1. Introduction
The high volume and variety of data in medical research offer several opportunities and challenges. Of these, Alzheimer’s disease (AD) is a particularly compelling case study because it is multicausal, involving genetic, biochemical, and environmental factors, and also involves complex clinical presentations. Despite the tremendous progress, few effective methods exist for diagnosing, treating, and preventing Alzheimer’s disease (AD). This knowledge gap is further exacerbated by the growing volume and fragmentation across various data modalities, including textual descriptions, clinical trial data, imaging studies, and molecular data. Traditional methods of synthesizing such a large volume of knowledge are ineffective; most have a single-modality approach, which may miss the insights obtained synergistically from integrated data. This gap in methodology underscores the need for a robust, unified framework that can leverage multiple modalities to enhance the retrieval process by making it more context-aware and reducing the retrieval of irrelevant or less pertinent information.
In this research, we describe a novel multimodal retrieval-augmented generation (RAG) application,
AlzheimerRAG (video demonstration), which integrates textual and visual modalities to improve contextual understanding and information synthesis from the biomedical literature. Our primary research objective in implementing multimodal RAG is to enhance context-aware retrieval capabilities by integrating heterogeneous data types, including textual data, images, and clinical trial information from PubMed articles. Existing methods [
1,
2,
3,
4] typically, focus on textual or visual data separately, leaving a gap for integrated multimodal solutions. Latest research in Context-Aware Retrieval [
5,
6,
7,
8,
9,
10] provides the foundation for RAG models by demonstrating how retrieval could enhance the generation capabilities of language models, particularly in knowledge-intensive tasks. Integrating RAG methodologies with multimodal inputs is a burgeoning area of research, as highlighted by Xia et al. [
11], who proposed a multimodal RAG system that enhanced data synthesis across text and image modalities. In light of these advancements, the novelty of our approach lies in the seamless integration and alignment of multimodal data during the cross-modal attention fusion process. The AlzheimerRAG framework combines rapid, accurate retrieval via object stores with specialized language models, enhancing its capability to address the nuances of multimodal information pertinent to Alzheimer’s disease. We utilize an optimized mechanism for fine-tuning by implementing Parameter-Efficient Fine-Tuning (PEFT) [
12] and inducing cross-modal attention fusion to facilitate the synergistic information flow between the text and image models. The fine-tuned models are then incorporated into a multimodal RAG workflow, developed as a web application with a user interface that allows end-users to retrieve context-aware answers from their queries. The target audience of this application includes biomedical researchers for synthesizing Alzheimer’s disease literature and identifying disease trends, clinicians to support diagnosis and treatment planning for AD, and healthcare institutions for clinical trial design and support.
Benchmark datasets such as BioASQ [
13] and PubMedQA [
14] have been instrumental in measuring the effectiveness of multimodal RAG systems [
15]. BioASQ, a large-scale biomedical semantic indexing and question-answering (QA) dataset, provides a robust framework for assessing models’ retrieval and QA capabilities. Similarly, PubMedQA offers insights into the accuracy of models in handling biomedical queries, making it an essential tool for evaluating AlzheimerRAG’s performance against existing benchmarks. In comparative studies, models that integrate multimodal data have been shown to outperform traditional single-modality systems. For instance, models like T5 [
16] have been evaluated in the context of biomedical question answering, demonstrating significant gains when multimodal inputs are utilized. This trend reinforces the need for AlzheimerRAG’s multimodal framework to enhance the understanding and treatment of AD.
In summary, our research contributions advance the multimodal RAG domain in AD in the following aspects:
Context-aware retrieval-augmented generation—Our framework enhances traditional RAG models by prioritizing the context relevance of domain-specific information, thereby increasing accuracy and utility in biomedical applications.
Advanced cross-modal attention fusion—AlzheimerRAG integrates multimodal data more effectively using transformer architectures and cross-modal attention mechanisms tailored to handle heterogeneous data types.
RAG user interface—Our system implements multimodal RAG as a web-based application using the latest state-of-the-art technologies like LangChain, FastAPI, Jinja2, and FaissDB to provide users with a robust interface for performing biomedical information retrieval tasks through the context-aware question-answering paradigm.
Comparable framework with state-of-the-art benchmarks—We evaluate the capability of the Multimodal RAG application with benchmark datasets like BioASQ and PubMedQA, along with other comparable LLM RAG models. We also study the effectiveness of our AlzheimerRAG against human-generated responses for different clinical scenarios in Alzheimer’s disease to gauge the accuracy and hallucination rates of the retrieved answers.
Although this research is rooted in the Alzheimer’s domain, the usage of cross-modal attention fusion in multimodal RAG makes it adaptable to any domain that requires alignment of textual data with visuals, audio, or structured data. The modular approach of AlzheimerRAG supports adapting it to answering capabilities on queries related to comparable medical domains linked to Alzheimer’s Disease. Additionally, the technical scalability is also enhanced by parameter-efficient fine-tuning techniques such as QLoRA, which enables efficient fine-tuning of LLMs (e.g., LlaMA and LLaVA) on niche datasets without full retraining.
2. Related Work
The AlzheimerRAG framework was developed within the rapidly evolving landscape of multimodal data integration and retrieval-augmented generation techniques, which are becoming increasingly crucial in biomedical research. Recent studies have demonstrated the importance of leveraging multiple data modalities to enhance diagnosis, treatment, and understanding of complex diseases, such as Alzheimer’s.
Existing research [
17,
18,
19,
20,
21] has highlighted the efficacy of attention mechanisms that span multiple modalities, which are instrumental in synthesizing heterogeneous information sources in medical contexts. For example, the effectiveness of multimodal token fusion for vision transformers [
22] has been demonstrated, which significantly improves the integration of visual and textual data in medical imaging [
23]. Similarly, cross-modal translation and alignment techniques [
24,
25] have been showcased that facilitate survival analysis, emphasizing the benefits of integrating diverse data types to yield richer insights. Additionally, recent developments in knowledge distillation have further enhanced model efficiency in healthcare applications, as demonstrated in the work by Hinton et al. and Gupta et al. [
26,
27], which involves transferring knowledge from a larger model (teacher) to a smaller model (student), thereby retaining performance while reducing computational costs. Various studies have adopted this methodology, notably, the work that discovered integrating imaging and genetic data improved predictive outputs in Alzheimer’s models [
2].
The application of AI in Alzheimer’s research has been underscored by studies [
28] which leverage multimodal inputs to improve early diagnosis and patient stratification. Other research, such as [
29,
30], has focused on using AI to manage Alzheimer’s disease symptoms, demonstrating that AI-driven solutions can provide valuable insights and recommendations for patient care. The BioBERT model [
31] represents a significant advancement in biomedical text mining, emphasizing the utility of transformer models fine-tuned for biomedical applications. This model has been foundational in developing various biomedical applications, including those focused on Alzheimer’s disease, where precision in information retrieval is critical. RAG methodologies [
32] have gained traction in biomedical research for efficiently synthesizing information from large datasets. The works [
5,
33] laid the ground for RAG models by demonstrating how retrieval could enhance the generation capabilities of language models, particularly in knowledge-intensive tasks. This has profound implications for healthcare, where accurate and timely information retrieval can guide clinical decisions.
Compared to these advancements, the AlzheimerRAG framework combines rapid, accurate retrieval via FaissDB with specialized language models, enhancing its capability to address the nuances of multimodal information pertinent to Alzheimer’s Disease.
3. Materials and Methods
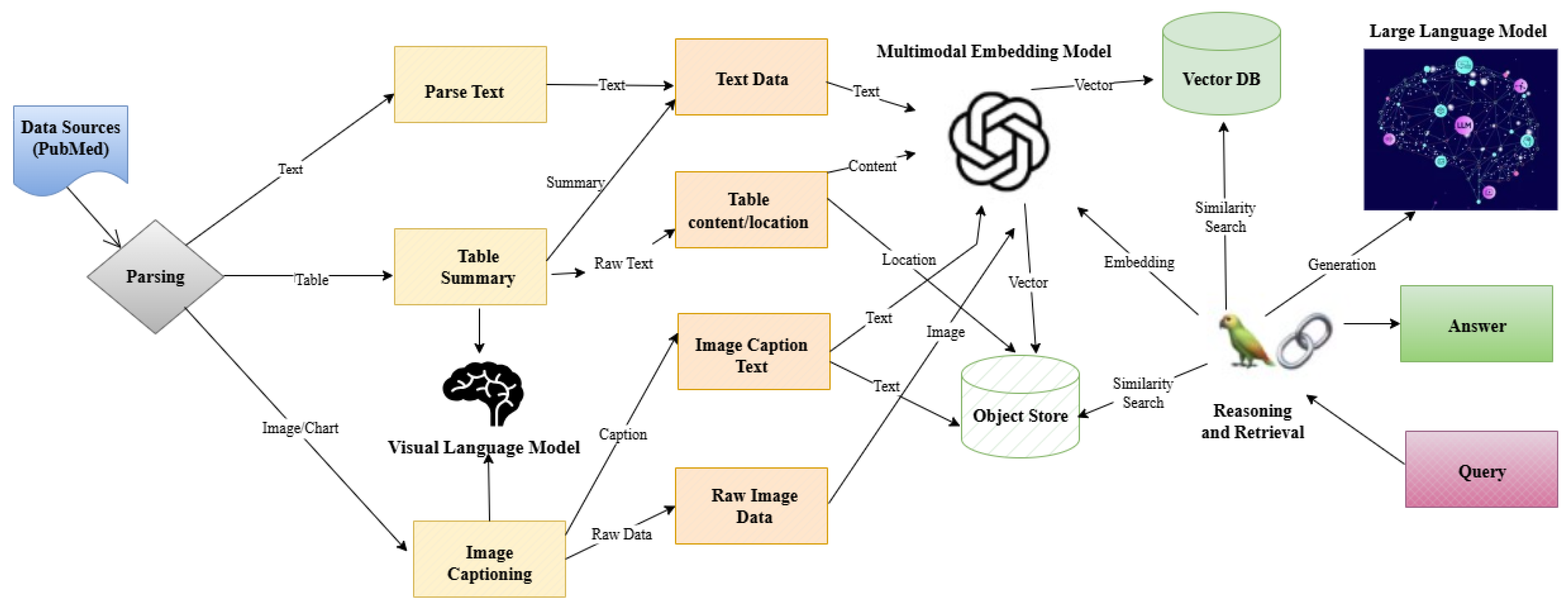
The overall architecture of AlzheimerRAG is described in
Figure 1. To simplify, the architecture diagram in
Figure 1 illustrates a multimodal RAG component for the biomedical literature (e.g., PubMed), where text, tables, and images are extracted through parsing and processed separately—text is parsed, tables are summarized, and images are captioned using a visual language model. These processed elements are then converted into embeddings through a cross-modal embedding fusion method and stored in an object store and a vector database. Upon receiving a user query, the system retrieves relevant information using similarity search and passes it to a large language model, which generates a context-aware answer by reasoning over the retrieved multimodal content.
In the subsequent sections, we describe each step in the architecture flow in more detail, followed by a demonstration of the application with the technical components.
3.1. Data Collection and Preprocessing
The first step of our process involved collecting relevant articles from PubMed. We accomplished this by writing a Python script [
34] that called the National Centre for Biotechnology Information (NCBI) Entrez Programming Utilities (E-utilities) API to fetch the top 2000 articles from the PubMed repository [
35] related to the “Alzheimer’s Disease” search term [
36]. The articles were fetched in batches per API request, adhering to NCBI API rate limits and sorted by relevance during the retrieval process. We parsed each document, collecting the full texts, abstracts, tables, and figures for textual and image retrieval. After that, we cleaned and normalized the data for the data preprocessing step to ensure consistency and usability. This involved removing hyperlinks, references, and footnotes. We also standardized the figures/diagrams format by converting them to a consistent image format for uniform processing.
3.2. Textual Data Retrieval
This step retrieves the clinical text data related to Alzheimer’s disease (AD) for textual and tabular data processing. In our workflow, for generating the text embedding, we fine-tuned the “Llama-2-7b-pubmed” [
37,
38] model by training it with the PubMedQA [
14] dataset from HuggingFace. The fine-tuning used parameter-efficient fine-tuning (PEFT) techniques like QLoRA [
39].
Table 1 outlines the QLoRA parameters and the training argument parameters used for fine-tuning.
Textual and Tabular Data Processing
The extracted data were chunked into structured text and table summaries. Then, a layout model (for tables) and titles were used for candidate sub-sections of the document (e.g., Introduction, Methods, etc.). Finally, post-processing was conducted to aggregate text under each title, and further chunking into text blocks was performed for downstream processing based on user-specific flags for each block. After that step, the text embeddings converted the smaller blocks into embedding vectors, which were used for cross-modal attention fusion.
3.3. Image Retrieval
For the generation of feature embeddings that capture image details from the PubMed articles, we fine-tuned the “LlaVA” (Language and Vision Assistant Model, version 2) [
40] model using the official LLaVA repo with the Llama-2 7B backbone language model [
41]. LLaVA combines pre-trained language models (such as Vicuna or LLaMA [
42,
43]) with visual models (such as CLIP’s [
44] visual encoder) by converting visual features into embeddings that are compatible with the language model. Its training has two stages: a pre-training stage, where image–text pairs align visual and language embeddings with only the projection matrix being trained [
45], and a fine-tuning stage, where the visual encoder remains frozen while the projection layer and language model are updated [
46]. Using the fine-tuned approach preserves the strengths of the large language model while lowering computational requirements, making it ideal for resource-limited environments and quick adaptation to new data. The hyperparameters used for fine-tuning are presented in
Table 2. QLoRA uses the 4-bit NormalFloat, which is explicitly designed for customarily distributed weights, thereby further reducing memory usage.
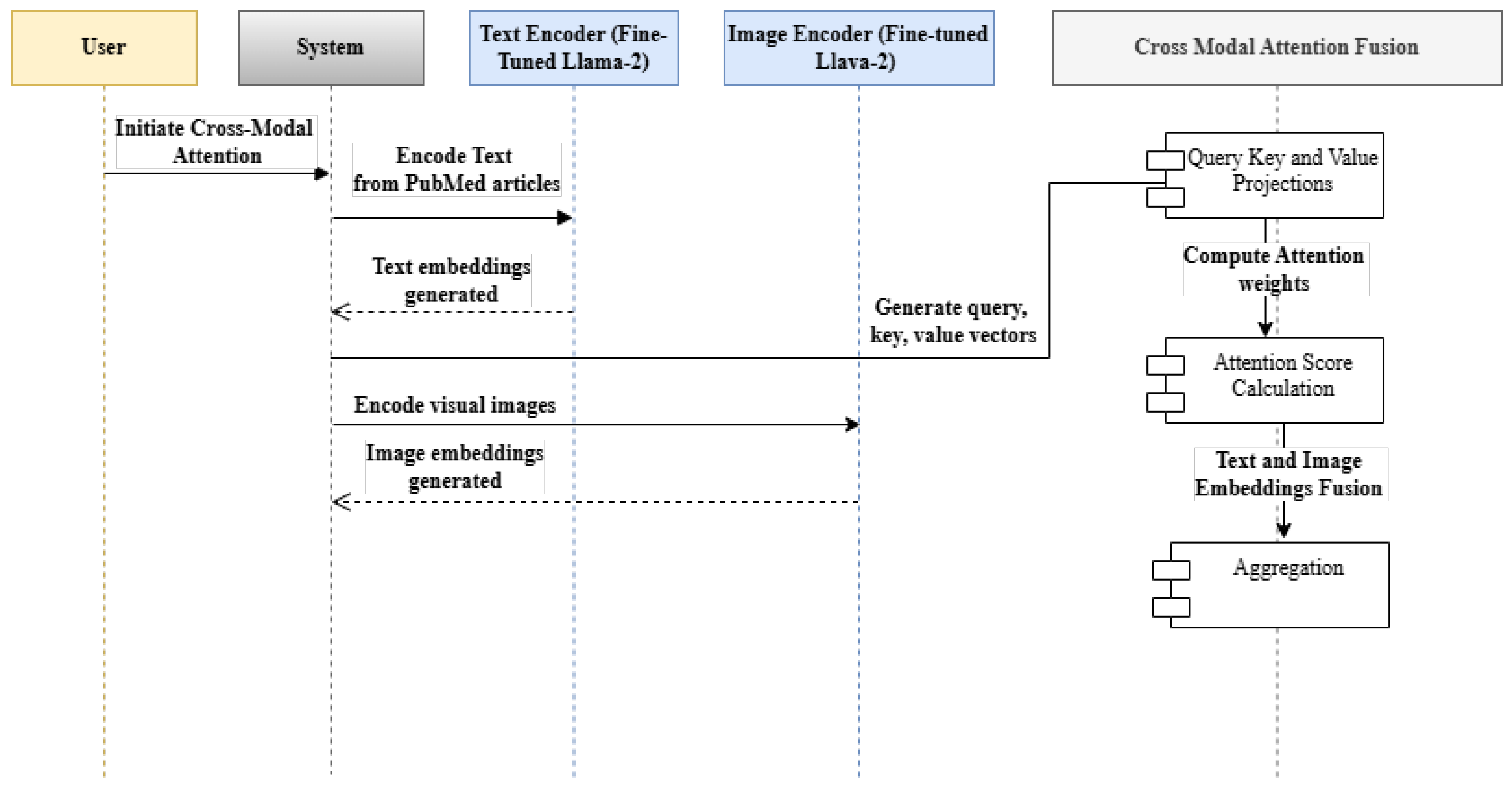
3.4. Cross-Modal Attention Fusion
Cross-modal attention fusion is a mechanism that facilitates interaction between different modalities, within our current scope, specifically between text and images. It allows a model to selectively focus on relevant parts of both modalities by computing attention weights. These weights are used to modulate the embeddings from each modality, enabling a richer and more comprehensive representation. In our context, the cross-modal attention fusion ensures that the integrated textual and visual data contribute meaningfully to medical information retrieval. The process steps of cross-modal attention fusion are detailed in
Figure 2 as a sequence diagram.
The three steps associated with this process are described below:
Generate query, key, and value vectors from the text and image embeddings from
Section 3.2 and
Section 3.3, respectively.
Compute the attention scores using the dot-product attention mechanism shown below:
where
- −
queries and keys are matrices of size , with n being the number of tokens and the dimension of each key.
- −
is the dimensionality of the keys used for scaling.
- −
scales the dot-product, helping to stabilize gradients in deeper networks.
Aggregate contributions from both modalities based on attention weights:
where
- −
is a matrix representing the attention scores, with dimensions , where n is the number of tokens, and m is the dimensionality of each value.
- −
values is a matrix of values corresponding to tokens, typically with dimensions , where d is the embedding size.
The resulting
is a combination of the values weighted by attention.
Finally, the combined feature embeddings are indexed as vectors in an object store, which allows quicker retrieval of multimodal data.
3.5. AlzheimerRAG Demonstration
3.5.1. System Walk-Through
AlzheimerRAG is implemented as a Python (version 3.8.x) Web Application utilizing FastAPI and Jinja2 Templates with LangChain integration. It provides a simple user interface
(Web Application) for leveraging efficient multimodal RAG capabilities related to AD. The application
(Source Code) is deployed in Heroku, a cloud-based Platform-as-a-Service (PaaS) solution that helps manage seamless continuous integration and deployment. It provides the functionality for information retrieval from user queries. The multimodal RAG component extracts context-aware relevant images as part of the output response. The demo video can be accessed from this link
(Video Demonstration).
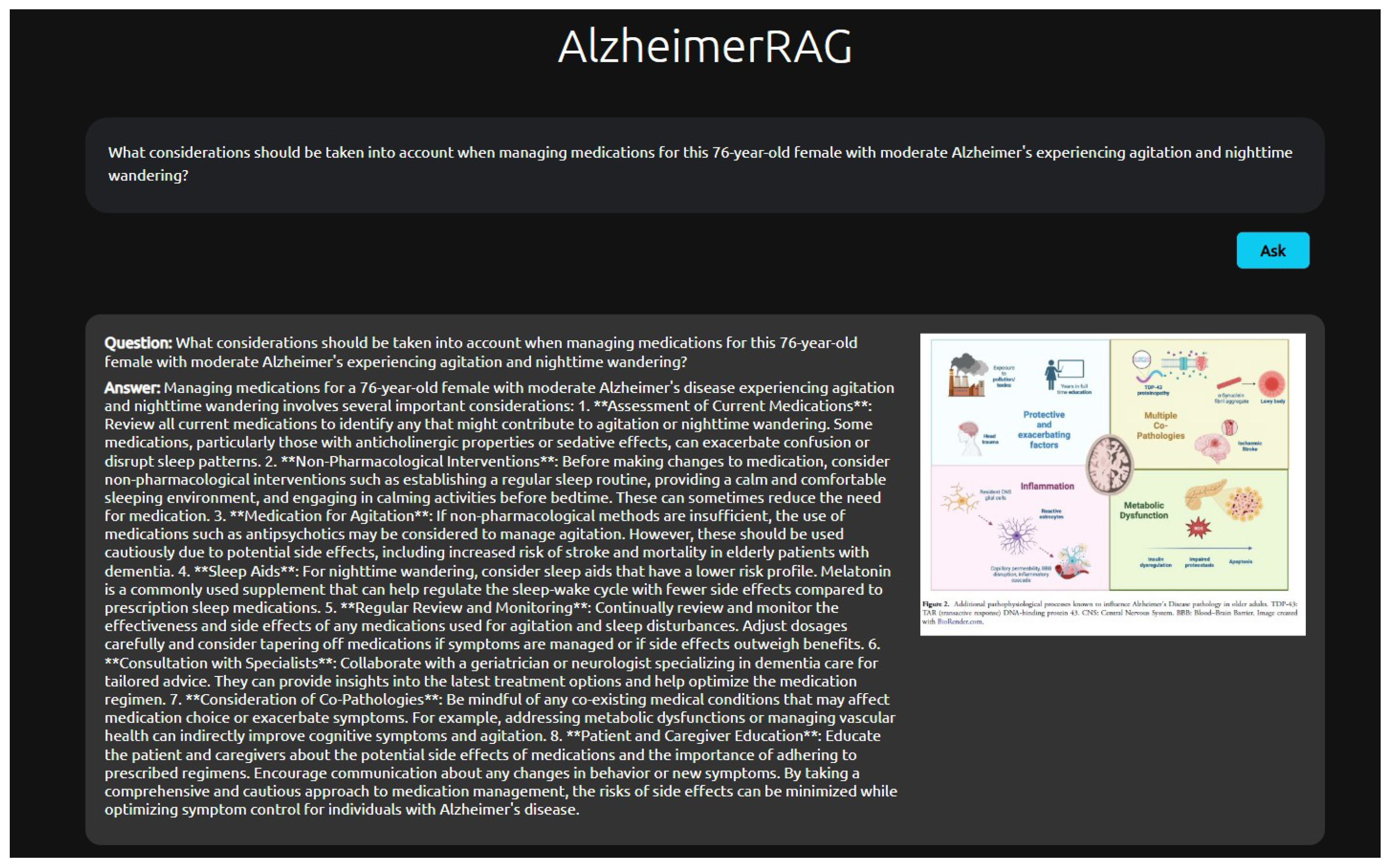
A sample response from the AlzheimerRAG user interface can be observed in
Figure 3, where relevant text and images are fetched for a particular user query related to Alzheimer’s disease from the embedded PubMed articles.
3.5.2. Key Technical Components
The key technical components are summarized below.
FastAPI for API development: FastAPI is a high-performance web framework for API development that provides an intuitive interface for API development and integrates seamlessly with Python’s async capabilities.
Jinja2 for template rendering: Jinja2 is a templating engine for Python that offers dynamic template rendering. It serves HTML content from backend data, enabling a seamless and interactive user experience.
FaissDB for embedding multimodal data: FaissDB [
47], a vector DB, is widely used for embedding multimodal data. Embedding is the process of converting content into a numerical representation (i.e., vectors) for large language learning models and is crucial for transforming preprocessed healthcare knowledge into individual vectors. The text and image embeddings are encoded into uniform, high-dimensional vectors and indexed for efficient similarity searches. When a query is made, the reasoning and retrieval component searches the vector space to extract relevant information. The benefit is that it uses an approximate nearest neighbor (ANN) search to quickly locate embeddings in high-dimensional space, which is essential for large-scale applications. The generation component uses the retrieved multimodal representations to produce outputs in various formats, such as text or images.
LangChain as a Retrieval Agent for multimodal RAG: The Retrieval Agent is a medium to pinpoint the most relevant knowledge in response to user queries. This process involves using the embedding model to convert the user query text into vectors, which are then searched through the vector storage to identify the closest matching vectors. The effectiveness of a Retrieval Agent is closely tied to the underlying framework upon which it is built. Therefore, we utilized the LangChain [
48,
49] framework, a premium existing open-source framework, along with LlamaIndex [
41] because of its significant advantages in (i) preservation of table data integrity, (ii) streamlining the handling of Multimodal data, (iii) enhanced semantic embedding. Together, LlamaIndex and LangChain enhance the context-awareness of extracted content, enabling efficient retrieval and synthesis of information and producing nuanced outputs.
6. Conclusions and Future Work
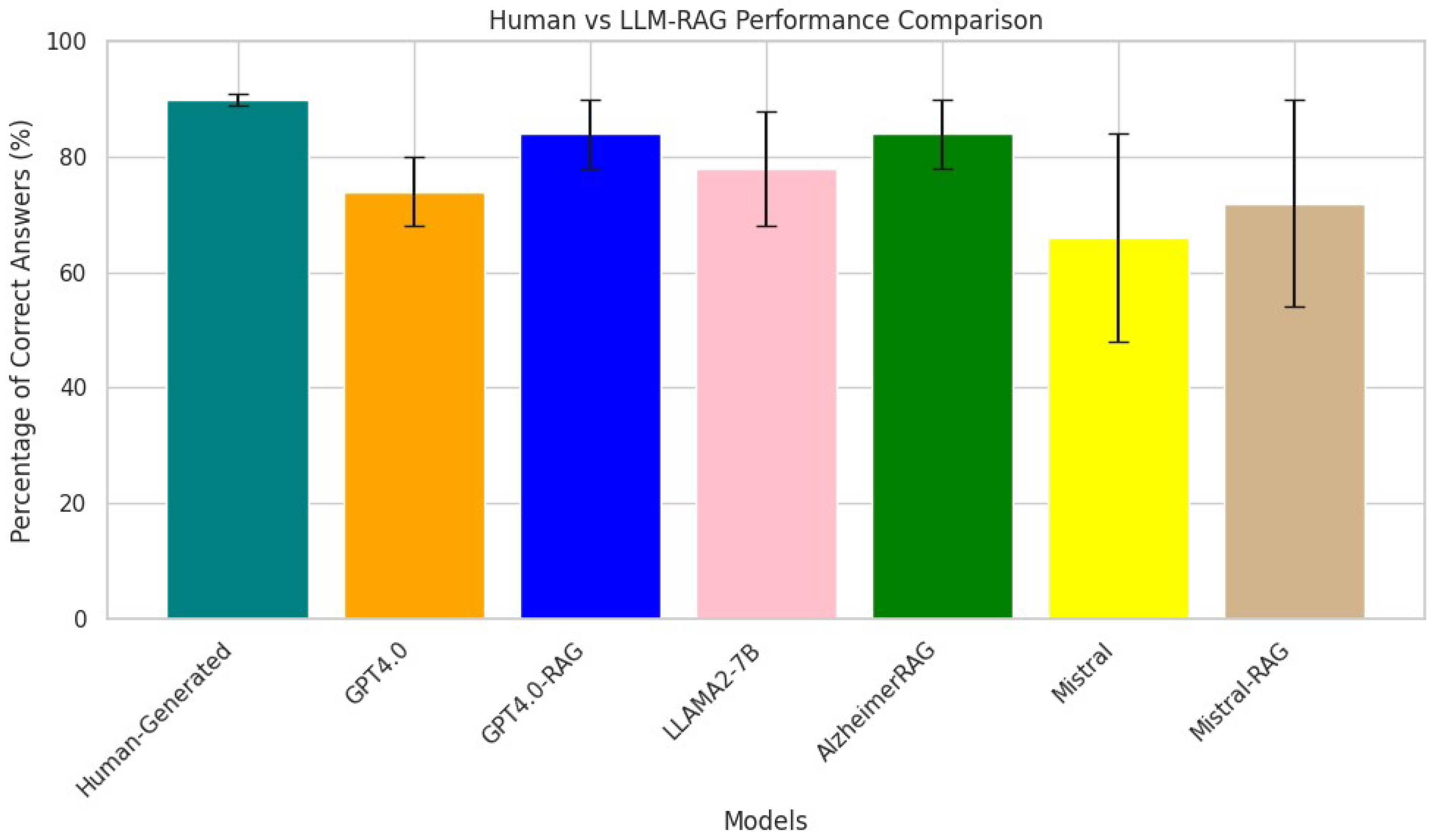
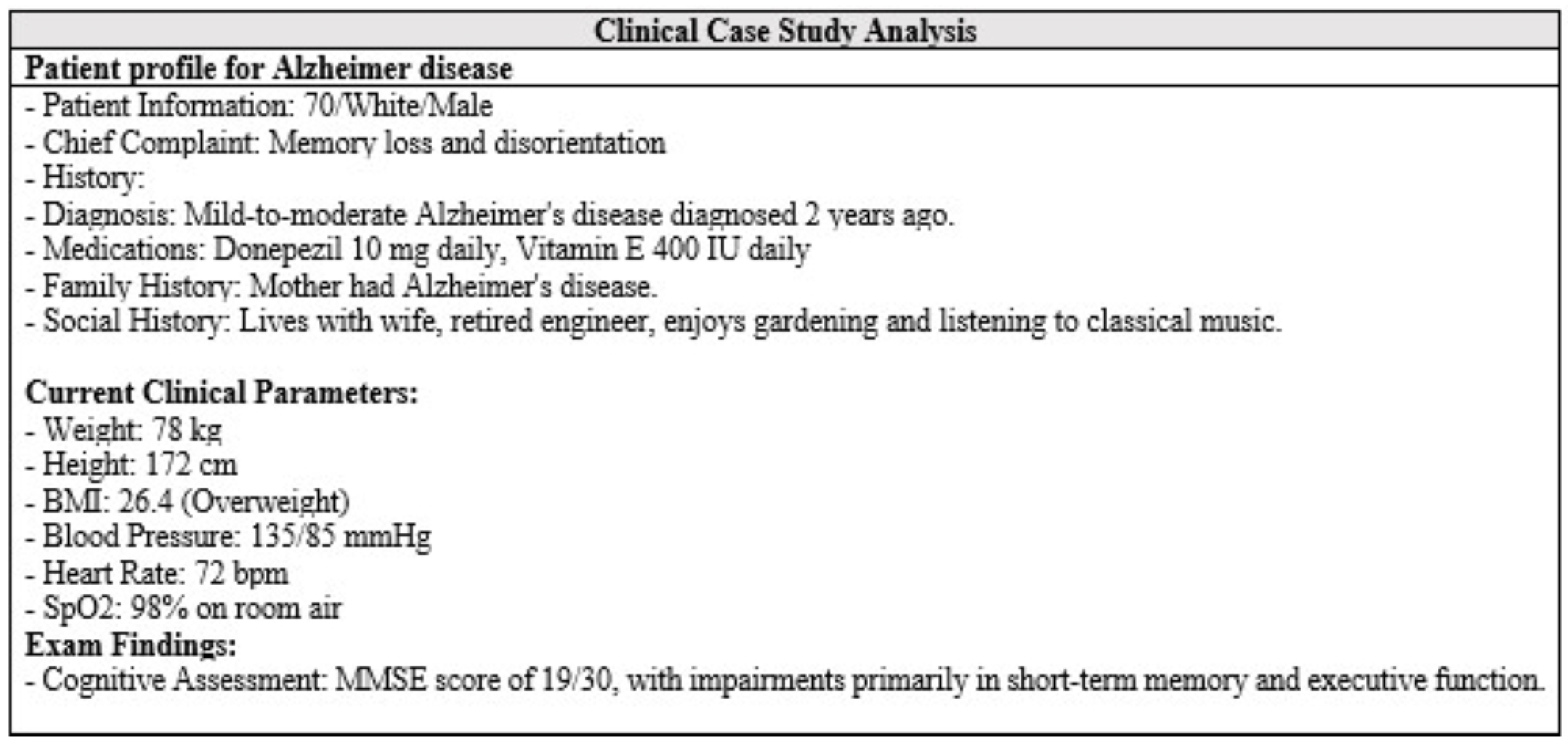
The AlzheimerRAG application represents a significant advancement in biomedical research, particularly in understanding and managing Alzheimer’s disease. By integrating multimodal data—including textual information from PubMed articles, imaging studies, and clinical trial scenarios—this innovative retrieval-augmented generation (RAG) tool provides a comprehensive platform for analyzing complex biomedical data. The use of cross-modal attention fusion enhances the alignment and processing of diverse data types, leading to improved accuracy in generating insights relevant to diagnosis, treatment planning, and understanding the pathophysiology of Alzheimer’s disease. The experimental results indicate that AlzheimerRAG outperforms existing methodologies in terms of accuracy and robustness, demonstrating the value of a multimodal approach in addressing the complexities inherent in Alzheimer’s disease research. While it exhibits low hallucination rates, the risks of generating misleading information in nuanced clinical scenarios remain; as discussed in
Appendix A and
Appendix B, it necessitates further research and clinical validation for real-world safety and applicability. Future enhancements could expand its scope to other neurodegenerative disorders, such as Parkinson’s, incorporate additional data sources (e.g., wearable devices, electronic health records (EHRs)), refine the user interface for improved interpretability, and optimize clinical trial support through enhanced patient recruitment and monitoring. Continuous improvements informed by user feedback will further enhance its utility and functionality for researchers and clinicians. In summary, while the AlzheimerRAG shows great promise in enhancing Alzheimer’s disease research, pursuing these outlined future directions will be essential for maximizing its impact in clinical settings.