biomed. Eng. 2020, 67, 399–410. [Google Scholar] [CrossRef]
- Tahirovic, A.A.; Angeli, D.; Tahirovic, A.; Strbac, G. Petri graph neural networks advance learning higher order multimodal complex interactions in graph structured data. Sci. Rep. 2025, 15, 17540. [Google Scholar] [CrossRef] [PubMed]
- Walke, D.; Micheel, D.; Schallert, K.; Muth, T.; Broneske, D.; Saake, G.; Heyer, R. The importance of graph databases and graph learning for clinical applications. Database 2023, 2024, baad045. [Google Scholar] [CrossRef]
- Shao, S.; Ribeiro, P.H.; Ramirez, C.M.; Moore, J.H. A review of feature selection strategies utilizing graph data structures and Knowledge Graphs. Brief. Bioinform. 2024, 25, bbae52. [Google Scholar] [CrossRef]
- Jarvis, R.A.; Patrick, E.A. Clustering Using a Similarity Measure Based on Shared Near Neighbors. IEEE Trans. Comput. 1973, C-22, 1025–1034. [Google Scholar] [CrossRef]
- Lin, Y.S.; Jiang, J.Y.; Lee, S.J. A Similarity Measure for Text Classification and Clustering. IEEE Trans. Knowl. Data Eng. 2013, 26, 1575–1590. [Google Scholar] [CrossRef]
- Liu, X. Entropy, distance measure and similarity measure of fuzzy sets and their relations. Fuzzy Sets Syst. 1992, 52, 305–318. [Google Scholar] [CrossRef]
- Gori, M.; Monfardini, G.; Scarselli, F. A New Model for Learning in Graph Domains. In Proceedings of the IEEE International Joint Conference on Neural Network (IJCNN), Montreal, QC, Canada, 31 July–4 August 2005; pp. 729–734. [Google Scholar]
- Zhou, J.; Cui, G.; Hu, S.; Zhang, Z.; Yang, C.; Liu, Z.; Wang, L.; Li, C.; Sun, M. Graph neural networks: A review of methods and applications. AI Open 2020, 1, 57–81. [Google Scholar] [CrossRef]
- Qiu, J.; Zhang, X.; Wang, T.; Hou, H.; Wang, S.; Yang, T. A GNN-Based False Data Detection Scheme for Smart Grids. Algorithms 2025, 18, 166. [Google Scholar] [CrossRef]
- Mateo, S.; Danijel, K.; Mršić, L. Graph Convolutional Networks for Predicting Cancer Outcomes and Stage: A Focus on cGAS-STING Pathway Activation. Mach. Learn. Knowl. Extr. 2024, 6, 2033–2048. [Google Scholar]
- Shao, J.; Zhang, H.; Mao, Y.; Zhang, J. Branchy-GNN: A Device-Edge Co-Inference Framework for Efficient Point Cloud Processing. In Proceedings of the IEEE International Conference on Acoustics, Speech and Signal Processing (ICASSP), Toronto, ON, Canada, 6–11 June 2021; pp. 8488–8492. [Google Scholar]
- Alawad, D.M.; Katebi, A.; Hoque, M.T. Enhanced Graph Representation Convolution: Effective Inferring Gene Regulatory Network Using Graph Convolution Network with Self-Attention Graph Pooling Layer. Mach. Learn. Knowl. Extr. 2024, 6, 1818–1839. [Google Scholar] [CrossRef]
- Mohanraj, P.; Raman, V.; Ramanathan, S. Deep Learning for Parkinson’s Disease Diagnosis: A Graph Neural Network (GNN) Based Classification Approach with Graph Wavelet Transform (GWT) Using Protein—Peptide Datasets. Diagnostics 2024, 14, 2181. [Google Scholar] [CrossRef]
- Hossain, M.E.; Uddin, S.; Khan, A. Network analytics and machine learning for predictive risk modelling of cardiovascular disease in patients with type 2 diabetes. Expert Syst. Appl. 2020, 164, 113918. [Google Scholar] [CrossRef]
- Xuan, P.; Wu, X.; Cui, H.; Jin, Q.; Wang, L.; Zhang, T.; Nakaguchi, T.; Duh, H.B.L. Multi-scale random walk driven adaptive graph neural network with dual-head neighboring node attention for CT segmentation. Appl. Soft Comput. 2023, 133, 109905. [Google Scholar] [CrossRef]
- Lee, Y.W.; Huang, S.K.; Chang, R.F. CheXGAT: A disease correlation-aware network for thorax disease diagnosis from chest X-ray images. Artif. Intell. Med. 2022, 132, 102382. [Google Scholar] [CrossRef] [PubMed]
- Zhang, T.; Shan, H.R.; Little, M.A. Causal GraphSAGE: A robust graph method for classification based on causal sampling. Pattern Recognit. 2022, 128, 108696. [Google Scholar] [CrossRef]
- Cardiovascular Diseases. Available online: https://www.who.int/health-topics/cardiovascular-diseases#tab=tab_1 (accessed on 17 January 2024).
- The DISCHARGE Trial Group. CT or Invasive Coronary Angiography in Stable Chest Pain. N. Engl. J. Med. 2022, 386, 1591–1602. [Google Scholar] [CrossRef]
- Mann, D.L.; Zipes, D.P.; Libby, P.; Bonow, R.O. Braunwald’s Heart Disease: A Textbook of Cardiovascular Medicine, 10th ed.; Elsevier Health Sciences: Amsterdam, The Netherlands, 2018; pp. 636–661. [Google Scholar]
- Reed, G.W.; Rossi, J.E.; Cannon, C.P. Acute myocardial infarction. Lancet 2017, 389, 197–210. [Google Scholar] [CrossRef]
- Moras, E.; Yakkali, S.; Gandhi, K.D.; Virk, H.U.H.; Alam, M.; Zaid, S.; Barman, N.; Jneid, H.; Vallabhajosyula, S.; Sharma, S.K.; et al. Complications in Acute Myocardial Infarction: Navigating Challenges in Diagnosis and Management. Hearts 2024, 5, 122–141. [Google Scholar] [CrossRef]
- Complications of Myocardial Infarction. Available online: https://emedicine.medscape.com/article/164924-overview (accessed on 21 July 2022).
- Alizadehsani, R.; Roshanzamir, M.; Sani, Z. Extention of Z-Alizadeh Sani Dataset Data Set; UCI Irvine-Machine Leaming-Repository: Irvine, CA, USA, 2013. [Google Scholar] [CrossRef]
- Golovenkin, S.E.; Shulman, V.A.; Rossiev, D.A.; Shesternya, P.A.; Nikulina, S.Y.; Orlova, Y.V. Myocardial Infarction Complications Data Set; UC Irvine-Machine Leaming-Repository: Irvine, CA, USA, 2020. [Google Scholar] [CrossRef]
- Olawade, D.B.; Soladoye, A.A.; Omodunbi, B.A.; Aderinto, N.; Adeyanju, I.A. Comparative analysis of machine learning models for coronary artery disease prediction with optimized feature selection. Int. J. Cardiol. 2025, 436, 133443. [Google Scholar] [CrossRef] [PubMed]
- Hashemi, M.; Komamardakhi, S.S.S.; Maftoun, M.; Zare, O.; Joloudari, J.H.; Nematollahi, M.A.; Alizadehsani, R.; Sala, P.; Gorriz, J.M. Enhancing Coronary Artery Disease Classification Using Optimized MLP Based on Genetic Algorithm. In Proceedings of the Artificial Intelligence for Neuroscience and Emotional Systems (IWINAC 2024), Olhao, Algarve, Portugal, 4–7 June 2024; p. 14674. [Google Scholar]
- Makhmudov, F.; Ravshanov, N.; Akhmedov, D.; Pekos, O.; Turimov, D.; Cho, Y.I. A Multitask Deep Learning Model for Predicting Myocardial Infarction Complications. Bioengineering 2025, 12, 520. [Google Scholar] [CrossRef]
- Bajaj, S.; Son, H.; Liu, J.; Guan, H.; Serafini, M. Graph Neural Network Training Systems: A Performance Comparison of Full-Graph and Mini-Batch. arXiv 2024, arXiv:2406.00552v3. [Google Scholar] [CrossRef]
- Tare, M.; Rattasits, C.; Wu, Y.; Wielewski, E. Harnessing GraphSAGE for Learning Representations of Massive Transactional Networks. In Proceedings of the 14th IAPR-TC-15 International Workshop, Graph-Based Representations in Pattern Recognition (GbRPR 2025), Caen, France, 25–27 June 2025; pp. 179–188. [Google Scholar]
- Badrinath, A.; Yang, A.; Rajesh, K.; Agarwal, P.; Yang, J.; Chen, H.; Xu, J.; Rosenberg, C. OmniSage: Large Scale, Multi-Entity Heterogeneous Graph Representation Learning. In Proceedings of the 31st ACM SIGKDD Conference on Knowledge Discovery and Data Mining (SIGKDD), Toronto, ON, Canada, 3–7 August 2025; pp. 4261–4272. [Google Scholar]


| Feature | Details |
|---|---|
| Demographic | Age, Weight, Sex, BMI (Body Mass Index), DM (Diabetes Mellitus), Current Smoker, Ex-Smoker, FH (Family History), CRF (Chronic Renal Failure), CVA (Cerebrovascular Accident), Thyroid Disease, CHF (Congestive Heart Failure), DLP (Dyslipidemia), etc. |
| Symptoms and examination | BP (Blood Pressure), PR (Pulse Rate), Edema, Weak Peripheral Pulse, Lung Rales, Systolic Murmur, Diastolic Murmur, Typical Chest Pain, Dyspnea, Function Class, Atypical, Nonanginal CP, Exertional CP (Exertional Chest Pain), Low Th Ang (Low Threshold angina). |
| ECG and vectorcardiogram | Rhythm, Q Wave, ST Elevation, ST Depression, T Inversion, LVH (Left Ventricular Hypertrophy), Poor R Progression (Poor R Wave Progression), LAD (Left Anterior Descending), LCX (Left Circumflex), RCA (Right Coronary Artery). |
| Laboratory and echo | FBS (Fasting Blood Sugar), Cr (Creatine) (mg/dl), TG (Triglyceride), LDL (Low-Density Lipoprotein), HDL (High-Density Lipoprotein), BUN (Blood Urea Nitrogen), ESR (Erythrocyte Sedimentation Rate), HB (Hemoglobin), K (Potassium), Na (Sodium), WBC (White Blood Cell), Lymph (Lymphocyte), Neut (Neutrophil), PLT (Platelet), EF (Ejection Fraction), Region with RWMAa (Regional Wall Motion Abnormality), VHD (Valvular Heart Disease). |
| Type of coronary artery disease | Yes or No |
| Feature | Details |
|---|---|
| The clinical description of myocardial infarction. | Demography, electrocardiogram, laboratory data, hospitalization records, medication records, etc. |
| The types of myocardial infarction complications. | Atrial fibrillation (AF), supraventricular tachycardia (ST), ventricular tachycardia (VT), ventricular fibrillation (VF), third-degree AV block (TA), pulmonary edema (PE), myocardial rupture (MR), Dressler syndrome (DS), chronic heart failure (CH), relapse of the myocardial infarction (RM), post-infarction angina (PA), lethal outcome (LO). |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, L.; Li, T.; Cui, H.; Zhang, Q.; Jiang, Z.; Li, J.; Welsch, R.E.; Jia, Z. A Novel Prediction Model for Multimodal Medical Data Based on Graph Neural Networks. Mach. Learn. Knowl. Extr. 2025, 7, 92. https://doi.org/10.3390/make7030092
Zhang L, Li T, Cui H, Zhang Q, Jiang Z, Li J, Welsch RE, Jia Z. A Novel Prediction Model for Multimodal Medical Data Based on Graph Neural Networks. Machine Learning and Knowledge Extraction. 2025; 7(3):92. https://doi.org/10.3390/make7030092
Chicago/Turabian StyleZhang, Lifeng, Teng Li, Hongyan Cui, Quan Zhang, Zijie Jiang, Jiadong Li, Roy E. Welsch, and Zhongwei Jia. 2025. "A Novel Prediction Model for Multimodal Medical Data Based on Graph Neural Networks" Machine Learning and Knowledge Extraction 7, no. 3: 92. https://doi.org/10.3390/make7030092
APA StyleZhang, L., Li, T., Cui, H., Zhang, Q., Jiang, Z., Li, J., Welsch, R. E., & Jia, Z. (2025). A Novel Prediction Model for Multimodal Medical Data Based on Graph Neural Networks. Machine Learning and Knowledge Extraction, 7(3), 92. https://doi.org/10.3390/make7030092






