Abstract
In vitro fertilization (IVF) is a well-established and efficient assisted reproductive technology (ART). However, it requires a series of costly and non-trivial procedures, and the success rate still needs improvement. Thus, increasing the success rate, simplifying the process, and reducing costs are all essential challenges of IVF. These can be addressed by integrating artificial intelligence techniques, like deep learning (DL), with several aspects of the IVF process. DL techniques can help extract important features from the data, support decision making, and perform several other tasks, as architectures can be adapted to different problems. The emergence of AI in the medical field has seen a rise in DL-supported tools for embryo selection. In this work, recent advances in the use of AI and DL-based embryo selection for IVF are reviewed. The different architectures that have been considered so far for each task are presented. Furthermore, future challenges for artificial intelligence-based ARTs are outlined.
1. Introduction
1.1. In Vitro Fertilization
Assisted reproductive technology (ART) is a term used for fertility treatments that handle eggs or embryos [1]. It encompasses techniques used to extract eggs from ovaries, inject them with sperm in a laboratory environment, and place them in the uterus of the treated woman for implantation to occur [2]. According to the CDC, about 2.3% of all children born in the U.S. every year are conceived using ART [3]. In the U.S., around 82% of the clinical pregnancies from ART cycles started in 2021 resulted in live birth delivery [4].
The most widely considered ART technique is in vitro fertilization (IVF). It involves several steps, from medication for ovarian stimulation, egg retrieval and sperm retrieval, egg fertilization, and embryo transfer to live birth delivery. IVF has a variable success rate, with many factors affecting the outcome, age being among the most important ones. So, naturally, medical professionals are constantly trying to improve every aspect of the IVF process, in a collective effort to improve the chances of success for the patients but also to improve other aspects of the process, like the costs for patients, the number of visits required, and improving the overall experience. This is where artificial intelligence comes in as a valuable support tool to improve many aspects of the IVF process, as discussed next.
1.2. Artificial Intelligence in ART
Artificial intelligence, through machine learning (ML) and deep learning (DL), has steadily evolved over the past 20 years to be an effective decision-support tool in the medical sciences. Many topics related to obstetrics and gynecology have benefited from the use of AI, such as fetal weight estimation [5], placental volume estimation [6], electronic fetal monitoring [7], and more [8]. In IVF, AI algorithms can be used as non-invasive tools [9] to evaluate embryo development and make predictions [10,11].
For the important task of embryo selection, AI algorithms take advantage of the developments in Computer Vision (CV) and image processing, to process embryo data gathered from patients, like time-lapse [12,13] or static images in grayscale or Red–Green–Blue (RGB) format, and perform tasks like embryo component segmentation, grading, live birth prediction, and more. Usually, the embryo data are combined with the patient’s clinical data to further improve predictions. In addition to the direct task of embryo selection, AI can also provide assistance for several aspects of the IVF cycle, like strategy selection, ovarian stimulation, and even quality assurance for the clinic. Of course, due to the interdisciplinarity of the topic, the development of such algorithms requires the cooperation of clinical personnel, medical practitioners, embryologists, biologists, physicists, and computer scientists.
It is clear from recent developments in the field that AI can bring significant positive changes to IVF if developed properly. This is why more and more research groups are focusing on this topic, with the topic gaining considerable attention in recent years. Indicatively, Figure 1 depicts a graph of publications in the area of AI-assisted ART over the last 20 years. This was extracted from Scopus, following a keyword search of “artificial intelligence” AND “in-vitro fertilization” in the title, abstract, and keywords. There are 233 results published from 2005 to 2023 with both keywords. The graph shows a very clear upward trend, as the previous decade has seen few contributions to this field, with a very steep increase over the last 5 years. This is indicative of the emerging nature of the field of AI and specifically DL in IVF. As seen in Figure 2, of these publications, the highest contributors are the United States with more than 70 articles, followed by the United Kingdom, Spain, China, India, and Australia, with the rest of the countries following, with six or fewer contributions not being shown.
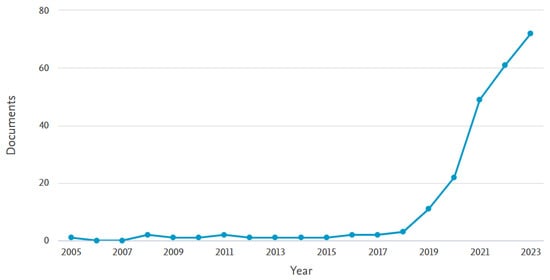
Figure 1.
Scopus results for publications with the keyword search “artificial intelligence” AND “in-vitro fertilization” in the title, abstract, and keywords, for the years 2005–2023.
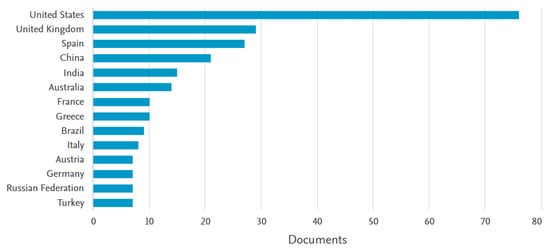
Figure 2.
Scopus results for contributing countries, with the keyword search “artificial intelligence” AND “in-vitro fertilization” in the title, abstract, and keywords, for the years 2005–2023.
Regarding the types of contribution, looking at Figure 3, as expected, the highest percentage refers to journal publications (50%) and conference papers (10%). However, fortunately, there is a high percentage of review articles (21%). This can be understood because this field is highly interdisciplinary and requires some contributions to serve as guidance for new researchers joining the field from different backgrounds.
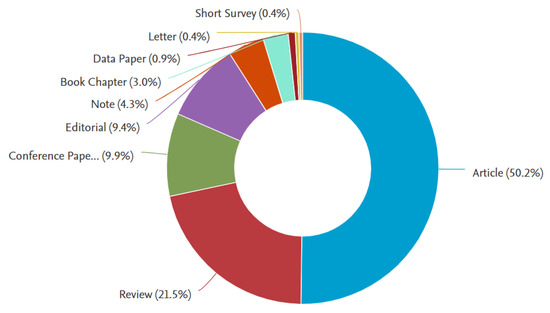
Figure 3.
Scopus results for paper publication types, with the keyword search “artificial intelligence” AND “in-vitro fertilization” in the title, abstract, and keywords, for the years 2005–2023.
1.3. Motivation and Contributions
Motivated by the increase in the use of AI-assisted applications in ART, this work provides a review of the latest advancements in IVF. The purpose of this review is to serve as a guide for all researchers in the field, providing a roadmap of current developments and identifying future challenges. Specifically, its contributions can be identified as follows:
- The different DL architectures are briefly outlined first.
- Several tasks are reviewed that cover IVF applications which can be addressed using AI techniques.
- An emphasis is given to more recent works, from 2021 onward.
- Emphasis is also given to DL techniques, as they constitute the state of the art in AI and ML methodologies.
- Future research directions and challenges are discussed.
Thus, this work consists of three parts, a brief outline of ML techniques, an extensive review of recent developments in AI-assisted IVF, and a discussion on the future of the field.
2. Review Methodology
Starting this review, it is important to define the conditions for choosing the works to be covered. The works included in this review were selected following the criteria below:
- The work should be published in a peer-reviewed scientific journal, presented at an international conference and included in its proceedings, or published as a book chapter in a collected volume. So, the works have already been submitted to a peer-review process.
- The publications are in English.
- Publications should have a digital object identifier (DOI).
- The works should be listed on an indexing service like Google Scholar, Scopus, or Web of Science.
The literature search was performed on Google Scholar and Scopus, following the keywords "artificial intelligence" and "in-vitro fertilization". By cross-checking the citations received from the works found in this way, the literature search was enriched.
Concerning notation and abbreviations, scalars are denoted by lowercase letters, matrices by bold capital letters, and vectors with lowercase bold letters. Math operators are denoted by capital letters. Abbreviations can be found in the Table at the end.
The rest of the work is structured as follows. Section 3 provides a brief overview of AI methodologies. Section 4 reviews the recent developments in AI-assisted IVF, covering different topics. Section 5 discusses the open challenges in the future of the field. Finally, Section 6 concludes the work, providing some final remarks.
3. Overview of AI Methodologies
In this section, the most common ML and DL architectures are briefly presented. Interested readers can find details of the respective references for each model. You can also refer to recent reviews in healthcare and medicine using reinforcement learning [14], federated learning [15], and self-supervised learning [16].
3.1. Regression Learning
Regression techniques encompass a class of statistical approaches to find the best rule that connects a series of input variables to one or more outputs; see Figure 4. Its simplest form is linear regression, where a linear rule between the input(s) and output(s) is considered, but several more exist, like logistic regression.
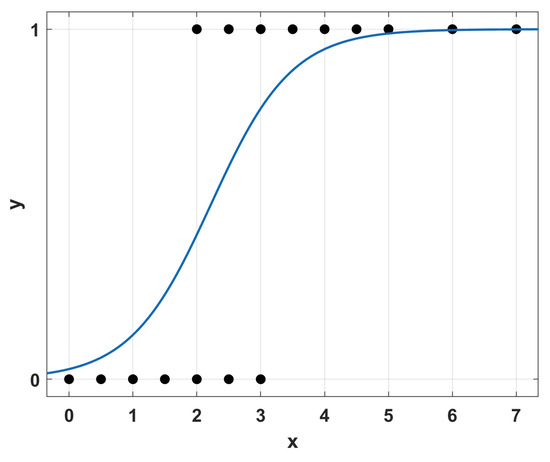
Figure 4.
Simple example of regression analysis.
3.2. Decision Tree Learning
Decision trees are based on the building of hierarchical graphs to categorize data based on a series of observations; see Figure 5. A tree can consist of multiple nodes, each divided into child nodes, based on the result of an observation. The end nodes (leaves) represent the end classes.
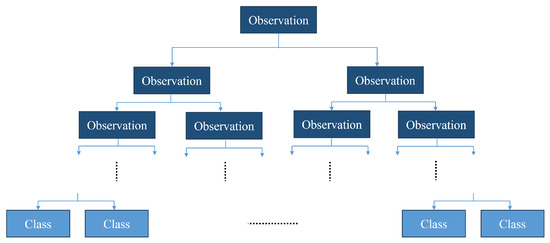
Figure 5.
Structure of a decision tree.
3.3. Artificial Neural Networks
The central part of any NN architecture is a simple structured computation unit, termed the artificial neuron. By combining multiple such artificial neurons in sequential groups, called layers, a neural network (NN) is formed. In this network, the k-th neuron takes as an input an N-dimensional vector , and computes an output as follows:
Here, the scalars are the weights of the neuron, and b is called bias. The weights of all neurons can be combined in a weight matrix . The function q is called the activation function, as a reference to the activation of biological neurons. Figure 6 shows a visual representation of the above operations that form the structure of the neuron.
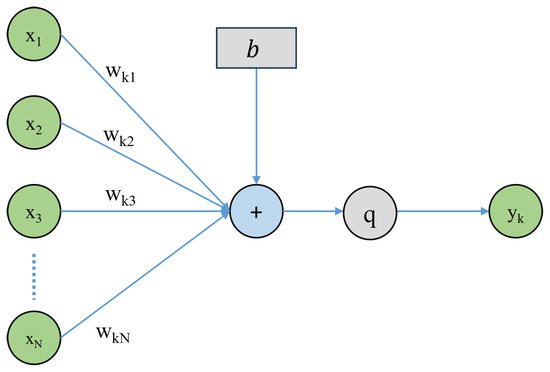
Figure 6.
Artificial neuron architecture.
In ML, each model consists of multiple neurons and layers, connected in different ways based on the model type. Model training is performed by updating the weight values in each iteration. The mechanism implementing this process is stochastic gradient descent (SGD). The most sufficient way to implement this is through error backpropagation, which is simply termed backpropagation. Backpropagation utilizes the principle of the chain rule of differentiation to compute the gradients through the entire network, starting from the end neuron and proceeding backwards toward the first neuron layer [17].
3.4. Deep Learning Methods
The DL techniques fall into two main types: DL networks for supervised or discriminative learning and DL networks for unsupervised or generative learning. Figure 7 provides a brief overview of DL methods for IVF applications. In the following, a brief description of the most common DL architectures is provided.
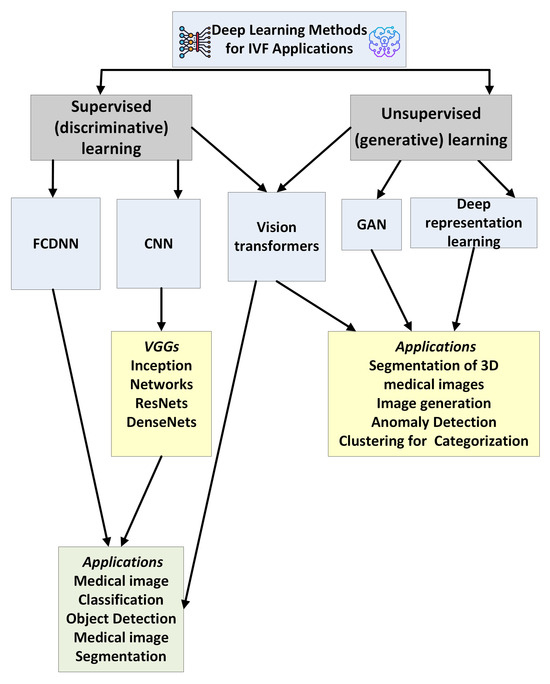
Figure 7.
A taxonomy of deep learning methods used in IVF applications.
3.4.1. Fully Connected Deep Neural Networks
Fully Connected Deep Neural Networks (FCDNNs) constitute the most basic DL architecture. Here, the neurons in each layer are connected to all the neurons in the next layer, as shown in Figure 8. In FCDNN models, the information is processed in a forward manner, as there are no loops or recursion operations. Their simpler structure makes training more straightforward, but they still face challenges in Computer Vision (CV) problems. Due to the full connection between the layers, with deeper nettworks, the number of parameters becomes high, resulting in inefficiencies and possibly overfitting. Moreover, FCDNNs are not suitable for exploiting the spatial correlation of image features. In other words, FCNNs are not translation-invariant, which means that they will perform differently under a given input image and its shifted versions.
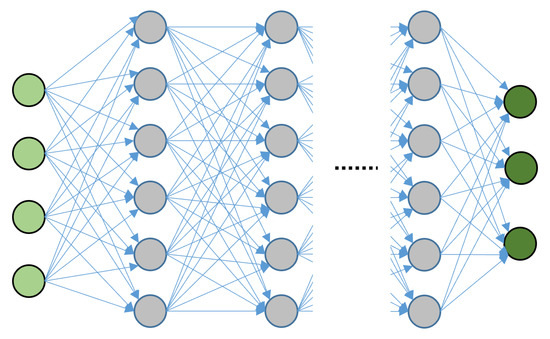
Figure 8.
Fully connected neural network. The light green nodes represent the inputs, the gray nodes are the hidden layers and the dark green nodes are the outputs.
3.4.2. Convolutional Neural Networks
Convolutional Neural Networks (CNNs) are especially suitable for CV applications due to their ability to process spatial data with a grid-like topology. As images and video frames are 2D data, where adjacent pixels are grouped to display information, CNNs are suitable for their processing. CNNs make use of the convolution operator instead of matrix multiplication, which requires fewer tuning parameters. A visual representation of the convolution operation is shown in Figure 9. In contrast to FCDNNs, the layers are sparsely connected, allowing for deeper architectures.
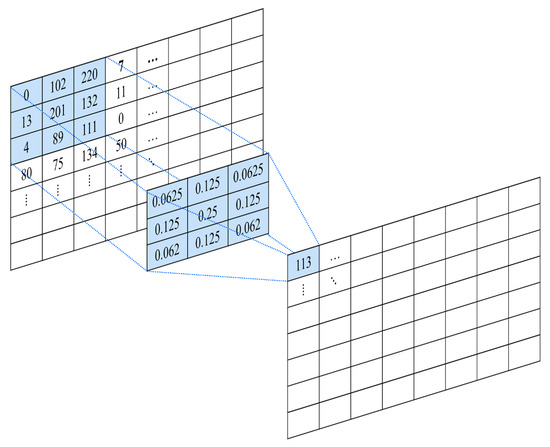
Figure 9.
Example of convolution operation.
A CNN model is built up by the following layers and operations:
- Input layer: The first layer of the network accepts the input data and, if required, transforms them in a format suitable for further processing. For example, RGB image data can be rearranged into multi-dimensional arrays.
- Convolution layers: The convolution layers are the distinct blocks of the CNN. They are used for feature extraction. In contrast to a fully connected layer, where each neuron receives input from all neurons in the previous layer, the neurons in the convolutional layer have a smaller receptive field. The receptive field indicates that every neuron receives input from only a restricted subset of the previous layer.
- Activation function: Most CNNs in the literature consider either a Rectified Linear Unit (ReLU) function or a variant of it. ReLU is defined as [17]:A variant of ReLU that has been successfully considered in many CV problems is Leaky ReLU, defined as:The parameter is usually taken as 0.01.
- Pooling layers: They are used to reduce the size of the incoming data by summarizing small groups of features using a computationally efficient method. For example, a max pooling layer will extract the maximum element from a feature region, like an image subregion, effectively reducing the feature data for the next processing step.
- Flattening: This operation reshapes the data into a 1D vector.
- Output layer: This is the end layer, which provides the model’s prediction.
There are several different archetypes of CNN architectures that are used in practice. The most common are listed below. Of course, variations and, more importantly, combinations of these can be considered which combine layers from different architectures.
- Visual Geometry Group models (VGGs) have a network ranging from 11 to 19 layers. They were initially proposed in order to demonstrate that deeper networks can outperform networks with fewer layers. Using a smaller size of convolutional kernels (), they can have fewer parameters and increased accuracy [18].
- The problem of overfitting can be avoided by Inception Networks that use modules consisting of multiple filters of varying sizes on the same level, effectively making the network ‘wider’. Here, the problem of the vanishing gradient is mitigated by alternating between fully connected layers and average pooling instead and also by adding auxiliary classifiers to the intermediate layers. Several improvements have been developed, like InceptionV2, InceptionV3, and InceptionV4 [19].
- Xception is an architecture built on the InceptionV3 model. Specifically, it replaces the inception modules with depth-wise separable convolutions, that is, a 2D convolution that is independent for each channel, followed by a 1D point-wise convolution. This architecture outperforms InceptionV3 in several image recognition tasks. Its parameter set is also reduced, leading to a decrease in learning latency [20].
- Residual Networks (ResNets) resolve the problem of degradation that can appear in several deep CNN architectures. Their implementation allows deeper networks to be trained and perform better. This is possible through the residual learning technique. Here, instead of using parameter layers to learn the relation between inputs and outputs, similar to VGG, they are used to extract the residual between inputs and outputs [21].
- Densely Connected Convolutional Networks (DenseNets) are inspired by ResNets. They establish maximum flow of information between layers by connecting all of them directly with each other with matching feature-map sizes. Therefore, DenseNet resolves the vanishing-gradient problem and underlines feature propagation and reuse. They also have a reduced set of parameters [22].
3.4.3. Attention-Based Models
Models based on attention only (Figure 10), that is, transformers [23], are widely used in the field of natural language processing (NLP) [24] and have been used effectively in CV problems [25]. In [25], emphasis is given to the analysis of the transformer model presented first, which is the basis for all the models developed afterwards.
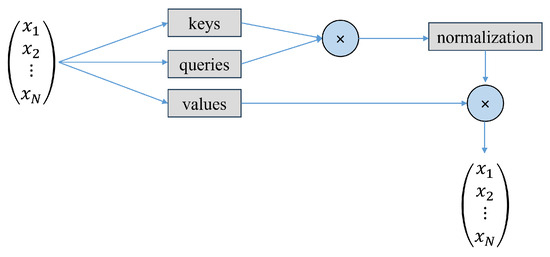
Figure 10.
Self-attention mechanism.
The transformer architecture follows an encoder–decoder structure. The encoder block maps the sequence of inputs to a sequence of vector representations . The decoder then takes this vector representation, which is the output of the encoder, and computes a sequence of outputs .
Vision transformers (ViTs) use transformer modules for VT tasks. Each module consists of an encoder transformer block.The ViT divides the image into smaller subimages and transforms them linearly to obtain a patch embedding. The position embedding is then added to form the input to the transformer encoder. The accuracy of ViTs is similar to that of CNNs. Vision transformers can be used for both supervised and unsupervised tasks.
However, it must be noted that an important drawback of transformers is the requirement of a large data size to achieve their maximum performance and compare to other state-of-the-art architectures. Moreover, current models can have more than a million parameters, making training and testing difficult and requiring time-consuming labor.
3.4.4. Generative Adversarial Networks
In the category of unsupervised DL models, generative models and specifically Generative Adversarial Networks (GANs) [26] (Figure 11) have found applications in medical imaging. Unsupervised methods try to identify patterns and structures in data that are not labeled [17]. For example, 3D-CNNs and K-means have been used in unsupervised segmentation of 3D medical images [27].
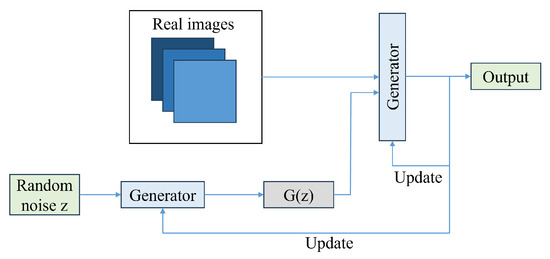
Figure 11.
GAN architecture.
To design a generative model, a dataset with samples drawn from a probability distribution P is required. The model must learn to represent an estimate of that distribution. The output can be an explicit estimation of a probability distribution Q, or the generation of samples from Q. GANs work in the second approach and employ the core idea of two adversarial entities, the generator block and the discriminator block. The input to the generator is random noise, and from this the generator tries to output sample data that come from the original training data distribution. The discriminator then examines the data that are output from the generator and judges whether they are real or fake.
4. DL-Empowered Embryo Selection for IVF Application
In this section, recent developments in several IVF tasks are reviewed. For each work, a brief outline is provided, with information on the models that were used, the main results, and the starting dataset information. Naturally, many of these works may include several more sub-problems studied and their corresponding volumes, but reporting each result would push this review to an unacceptable length. So, only the most important results are reported here. Interested readers are encouraged to look up each work for further information. Note also that some works may address problems that cover multiple tasks (for example, component segmentation and implantation prediction), but they are only reported in one subsection.
A general outline of an ML architecture used for an IVF task is shown in Figure 12. Starting from a given dataset, an initial preprocessing is performed to homogenize the data, discard unusable data, and possibly address any missing information in them. Data could be embryo images, clinical information, or both. These data are then used to train and validate an ML model, which can use either one or both of these types of data to make a decision. This decision could be a suggestion for an action to be taken, a grading, or a prediction. Figure 13 shows the IVF applications that we will discuss in the following sections. Brief descriptions of each are provided below.
- Strategy Selection: DL is used as a support tool in medical decision-making.
- Embryo Development Annotation: In this case, the researchers develop an automated annotation tool for human embryo development in time-lapse devices based on image analysis.
- Intracytoplasmic Sperm Injection: In this case, DL is being implemented in intracytoplasmic sperm injection (ICSI) procedures to improve the selection, analysis, and ultimately success rates of fertilization. This includes the creation of models that can be used to identify high-quality sperm, evaluate DNA fragmentation, and even monitor sperm movement during the procedure.
- Component Segmentation: Semantic segmentation of images in combination with an object detection technique can support the further processing of embryos for tasks like grading or outcome prediction.
- Embryo Grading: It involves a classification task in which embryo images are classified according to a specific grading system that evaluates the quality and developmental potential of embryos.
- Ovarian Stimulation: Ovarian stimulation is a critical stage in IVF technologies, requiring the formulation of numerous decisions regarding drug protocols, dosing, and timing that can be customized to the individual profile of each patient. DL has the potential to help fertility physicians recommend personalized treatment plans, optimize the number of retrieved oocytes, and improve patient outcomes by analyzing extensive datasets from previous IVF cycles.
- Predicting Retrieved Oocytes: It involves ML methods to predict the number of retrieved oocytes.
- Pregnancy and Live Birth Prediction: ML has been extensively employed to evaluate the prospective maternal risks during pregnancy and predict the mode of childbirth
- Intrauterine Insemination (IUI): In this case, ML methods are applied for predicting clinical pregnancy outcomes from intrauterine insemination (IUI) and identifying significant factors affecting pregnancy.
- Sperm Analysis: ML has the potential to improve intracytoplasmic sperm injection by assisting clinicians in the objective selection of sperm. This is a classfication task.
- Quality Assurance: DL is applied as an assistive quality assurance tool to identify perturbations in the embryo culture environment that may affect clinical outcomes.
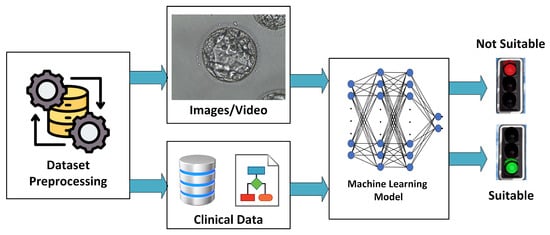
Figure 12.
Outline of an ML architecture used in IVF.
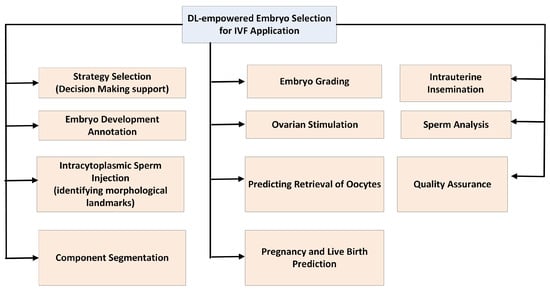
Figure 13.
The IVF applications discussed in this review.
4.1. Reviews on the Topic of AI in IVF
Given the interdisciplinarity and complexity of the topic of AI in IVF, there have been several reviews, as well as discussion articles on the future promise of the field [28]. In each of these reviews, some common conclusions are drawn regarding the challenges in the field. In addition to these similarities, based on the different backgrounds, experience, and specializations of the authors in each paper, the interpretations which are the most important contributions of AI to IVF or which are the most important future challenges show variations.
The work [29] reviews the use of AI in IVF and healthcare in general, giving examples of successful and unsuccessful commercial implementations.
The work [30] discusses several aspects of improving IVF outcomes from the introduction of new technologies into laboratories. With respect to the use of AI, the potential impacts include increased fertilization rates as a result of accurate identification of the most viable spermatozoa, decreased time to pregnancy, consistency in embryo grading, and improved laboratory quality management. For all tasks, embryologists will always be an integral part of the lab and help ensure the success of ART.
In [31], the authors review the progress made towards automation in ART. Some of the topics covered are data management, patient treatment pathways, trans-vaginal oocyte retrieval, oocyte selection, semen analysis and preparation, insemination and Intra-Cytoplasmic Sperm Injection (ICSI), embryo culture and selection, Preimplantation Genetic Testing (PGT) and Metabolomics, endometrial evaluation for personalized embryo transfer, cryopreservation, and cryostorage. For the future, the authors speculate if the role of the embryologist will switch from repetitive manual tasks to precise critical thinking for decision-making.
In [32], the authors review the use of DL-based machine vision in IVF. Four important tasks are identified where DL can be of service. The first is embryo development annotation, which refers to identifying and annotating the development stage of an embryo. The second is embryonic cell detection and tracking, where the goal is to authorize the automatic annotation of the embryo development stage through images and locate each cell in an image or video. The third is IVF cycle outcome prediction. The fourth is embryo grading and selection decisions. The authors note here an important issue in IVF, namely the lack of correlation between the predicted and actual outcome of a pregnancy. They also remark that clinical implementations of AI/DL techniques in the future are important to test their potential.
In [33], the authors review the developments of AI in IVF and embryology and identify three main topics. These are the authomatic annotation of embryo development, embryo grading, and embryo selection for implantation. They also identify the potential in predicting ploidy, miscarriages, as well as other topics related to infertility, like measuring ovarian reserve by antral follicle count and the evaluation of the endometrium and contour of the uterus. The authors foresee that the use of AI in the future will encompass many more topics in reproductive medicine.
The work [34] reviews the developments in AI-assisted IVF in the years 2010–2023. The tasks covered include the assessment of oocyte quality, sperm selection, fertilization assessment, embryo assessment, ploidy prediction, embryo transfer selection, cell tracking, embryo witnessing, micromanipulation, and quality management. The importance of removing human subjectivity through AI is noted.
The work [35] discusses the use of AI technologies in IVF for 2023 and beyond. The topics covered include prestimulation testing, outcome prediction, initial dose of gonadotropin for ovarian stimulation, monitoring schedule and workflow during ovarian stimulation, assignment of the trigger day option, and prediction of the ovarian response. Three important topics are also identified for the future. First, the assessment of new tools, such as large language models (Chat-GPT), in clinical care. Second, the clinical culture for the adoption of new clinical tools. Third, the development of guidelines and criteria for publishing results of AI studies in the clinical literature.
In [36], the use of AI in time-lapse systems is reviewed. Some limitations identified include model interpretability, acquisition of standardized datasets, the problem of sharing data among clinics, and the necessity for external validation. Among the future challenges reported are the development of explainable AI algorithms to understand their decision making process, the study of algorithm robustness through large studies in many clinics with a diverse patient population, the use of collaborative federated learning [37] among clinics to allow data sharing without compromising privacy, and the participation of clinicians in the development and refinement of AI models.
The work [38] reviews the use of AI in ovarian stimulation. Some of the aspects covered include the development of decision support systems, outcome prediction, the selection of doses and protocol, and scheduling. The authors underline the importance of maintaining the contact between patients and healthcare practitioners, so the AI developments should not build a distance between the two groups. AI technologies have the prospect of democratizing access to ART care by increasing its capacity for clinical service.
The review [39] focuses on the problem of embryo selection. It was observed that AI techniques outperformed the clinical teams in all the studies that focused on embryo morphology and clinical outcome prediction during the embryo selection assessment. Of the articles reviewed, the median accuracy of AI systems was 75.5%, with a range of 59–94% on predicting the embryo morphology grade. When predicting clinical pregnancy through patient clinical treatment information, the median accuracy was 77.8%, with range of 68–90%. When both image and clinical data were provided, the median accuracy rose to 81.5%, with a range of 67–98%. These results are very promising but have limitations. For example, the authors note that many works used their own datasets, which were gathered from the local population, thus creating subjectivity in the ground truth for each study.
The short review [40] underlines the significance of time lapse imaging in embryo selection. Although there is not a single kinetic parameter that can provide predictions with absolute accuracy, the use of time-lapse imaging has helped in gathering large data and providing a ground for ML techniques to be developed.
The work [41] also reviews the use of time-lapse imaging systems, spent embryo culture media, and morphological criteria for the non-invasive evaluation of embryo quality and transfer selection. The fusion of complementary methodologies can be a key in enhancing their effectiveness.
The work [42] reviews the use of CNN models in embryo evaluation using time-lapse monitoring. Three tasks were considered, successful in vitro fertilization, blastocyst stage classification, and blastocyst quality. Of the articles reviewed, most of them reported an accuracy greater than 80%, and in certain works the models performed better than the practitioners. Observing the heterogeneity of the studies with respect to the DL architectures, reference standards, datasets used, and final results, the authors underscore the importance of sharing databases between laboratories, as well as standardizing the reporting.
In [43], the authors review DL approaches for blastocyst grading. Among the challenges, they identify the importance of having large datasets to train the models. They also highlight the importance of spatial and temporal information obtained from time-lapse imaging to improve the model’s efficacy. The authors note that integrating the patient’s clinical data and the outcome of IVF into the prediction model could help recognize scenarios of increased or decreased pregnancies.
The work [44] reviews AI techniques for embryo ploidy status non-invasive prediction. The importance of mosaicism inclusion for future studies is underlined. The integration of AI techniques into microscopy equipment and embryoscope platforms will be key for the wide use of non-invasive genetic testing. The predictive power of algorithms will be increased by optimizing the use of clinical considerations and incorporating the minimally necessary covariates.
The work [45] provides a review of DL applications for embryo classification and evaluation. The work outlines the problem tasks and architectures used and gives a short comparison between some recent works, pointing to the ones that yielded the highest accuracy, although each work was tested on different datasets.
The work [46] reviews the task of embryo selection and AI. The authors foresee that the use of AI methods will support successful IVF clinic business models, and its use will eventually become standardized in IVF laboratories. As the grading and ranking of the embryos is a time-consuming task, automating it in an objective and reproducible way through AI would greatly allow the reallocation of the researchers efforts to other tasks. It will also help reduce inter/intra-observer variability in grading, which is an important issue [47]. Moreover, it will further support data sharing between laboratories, a topic discussed in Section 5.3.
The short reflection [48] takes note of the importance of finding a balance between the use of AI techniques and human experts for the progression of the field. Also, when considering the use of a new algorithm, it is important to consider not only its performance but also its practical feasibility for being implemented in a clinical setting.
The work [49] reviews the developments of AI techniques in the prediction of the best embryo for transfer. The authors take note that despite many AI prediction models slightly outperforming the embryologists in the studies covered, they are still too early in their development stage to claim superior performance over the embryologists’ assessments.
The work [50] reviews the use of AI in the embryology laboratory. The topics covered include spermatozoa analysis, ovarian stimulation management, oocyte analysis, pronuclear-stage embryos, cleavage-stage embryos, blastocyst-stage embryos, time-lapse microscopy image analysis, static image analysis of blastocysts, automated annotation of blastocysts, implantation prediction, and non-invasive ploidy screening. The authors note that although the use of AI in the improvement of outcomes is not yet fully proven and established, it has the potential to help address many persistent problems in the field of reproductive medicine.
In [51], the developments of deeptech and femtech for IVF are reviewed. The DIY IVF cycle is discussed, with its potential leading to reduced treatment costs and, subsequently, democratized access to this service. However, the importance of human interaction with physicians for patients undergoing IVF is underlined, as well as the need for data security. The need for inclusive and diverse population datasets is also noted.
The work [52] reviews the use of AI in sperm selection, a task highly relevant to ART. The topics discussed are sperm morphology, DNA integrity, and motility. The authors denote the importance of sample size and representation in the data used, as well as the ethical concerns that span from automating the sperm selection process.
The work [53] reviews the use of quantitative models in clinical reproductive endocrinology. They identify examples where AI models can be used to support decision making in endocrinological interventions, like the selection of gonadotropin doses for ovarian stimulation, the prevention of premature ovulation, and the induction of oocyte maturation.
In [54], the use of predictive models for the initial dose of gonadotropin in controlled ovarian hyperstimulation is reviewed. For future research, the authors underscore the need to have some common consensus on study parameters, for example, having the same range of the number of retrieved oocytes in patients with normal responses and considering MII oocyte number, MII oocyte rate, or follicle output rate as outcome indicators. In addition, the need to include potential hidden variables, such as a history of pelvic surgery or chronic pelvic inflammatory disease, that can affect the predictive model.
In [55], the use of AI in ultrasound is reviewed, with applications like monitoring follicles, assessing endometrial receptivity, and predicting the pregnancy outcome of IVF and embryo transfer. Current limitations are discussed, such as ethical and liability concerns; dataset issues such as sample size, quality, and diversity; and the need for systematic thinking that only clinicians can provide. It is suggested that proper use of AI should focus on early detection and detection.
In [56], the authors provide an ethical assessment on the use of AI in IVF. They underline the importance of transparency in the algorithms being developed to avoid bias and understand their decision making.
Finally, as a topic generally related to ART technology, the work [57] reviews the use of deep technology to optimize cryostorage.
4.2. Strategy Selection
Prior to designing DL techniques for IVF, AI methods can also be used as a support tool in medical decision-making. For example, in [58], several machine learning models were considered to support the decision according to the type of treatment strategy used by medical professionals. Seven strategies were considered, the long strategy, short strategy, antagonist strategy, ultralong strategy, ovulation induction with clomiphene citrate, letrozole microstimulation for ovulation, and ovulation induction in the luteal phase, with focus on the first four strategies. Ten different algorithms were tested, logistic regression, random forest, k-nearest neighbor, NN, support vector machine (SVM), Bagging, AdaBoost [59], gradient boosting decision tree, extreme gradient boosting, and light gradient boosting machine [60], giving varying precisions for different treatment strategies and age groups. The dataset consisted of 95,868 records of couples who had gone through IVF-ET in the Reproductive Medicine Center of Tongji Hospital, Tongji Medical College, affiliated with Huazhong University of Science and Technology (2006–2019).
4.3. Embryo Development Annotation
Development annotation is a fundamental task for embryo image evaluation and can be a basis for further processing actions.
In [61], the problem of early stage development classification was considered using time-lapse videos. A multi-task deep learning with dynamic programming (MTDL-DP) architecture was developed. Initially, MTDL classifies each frame of the time-lapse video as a development stage. Then, the DP part optimizes the stage sequencing so that it is monotonically non-decreasing. The ResNet50 model [21] was considered as a basis, pretrained on ImageNet [62]. One-to-many, many-to-one, and many-to-many architectures were considered. The one-to-many and many-to-many models had the best results, with accuracies up to 85–86.9%, but the one-to-many model obtained the best balance between performance and computational cost. The dataset consisted of 170 time-lapse videos, obtained using an EmbryoScope+ microscope, with 59,500 frames overall, obtained from the Reproductive Medicine Center of Tongji Hospital, Huazhong University of Science and Technology, Wuhan, China.
In [63], a model was developed for predicting blastocyst development from oocytes. The architecture considered was EfficientNet B-7 [64], pretrained on ImageNet [62], which performed a binary classification (positive/negative blastocyst prediction). The model achieved an AUC of 0.64 and 0.63 on the test and external data. The dataset consisted of 37,133 oocyte images from eight clinics in six countries (Canada, USA, UK, India, Spain, Czechia) between 2014 to 2022. An additional 12,357 pieces of image data were also used for validation, from two clinics in two coutnries (2017–2022).
In [65], the recognition of ploidy status was considered using time-lapse videos. The DL distinguished between aneuploid embryos (group 1) and other types (group 2, euploid, and mosaic). The model was the Two-Stream Inflated 3D ConvNet (I3D) [66], pretrained on the ImageNet [62] and Kinetics [67] datasets. The model used RBG and optical flow data in two I3D models, RGB-I3D and Optical flow-I3D, whose predictions are averaged into the final output. The AUC achieved was 0.74. The dataset was gathered from 108 patients undergoing 119 PGT-A cycles at the Lee Women’s Hospital, Taichung, Taiwan, and included 144,210 images from 690 videos.
In [68], oocyte classification was considered. The architecture consisted of two main parts, a DeepLabV3Plus [69] model for image analysis, used to extract oocyte regions, pretrained on ImageNet [62], and a network inspired by SqueezeNet [70], used for classification. This network was improved by a genetic algorithm in order to have a better generalization and reduce the number of learnable parameters, FLOPs, and inference time. The classification was performed in three classes, metaphase I meiotic division (MI), metaphase II meiotic division (MII), and prophase I meiotic division (PI). The mean accuracy coefficient achieved was 0.957 on the test set. The dataset consisted of 766 images, of which 44 were MI oocytes, 663 MII oocytes, and 59 PI oocytes, collected from 100 patients undergoing intracytoplasmic sperm injection (ICSI).
In [71], six pretrained models were studied for embryo development annotation in time-lapse videos, with 14 classes considered, based on their morphological differences. The highest accuracy achieved was 67.68%, from EfficientNet-B6 [64]. The dataset consisted of 163 embryo cycles and 15,831 embryo images, obtained from Morula IVF Jakarta Clinic, Jakarta, Indonesia.
In [72], a model was developed for anomaly detection in embryos using time-lapse images. The architecture designed consists of a local binary CNN [73] in series with an LSTM. The model can achieve an early (72 h) detection of abnormalities with a precision of 82.8%, outperforming other architectures. The dataset comprised 8 non-healthy embryos and 12 healthy embryos, with 120 monitoring hours, from Istanbul Aydin University, Turkey.
4.4. Intracytoplasmic Sperm Injection
In [74], the problem of identifying morphological landmarks in images of embryos in the cleavage stage was considered. Two problems were addressed. First, a CNN-ICSI model was developed to identify the optimal location for intracytoplasmic sperm injection through polar body identification. The model achieved a 98.9% accuracy. The second model, CNN-AH, was developed to identify the optimal location for assisted hatching on the zona pellucida, with a maximum distance from healthy blastomeres. This model achieved a 99.41% accuracy. The models classified images into 12 classifications, with each class resembling a location, similar to the 12 h digits on a clock, equally spaced by 30 degree angles. The dataset consisted of over 19,000 oocyte images and 19,000 cleavage stage embryo images collected from the Massachusetts General Hospital (MGH) Fertility Center in Boston, Massachusetts.
In [75], a CNN model was developed to identify the stages of intracytoplasmic sperm injection from video sequences. Two classes were considered for each frame, the selection stage and the injection stage. The CNN architecture consisted of four convolution layers, one max-pooling layer, and a flattening layer. The accuracy was over 99%. The dataset consisted of 50 videos at 15 frames per second, with 2550 frames in total, obtained from 10 clinics (2021–2022).
4.5. Component Segmentation
Component segmentation in embryo images is a fundamental task, as it can support the further processing of embryos for tasks like grading or outcome prediction.
In [76], the segmentation of two blastocyst components, trophectoderm (TE) and inner cell mass (ICM), was considered using texture analysis. The algorithms utilized include k-means and watershed segmentation. The accuracy achieved is 86.6% for TE and 91.3% for ICM. The work also introduces a novel open-source dataset. The dataset consists of 211 Hoffman Modulation Contrast (HMC) blastocyst images from the Pacific Centre for Reproductive Medicine (2012–2016).
In [77], implantation outcome prediction was considered using single-blastocyst images. The design has two parts. The first part is a blastocyst component segmentation. This architecture uses Dense Progressive Sub-Pixel Upsampling (DPSU), inspired by [78], in combination with DeepLabV3 [79]. The second part is a multi-stream cross-modality classification utilizing a Compact–Contextualize–Calibrate (C3) feature extraction technique for predicting the outcome of implantation. The mean accuracy achieved was 70.9%. The C3 unit was also incorporated into the ResNet50 [21] and InceptionV3 [80] models and also improved their performance. The dataset consisted of 578 blastocyst images from the Pacific Centre for Reproductive Medicine (PCRM) (2012–2018).
In [81], blastocyst segmentation into blastocoel (BC), zona pellucida (ZP), inner cell mass (ICM), trophectoderm (TE), and background (BG) components is studied. A sprint semantic segmentation (SSS-Net) architecture is proposed, based on a fully convolutional semantic segmentation scheme. The model uses a sprint convolutional block (SCB) that uses asymmetric kernel and depth-wise separable convolutions. Residual and dense connectivity models are considered, with a mean Jaccard index of 85.93%, and 86.34%, respectively. The model outperforms other architectures such as UNet-Baseline [82], TernausNet U-Net [83], PSP-Net [84], DeepLab V3 [79], and Blast-Net [85] and is also more computationally efficient, having 4.04 million parameters compared to the rest, which range from 10 to 40 million. The dataset was obtained from the work [76].
In [86], the same problem is considered for these five components: BC, ZP, ICM, TE, BG. The model is termed embryo component segmentation network (ECS-Net). It consists of two streams that use a convolutional block and a depth-wise separable convolutional block and also considers dense connectivities [22]. The model achieves a mean Jaccard index of 86.46% and outperforms other architectures such as UNet-Baseline [82], TernausNet [83], PSP-Net [84], DeepLab V3 [79], and Blast-Net [85] and is more computationally efficient, with 2.83 million parameters compared to the other models, which range from 10 to 40 million. The dataset used was [76], as in [81].
In [87], five-component segmentation is considered again. The architecture developed is termed the feature-supplementation-based blastocyst segmentation network (FSBS-Net). The architecture consists of convolutional layers, batch normalization layers, strided convolutional layers, transposed convolutional layers, ascending channel convolutional blocks, pixel classification layers, and feature supplementation mechanisms. The model achieved a mean intersection over a union value of 87.26%. It outperformed other architectures like UNet-Baseline [82], TernausNet U-Net [83], PSP-Net [84], DeepLab V3 [79], Blast-Net [85], SSS-Net Residual [81], SSS-Net Dense [81], and ECS-Net [86] with respect to segmentation accuracy and computational efficiency, as it consisted of 2.01 million trainable parameters, significantly fewer than the other models (2.83 million to 31.03 million). The dataset was obtained from the work [76], as in [81,86].
In [88], blastocyst segmentation was considered from its background using U-Net models [82]. Two models were developed. In the first model, the original U-Net encoder or contraction section was replaced by a pretrained DenseNet121 architecture [22]. The second model was developed by replacing all convolutional blocks in the U-Net by dense blocks, keeping the symmetry unchanged. Both models outperformed the basic U-Net, and the second model, termed Densely U-Net, achieved the best accuracy of 99.8%. The dataset consists of 327 images from the Indonesia Medical Education and Research Institute (IMERI) [89]. Each image includes two to three embryos.
In [90], a clustering-based system was developed for localizing and counting blastomeres, from day-2 and day-3 images. The model has a preprocessing, a segmentation, and a hierarchical clustering-based module for the segmented blastomeres, using an agglomerative hierarchical clustering algorithm. This clustering module combines the results of different experiments to improve the overall performance. The average precision was 87.9%. The dataset consisted of 50 images from the Assisted Reproduction Technology (ART) Unit, International Islamic Center for Population Studies and Research, Al-Azhar University, Cairo, Egypt.
In [91], the segmentation of the oocytes was studied using deep NN. There were 14 areas considered, including the background, and 71 different deep neural networks were tested, based on DeepLab v3+ [69], Fully CNNs, SegNet [92], and U-Net [82]. The best performance was achieved by a DeepLab-v3-ResNet-18 variant, with 79% validation accuracy. The dataset consisted of 334 oocyte images from 60 patients.
4.6. Embryo Grading
The grading and selection of embryos is another fundamental task at the core of IVF [93]. This task can be assisted by AI, as reviewed in [46], and is greatly supported by image processing architectures.
On the topic of data utilization, DL models generally perform better when working with numerical data rather than categorical data. As some aspects of embryo characterization are categorical, the work [94] addressed the task of converting them to numerical values to facilitate statistical analysis and understanding of the contribution of embryo quality to the cycle outcome. The Gardner embryo grading scale was converted to the numerical embryo quality scoring index (NEQsi), with values of 2 to 11.
In [93], the KIDScoreTM D5 v3 algorithm for grading was considered. The authors found the tool useful as a support tool for embryologists in selecting blastocysts for embryo transfer. It can improve consistency in embryo selection and also improve the embryologists’ workflow. The dataset consisted of 12,468 embryos from 1678 patients from IVI Valencia in Spain (2018–2020).
In [95], the problem of grading day-3 embryos was studied. Five grades were considered (A–D, and compacted). Two models were considered for this, ResNet50 [21] and Xception [20], pretrained on ImageNet [62], with some additional fully connected layers. The Xception model was the best, achieving an accuracy of up to 98%. The dataset consisted of 152 images from around 20 patients from the Vistana Fertility Center in Kedah, Malaysia.
In [96], the problem of blastocyst grading was studied. Four grades were considered, excellent, good, average, and poor. Two models were developed, a CNN, and a VGG-16 [18], with appropriate new classification layers. Both models achieve good results, with a testing accuracy of 90% for the CNN and 94% for the VGG-16. The dataset consisted of 110 blastocyst images, from Vistana Fertility Center, in Kedah, Malaysia.
In [97], a novel model for binary embryo classification (good/bad) was developed. The architecture combined InceptionV3 [98] and DenseNet201 [99] with a fusion of features from each model to obtain the classification. The models were pretrained on ImageNet [62]. The model achieved an accuracy of 95.83% and outperformed other architectures like ResNet50 [95], VGG16 [96], DenseNet201 [99], InceptionV3 [98], and InceptionResNetV2 [98]. The dataset consisted of 840 images from day 3 and day 5, from the fourth international competition of AI and data science on embryo classification on microscopic images by Hung Vuong Hospital [100].
In [101], a deep CNN with a morphology attention module (MAM) was applied to embryo grading. The design (LWMA-Net) consisted of six blocks and a classification head. Pre-training was performed using ImageNet [62]. The model achieved an AUC of 96.88% and 97.58% in four- and three-category gradings. More experiments showed that embryologists using LWMA-Net achieved an improved embryo grading performance on the four- and three-category grading tasks. The dataset used consisted of 4290 embryo images from 2639 couples (2016–2021).
In [102], the authors studied the sensitivity of automatic embryo grading to different focal planes, with the goal of ensuring consistency. They concluded that by performing test-time augmentation and ensemble modeling can reduce this sensitivity. ResNet18 and EfficientNet-b1 models were considered. The dataset consisted of blastocyst images from 11 IVF clinics in the US (2015–2020).
In [103], time-lapse images were used for blastocyst grading. The model predicts inner cell mass (ICM) and trophectoderm (TE) grades. The architecture consists of a CNN that extracts features from frames, coupled with an RNN that uses temporal information by combining image features over consecutive frames. The model performed on the same level as that of human embryologists. The dataset included 4032 treatments undergoing IVF (1169), intracytoplasmic sperm injection (ICSI) (2534), or a mixture of both (329). In total, 8664 embryos having reached the blastocyst stage were analyzed. Data were collected from four clinics.
The work [104], considered embryo grading following Gardner’s system, for blastocyst development (rank 3–6), ICM (A, B, C), and TE (a, b, c). The ResNet50 architecture [21] was considered, pre-trained on the ImageNet dataset [62]. It achieved a 96.24% accuracy for blastocyst development, 91.07% for ICM quality, and 84.42% for TE quality. The dataset consisted of 171,239 images of 16,201 embryos, from 4146 IVF cycles obtained from the Stork Fertility Center [105] (2014–2018).
In [106], the problem of embryo grading was addressed using a combination of CNN and LSTM networks. A CNN was used to extract features from embryo images, and then the features were used as input to LSTM to perform the classification. Various pretrained CNN models where considered, many of which gave a 100% validation accuracy score, like VGG16-LSTM, VGG19-LSTM, MobileNetV2-LSTM. Instead of the LSTM layer, a VGG16-GRU was also considered, which again gave 100% validation accuracy. The grading consisted of five classes (A—best, B—good, C—fair, D—poor, E—Non-viable). The dataset was obtained from Nova IVF Fertility, Ahmedabad, Gujarat, India [107], and consisted of 803 labeled samples from 60 patients, with 5458 image frames used overall.
In [108], blastocyst grading was considered using multifocal images. Three models were considered, using VGG-16 as a basis, appended by average pooling and fully connected layers. The first model used an ensemble network structure inspired by [109]. The second model used a voting mechanism for the classification. The third model only used the three images with the highest sharpness out of the inputs. The classification was performed in two classes, good and poor. The highest AUC was 0.936, achieved by the third model. Moreover, using Grad-CAM [110], a color-graded version of the blastocyst images was generated, to highlight the important image parts. The dataset consisted of 1025 embryos from the Center for Reproductive Medicine of the Affiliated Drum Tower Hospital of Nanjing University Medical School in China, with 11,275 images overall (2017–2018).
In [111], CNNs were considered to judge the consistency of grading. Five classes are considered (1—poor, 2—fair, 3—good, 4—great, 5—excellent). Although embryologists had a high degree of variability in their gradings, the DL model had a consistency of 83.92% in selecting blastocysts for biopsy and cryopreservation. The training dataset consisted of 3469 embryo images recorded at 70 and 113 h post insemination from the Massachusetts General Hospital (MGH) Fertility Center in Boston, Massachusetts. The dataset used for evaluation consisted of 748 embryo images at 70 h post insemination and 742 at 113 h.
In [112], embryo classification was studied on the third day, the cleavage stage. A network of eight layers, five convolution and three fully connected, achieved 75.24% accuracy in predicting the embryo destiny (discard or transfer). Considering the batch context, where embryos are predicted with relation to other ones from the same batch, the accuracy increased to 84.69%. The predictions made by the model were also related to clinical implantation rates. The dataset consisted of 38,000 records from UZ Leuven Hospital, each having multiple images at various focal depths, for a single embryo.
In [98], the grading problem for embryo images at 113 hpi was studied. The model had five grades, for non-blastocysts (grades 1–2) and blastocysts (grades 3–5). A collection of architectures were considered, like Inception V3 [80], ResNET-50 [21], Inception-ResNET-V2 [19], NASNetLarge [113], ResNeXt-101 [114], ResNeXt-50 [114], and Xception [20], pretrained on ImageNet [62], with the Xception model outperforming the rest. Data were obtained from the Massachusetts General Hospital (MGH) Fertility Center in Boston, Massachusetts, and consisted of 3469 embryo videos from 543 patients. The evaluation dataset included 2440 images at 113 hpi.
In [115], the selection problem for embryos with single-timepoint images collected at 113 hpi was studied. The architecture consisted of an Xception network from [98] paired with a genetic algorithm for rank ordering embryos by generating unified scores. The CNN was pretrained using ImageNet [62]. The accuracy for choosing the best embryo reached 90%. In addition, on the problem of evaluation of implantation potential, on a set of 97 euploid embryos capable of implantation, a CNN model outperformed a group of embryologists. The dataset included 3469 videos from 543 patients, from the Massachusetts General Hospital (MGH) Fertility Center in Boston, Massachusetts. The evaluation dataset had 2440 static human embryo images at 113 hpi, The test dataset included 97 patient cohorts and 742 embryo images.
In [116], an experiment was designed to test the aid of AI in embryo selection. Embryologists were given a set of embryos to choose the best embryo to transfer. The embryo images were first given without any further notes, and then, they were provided again, but this time accompanied by a suggestion made by the AI system as to which embryo should be transferred. With the aid of the algorithm, embryologists could identify successfully implanted embryos in 73.6% of the cases, compared to 65. 5% for the case in which no AI suggestion was provided. The model used was based on Xception described in [115], pretrained on ImageNet [62]. The dataset consisted of time-lapse images from 400 embryos and 160 patients, from the Massachusetts General Hospital Fertility Center (2014–2018).
In [117], the authors considered embryo grading at the blastocyst stage, under the scope of developing a model that can be equally applicable to all types of embryo data, regardless of microscope type, image capture day, and cycle type. Images were collected on days 5, 6, or 7 prior to transfer, biopsy, or freezing, using inverted microscopes, stereo zoom microscopes, or time-lapse incubation systems. A deep CNN, ResNet-18 [21] with dropout, was trained to rank images based on the possibility of reaching clinical pregnancy. In total, 5100 blastocysts from fresh, frozen, and frozen euploid transfers were considered and were matched to pregnancy outcomes, as well as 2,900 blastocysts, corresponding to aneuploid PGT-A results. The AUC achieved was 0.72 for all embryos. The model also outperformed manual grading for euploid transfers. Data were gathered from 11 IVF clinics in the US (2015–2020).
In [118], emphasis is given on applying image filters at the pre-processing stage, for day-3 embryo images for grading (excellent, moderate, poor). The images are passed through filters, and after a selection process, the most suitable are used as input to a CNN (VGG-Net [18]). The filters considered are Blur, Gaussian, Sharpening, Laplacian, Vertical-Sobel, Horizontal-Sobel, and Median. This model outperformed the models that used no filters, all the filters, or a random collection of them, by reducing the test error by over 8%. The dataset consisted of 1386 embryo images from 238 patients, which were obtained from an infertility clinic in Indonesia (2016–2018).
The authors of [99] considered the prediction of blastocyst formation and quality by time-lapse monitoring from the first three days. The model considered is termed the spatial-–temporal ensemble model (STEM), and predicted blastocyst formation. The STEM model uses a weighted average between a temporal stream (LSTM) model and a spatial stream (gradient boosting classifier) one. DenseNet201 [119] was implemented in the design. The accuracy was 78.2% for blastocyst formation prediction. The STEM+ model also performed well in usable blastocyst prediction, with 71.9% accuracy. The initial dataset consisted of 26,113 embryos, from 2594 IVF and intracytoplasmic sperm injection (ICSI) cycles, cultured in TLM incubators (2014–2017).
In [120], the grading of blastocysts was considered using multi-focus images. A ResNet-50 [21] architecture is used in combination with an attention module to use the high level features from the images. It was observed that the use of multi-focus images improved performance over the single image case. The model also outperformed embryologists for all three grading categories (development stage, ICM, TE). The dataset consisted of images of 4997 blastocysts, cultured in a time-lapse incubator (EmbryoScope, Vitrolife, Sweden) at CReATe Fertility Centre, in Toronto, Canada.
In [121], a euploid prediction algorithm (EPA) was developed using time-lapse imaging, embryo data, and clinical data. The algorithm has three main modules, one for extracting features from the embryo image sequences, one for normalizing the embryo and clinical data features, and one for fusing these features and making the prediction. For processing the image sequences, a 3D-ResNet50 model [122] is utilized as a basis. A fully connected layer network is used at the fusion and prediction part. The AUC achieved was 0.80. The dataset consisted of 469 PGT cycles and 1803 blastocysts, from the Reproductive Medicine Center of Tongji Hospital, Huazhong University of Science and Technology, Wuhan, China (2018–2019). In total, 155 PGT cycles and 523 blastocysts (2019–2020) were also used for verification.
In [123], blastocyst grading was considered for time-lapse images. The developed architecture was termed STORK, which is based on Google’s inception-V1 model [124], pre-trained on ImageNet [62]. The model predicted blastocyst quality with an AUC of over 0.98, and outperformed individual embryologists. It was also tested on datasets from two different clinics, from the Institute of Reproduction and Developmental Biology of Imperial College, London, UK, and Universidad de Valencia, Valencia, Spain, and achieved an AUC of 0.90 and 0.76, indicating its robustness. A descision tree was also designed that combined embryo quality with patient age for pregnancy likelihood prediction. The chances of pregnancy based on individual embryos ranges from 13.8% for poor quality embryos and ages over 41, up to 66.3%, for good quality embryos and ages below 37. The dataset included 10,148 embryos, obtained from the Center for Reproductive Medicine at Weill Cornell Medicine (2012–2017). Images were collected using the EmbryoScope (Vitrolife, Sweden) time-lapse system. There were 50,392 images used.
In [125], a model termed STORK-A was developed for predicting embryo ploidy status. The model uses image data from time-lapse microscopy, in combination with clinical data, like maternal age, morphokinetic parameters, and morphological assessment. STORK-A is based on ResNet18 [21] pretrained on ImageNet [62]. Three classification problems were considered. The accuracy for predicting aneuploid/euploid was 69.3%. The second case predicted complex aneuploidy versus euploidy and single aneuploidy, with accuracy of 74.0%. A third case was to predict complex aneuploidy versus euploidy, with accuracy of 77.6%. The dataset consisted of 10,378 human blastocysts at 110 h after intracytoplasmic sperm injection, from 1,385 patients at the Weill Cornell Medicine Center of Reproductive Medicine, New York (2012–2017). The model was also tested on two independent datasets for aneuploid versus euploid classification, one from the Weill Cornell Medicine Center using EmbryoScope+ machines, with 63.4% accuracy, and one from IVI Valencia, Health Research Institute la Fe, Valencia, Spain, with 65.7% accuracy, and thus showed generalizability.
In [126], a model was developed for embryo euploidy prediction from static images of day-5 blastocysts. The model was an ensemble one, following [127], and included ResNet [21] and DenseNet [22] architectures, along with techniques like data cleansing [128] and distillation [129]. The model’s accuracy was 65.3%, which increased to 77.4% once the dataset was cleaned of some poor and mislabeled images. The model can also be used for evaluating day-6 embryos. Using additional data from independent clinics, it was observed that the model was also generalizable to different patient demographics and it could be applied to images from multiple time-lapse systems. The initial dataset consisted of 15,192 images of embryos at the blastocyst stage from 10 clinics in the USA, India, Spain, and Malaysia.
In [130], two models were used to predict aneuploidy and mosaicism in IVF-conceived embryos. The data used for prediction were general, maternal, paternal, couple related, IVF cycle related, and embryo related. From the many algorithms considered, the random forest algorithm was the best in both cases, with an AUC of 0.792 for aneuploidy and 0.776 for mosaicism. The most important predictive variable for aneuploidy was maternal age, and after this was paternal and maternal karyotype and embryo quality. For mosaicism, the highest importance variable was the technique used in preimplantation genetic testing for aneuploidies and the embryo quality, followed by maternal age and the day of biopsy. The dataset consisted of 6989 embryos taken from 2476 cycles, from Instituto Bernabeu (2013–2020).
The work [131] considered grading for blastocysts and cleavage stage embryos. The classification was binary, with discarded embryos labeled as poor, and transferred or frozen labeled as good. Four models are considered, an EfficientNet variant [64] termed EfficientNet-L2 with Noisy Student Training [132], Swin Transformer [133], STORK [123], and AlexNet [134]. The models were pretrained on ImageNet [62]. The best performance was achieved by the Swin Transformer for both day-3 and day-5 embryos, with an accuracy of around 99.5%. The dataset was obtained from the Xinan laboratory, using time-lapse incubators, and consisted of 4543 videos, from 1037 patients. Overall, 21,915 images for day-5 embryos and 24,489 images for day-3 embryos were used.
In [135], DL was considered for predicting the blastocyst survival after thawing. The model was termed EmbryoNeXt, which was an ensemble of models, ResNet-18, ResNet-34, ResNet-50 [21], and DenseNet-121 [22], pretrained on ImageNet [62]. The ensemble model is averaging the rank predictions of these models. The model, and a team of embryologists, made predictions from images obtained at 0.5 h increments, starting from 0 h to 3 h post thaw. The model achieved an AUC of 0.869 at 2 h and 0.807 at 3 h, while the embryologists achieved an average of 0.829 at 2 h and 0.850 at 3 h. By combining predictions from both the model and the embryologists though, an AUC of 0.880 at 2 h, and 0.860 at 3 h was achieved. The dataset consisted of 652 time-lapse videos of freeze–thaw blastocysts from 119 patients from the Center for Reproductive Health at University of California, San Francisco (2019–2020).
In [136], a model to predict ploidy was developed, termed the Embryo Ranking Intelligent Classification Algorithm (ERICA). The model uses fully connected layers for classification. The model achieved an accuracy of 0.70. It also ranked a euploid blastocyst first in 78.9% of the cases, and at least one euploid embryo in the top two positions in 94.7% of the cases. The dataset of static images consisted of 1231 blastocyst micrographs from three New Hope Fertility Centers in Mexico City, Guadalajara, and New York City (2015–2019). Images were gathered within 5 or 6 days after fertilization and prior to any other intervention.
In [137], a model is developed for embryo grading. A Blast-Net segmentation model is used, and a CNN and VGG-16 are considered for classification. The VGG-16 model had an accuracy of 94%. This architecture is also implemented as a web application, for ease of use. The dataset was provided by the Vistana Fertility Center.
In [138], the authors considered blastocyst grading under a heavily imbalanced dataset, as only 1% of the samples belonged to the minority class. The authors made sure that these samples were included in training, validation, and test phases. The proposed model was VGG16-Multi-Label, which simultaneously grades ICM, TE, and ZP by having some shared convolution and pooling layers between the three labels. The model achieved an accuracy of 73.9% for ICM, 67.3% for TE, and 81.8% for ZP grading, outperforming ResNet50 [21], InceptionV3 [80], and VGG16 [18]. The models were pretrained on ImageNet [62]. The dataset consisted of 704 images provided by the Pacific Centre for Reproductive Medicine (PCRM) in Burnaby, BC, Canada (2012–2018), with some images being from online sources.
In [139], the iDAScore v1.0 was evaluated for time-lapse images. The AUC for euploidy was 0.60 and for live-birth prediction 0.66, which was almost on par with the embryologists. Also, iDAScore v1.0 placed euploid blastocysts as top quality in 63% of cases with one or more euploid and aneuploid blastocysts from the same cohort. When at least two euploid blastocysts were identified within each cohort, the embryologists and the model were both successful in prioritizing the euploid competent blastocyst for transfer in 52% of the cases. The dataset included 3,604 blastocysts and 808 euploid transfers from 1232 cycles, gathered from a private clinic Clinica Valle Giulia, GeneraLife IVF, Rome, Italy (2013–2022).
In [140], day-3 embryos were classified in four categories. An ensemble learning model was proposed (DenseNet169, Inception V3, ResNet50 and VGG19) and achieved an average 74.14% accuracy. When categories 1 and 2 were combined, the accuracy increased to 89.16%. The model outperformed DenseNet121 [141], DenseNet169 [141], InceptionV3 [142], ResNet50 [143], VGG16 [144], and VGG19 [144] in both cases. The models were pretrained on ImageNet [62]. The model also performed better than the embryologist average in both cases in an independent test cohort. The dataset consisted of 3601 images of day-3 embryos from 1800 couples (2016–2018). An independent test set of 699 images form 350 couples (2018) was also used to test the models.
The work [89] considers day-3 embryo grading after IVF. The grading was performed on five classes, based on the Veeck criteria [145]. Several DL architectures were considered, ResNet18, ResNet34, ResNet50, ResNet101 [21], DenseNet121, DenseNet169 [22], Xception [20], and MobileNetV2 [146], developed in fastai [147]. The ResNet50 model achieved the highest accuracy of 91.79%. The dataset included 1084 images from 1226 embryos of 246 IVF cycles captured at the third day after fertilization, obtained from the Yasmin IVF Clinic, Jakarta, Indonesia.
In [148], the authors developed two platforms for embryo image capture and evaluation. The first is a standalone device which can be controlled wirelessly though a smartphone, with a development cost of around USd 85. The second device is attachable to a smartphone and had a cost of around USD 3. The Xception architecture [20] was used, pretrained on ImageNet [62]. The accuracy for classifying between blastocysts and non-blastocysts captured from the stand-alone system was 96.69%, and for the smartphone it was 92.16%, so both systems performed well. The dataset consisted of over 2450 embryos collected using a commercial time-lapse imaging system, along with additional images captured with the devices and gathered from the Massachusetts General Hospital’s fertility center. So training was performed using both the high-quality images and the images captured from the devices separately.
The work [149] studied DL as an early detection system for possible adverse outcomes and to monitor performance of embryologists performing intracytoplasmic sperm injection. The model considered was Xception [20], pretrained on ImageNet [62]. The dataset consisted of EmbryoScope videos from 2366 embryos obtained from a fertility center in Boston, MA, USA.
In [150], a model was developed for predicting the euploidy of blastocysts and live birth in PGT-A treatments, combining morphokinetic and morphological characteristics of blastocysts, as well ass the patient’s clinical parameters. The methodology combined t2, t3, t5, tB, KIDScore, Gardner grade, female age, and the number of embryonic frozen days with two-logistic regression to predict the euploidy of blastocysts. The AUC for euploidy prediction was 0.879, which was a positive outcome, but the model was not successful in predicting live birth after frozen embryo transfer. The study consisted of 403 patients who underwent PGT-A treatment at the Reproductive Medicine Centre of Xuzhou Maternal and Child Health Care Hospital (2019–2022).
In [151], several models were tested for ploidy prediction using morphokinetic, embryological, and clinical data. The models considered were mixed-effects multivariable logistic regression, random forest classifier, extreme gradient boosting, and a deep learning architecture. The logistic regression model with 22 predictors was the best performing one, with an AUC of 0.71. When only morphokinetic predictors were used, the AUC was 0.61. The dataset consisted of 8147 biopsied blastocysts from 1725 patients, gathered from nine IVF clinics in the UK (2012–2020).
The work [152] considered embryo ploidy status classification. Two cases were considered, the first was a classification between euploid/aneuploid/mosaic, while the second was between euploid/(aneuploid + mosaic). In case study 1, the models considered achieved low accuracy scores. For the second case, a gradient boosting algorithm with histogram of oriented gradient and principal component analysis performed the best, with an accuracy of 0.74. The second best model was a decision tree with histogram of oriented gradient and principal component analysis, which achieved an accuracy of 0.70. Other models like DenseNet [22] had inferior results. The dataset consisted of 1123 embryo samples from 483 couples, obtained from the Morula IVF Jakarta Clinic, Jakarta, Indonesia.
The work [153] studied the prediction of oocyte maturation in the GnRH antagonist flexible IVF protocol. The algorithm considered was XGBoost [154], which achieved an accuracy of 75% for the prediction of a high oocyte maturation rate. The most important parameters for prediction were the peak estradiol level on trigger day, the estradiol level on antagonist initiation day, the average dose of gonadotropins per day, and the progesterone level on trigger day. The dataset consisted of 462 patients (2005–2015) of age less than or equal to 38 years on their first IVF-ICSI cycle using a flexible GnRH antagonist protocol.
In [155], the problem of blastocyst classification into two classes, suitable/unsuitable for transfer, was considered. A DL ensemble model was developed based on AlexNet [134], VGG11 [18], and a variant of VGG with no dropout layers, in contrast to the AlexNet and VGG11. The ensemble model has an accuracy of 78%, more than the individual models. By further implementing a voting classifier from five instances of the ensemble model, the accuracy reaches 81%. The dataset consisted of 2269 images.
In [156], Small Non-Coding RNAs were studied as biomarkers for the prediciton of embryo quality. A prediction pipeline model was developed for this, utilizing an XGBoost model, a Lasso model, extra random trees, and a voting mechanism. Two miRNAs and five piRNAs could be used to predictively distinguish a high-quality embryo from a low-quality one, with an average accuracy of 86%. The dataset consisted of spent blastocyst medium samples from 60 patients with idiopathic infertility.
A variation of the problem of embryo grading, in [157] the problem of patient identification was considered from cleavage and blastocyst stage embryos. The model was initially fed embryo images on day 3 and day 5 from each patient. It could then evaluate embryos at a different timepoints on day 3 and 5 and match them to the same patient. Such a task can be of high importance for specimen tracking in a lab. The CNN model was based on [98,158], in combination with a genetic algorithm [115], which computed the unique identification scores for the embryos. The model achieved 100% accuracy for patient identification when choosing from random pools of eight patient embryo cohorts. The dataset consisted of TLI data from 400 patients from the Massachusetts General Hospital (MGH) Fertility Center in Boston, MA.
In [159], a random forest algorithm was developed to predict blastocyst ploidy, using copy number variation patterns. The ensemble learner used multiple decision trees and a bagging strategy. The embryos were categorized in three grade classes, based on their euploidy probability. Overall, the A- and B-grade embryos had better outcomes compared to the C-grade ones. The initial dataset consisted of 345 blastocyst embryos, and the system was validated on 266 patients.
In [160], a model was considered for binary classification of embryos (good/poor). The model was based on Inception-V121, pretrained on ImageNet [62]. The accuracy achieved was 93.8%. A dataset of 68 images was used, obtained from GitHub.
In [161], a pipeline termed BlastAssist was developed for measuring features of embryos. The features considered included fertilization status, cell symmetry, developmental timing, pronuclei fade time, degree of fragmentation on day 2, and blastulation time. The pipeline consisted of six neural networks for different tasks, building upon results of previous works [162,163,164]. The model can perform on par or outperform embryologists. The dataset consisted of time-lapse image data of 32,939 embryos, with 67,043,973 images overall, with clinical annotations, from the IVF unit of the Tel Aviv Sourasky Medical Center in Israel (2012–2017).
The work [165] considered the generation of predictive embryonic development videos. The Siamese model is proposed. Its first model, the overview diffusion model, learns to predict frames from one or more real images and generates an overview fake video. The second model, the fill diffusion model, generates images that are interpolated between the frames of the overview fake video. The model can generate videos up to 197 frames. The dataset used is a public dataset from [166], which includes 704 videos of different focal planes, with 2.4 million images overall.
4.7. Ovarian Stimulation
Ovarian stimulation is another important task in the IVF process. The authors of [167] studied the day-to-day support of IVF, considering four important decisions. The decisions were to stop or continue the stimulation, trigger or cancel, the number of days to follow-up, and whether dosage should be adjusted. A hybrid algorithm was developed, incorporating classification and regression trees, random forests, support vector machines, logistic regression, and neural networks. The accuracies for the four decisions were 0.92, 0.96, 0.87, 0.82. The database consisted of 2603 cycles, of which 1,853 were autologous and 750 donor.
In [168], a model was developed for selecting an individualized starting dose of gonadotrophin during ovarian stimulation. To achieve this, for a given patient cycle, a set of the 100 most-similar patients was identified using K-nearest neighbours, based on some parameters like age, body mass index, baseline anti Müllerian hormone, and baseline antral follicle count. Using these, a dose–response curve was generated, relating the starting dose to the mature oocytes retrieved. From these curves, patients were categorized as dose-responsive if their curve showed a region that maximized MII oocytes and flat-responsive otherwise. For the dose–response category, the outcomes between people receiving a dose from the optimal range and those receiving a dose outside the optimal range were compared. Those in the optimal range had indeed better outcomes that were achieved with lower starting and total follicle-stimulating hormone (FSH) dose. For the flat-response category, the outcomes between people receiving a low or high dose were compared. Those receiving lower starting and total FSH doses had slightly better outcomes. The dataset included 18,591 cycles, gathered from three IVF clinics in the USA (2014–2020).
In [169], a model was used to predict the best day of trigger. The goal was to optimize the fertilized oocytes (2PNs) and the total usable blastocysts. The descision that the model made was whether to start the triggering on that day or wait another day. A T-learner with bagged light gradient boosting machine (LightGBM) base learners was used for this. The predicted treatment effect was computed as the difference between the treatment and control estimates. The model indeed provided improvements in the outcome of the fertilized oocytes (2PNs) and the total usable blastocysts over the physician decisions. The most significant decision features for the algorithm were the number of follicles 16–20 mm in diameter, the number of follicles 11–15 mm in diameter, and the estradiol level. The dataset consisted of 7866 ICSI cycles, from the University of California San Francisco Center for Reproductive Health (2008–2019).
In [170], a model was developed for predicting the best day for trigger during ovarian stimulation. The optimization was performed with respect to the average number of MII oocytes, the fertilized oocytes (2PNs), and the usable blastocysts. The developed model used linear regression to predict MII oocytes retrieved if triggered today or tomorrow, using the follicle counts and E2 levels measured on the day of the trigger and the previous day. A model was also used to predict the next-day E2 levels from follicle counts and E2 levels measured on the previous day. When applying this model for daily predictions on the considered dataset, probable early triggers were identified in 48.7% of the cycles and late triggers in 13.8% of cycles. So the patients who had matched with timely triggers had on average higher MII oocytes, 2PNs, and usable blastocysts compared with patients with early and late triggers. The dataset consisted of 30,278 cycles from three ART centers in USA (2014–2020).
In [171], the optimal day of triggering was considered in antagonist protocol cycles. The goal was to maximize the number of total and mature oocytes to be retrieved. The architecture used was XGBoost [154], and the model suggested one to three days as options for trigger. From the tests performed on three quality groups, there was an overall improvement in oocytes for the concordant group; that is, the group where the day chosen by the physician was among the algorithms suggested. The algorithm also performed outlier detection. The dataset consisted of 9622 cycles from the IVF unit of Herzliya Medical Center in Israel (2018–2022).
In [172], a linear regression model was developed to recommend the optimal first dosage of follicle-stimulating hormone for ovarian stimulation. The predictor variables used were the age, BMI, AMH, AFC, and previous live births. Using a performance score which was based on the number of MII oocytes retrieved and the dose received, the model outperformed clinicians. The dataset included 2713 patients from five private reproductive centres from two countries (2011–2019) and an extra 774 cycles (2020–2021) for validation.
4.8. Predicting Retrieval of Oocytes
Predicting the number of retrieved oocytes is another piece of important information that can be extracted using ML. The study in [173] developed three models for predicting the number of oocytes retrieved from controlled ovarian hyperstimulation. The Substra framework [174] was used for the analysis, ensuring data security. The models were developed using the light gradient boosting machine (LightGBM) algorithm [60]. One model predicted the number of oocytes retrieveed directly and achieved a mean absolute error of 4.21 oocytes, surpassing a linear regression model. The other two models predicted in which bin the number fell into, with two clinicians providing the bins. The first of two models achieved a mean absolute error of 0.73 for the bins of the first clinician, and the second achieved 0.62 for bins of the second clinician, both surpassing a logistic regression model. The features with the highest importance were antral follicle count and then basal anti-mullerian hormone (AMH) and basal FSH. The dataset consisted of 11,286 cycles from a center in France (2009–2020).
In [175], a model was developed using three-dimensional ultrasound scanning for predicting the number of mature oocytes that are retrieved and optimizing HCG trigger timing. The prediction of ovarian hyper-response was also considered. The architecture considered is C-rend [176], based on three-dimensional U-Net [82]. This model obtained a follicle volume biomarker. The prediction for the number of retrieved mature oocytes was better compared to using two-dimensional diameter measurement data. The dataset consisted of 515 cases (2019–2020).
In [177], two algorithms were studied for predicting the retrieval of oocytes, an NN and a support vector regression, with the NN being the better one. The most important feature for prediction is the antral follicle count, and next are the E2 level on the human chorionic gonadotropin day, the age, and the anti-Müllerian hormone. The dataset consisted of 1365 women from the Renming Hospital of Wuhan University (2019–2020).
4.9. Pregnancy and Live-Birth Prediction
Pregnancy prediction is possibly the most essential prediction that can be made using the available data from the IVF cycle.
In [178], the problem of interobserver variability when evaluating implantation probability was considered. For this, 39 embryologists from 13 different countries were used in the study and were asked to provide implantation probability grades (scale 1 to 5). A DNN model was also developed to provide implantation probability grades. This model was an ensemble of two DNNs, utilizing EfficientNetV2 [179] and Seg-Net architectures [92]. The average implantation prediction accuracy for the embryologists was 51.9%, while for the DNN model it was 62.5%. The interobserver agreement for the embryologists was in general moderate, but it was higher for embryos in the poor- and top-quality groups. The dataset consisted of 136 TLI videos from two different IVF clinics (2018–2019).
In [11], the problem of embryo selection was considered using light microscopic images and taking into account factors like the patient age, embryo development stage, the quality of inner cell mass (ICM), and trophectoderm (TE), following the Istanbul grading system [180]. The architectures considered were ResNet-34 [21], EfficientNet-B0 [64], CoAtNet-2 [181], Xception [20], and ViT-Tiny-S16 [25]. The models were first pretrained on the ImageNet dataset [62] and fine-tuned afterwords with the considered dataset in order to make a binary prediction (pregnant or non-pregnant). The best-performing model was EfficientNet-B0, which obtained an accuracy of 65%, sensitivity of 74.29%, and AUC score 0.72. The additional features indeed increased the model performance. The data consisted of 1099 entries, 747 non-pregnant and 352 pregnant, gathered from the Reproductive biology unit, Faculty of Medicine, Chulalongkorn University (2018–2022).
In [182], the problem of predicting clinical pregnancy from analyzing day-5 blastocyst stage embryos was considered. The model used was the one in [127]. It was shown that the model scores had a positive linear correlation with the pregnancy outcomes, among other observations about correlative relations. The dataset consisted of 9359 images from 4709 women, obtained from 18 IVF clinics in six countries (2011–2021).
In [183], the iDAScore v1.0 model was developed, which is a fully automated system for embryo scoring using time-lapse images. The model performed binary classification of the positive and negative fetal heartbeat (and discarded embryos). The model uses an inflated 3D (I3D) CNN architecture [66], in series with a bidirectional LSTM, and a fully connected layer. The model was extensively tested to evaluate its robustness and generalizability, with positive results. The model also performed better than the KIDScore D5 v3 model [184]. The dataset was of significant size, consisting of 115,832 embryos from 18 IVF centers worldwide (2011–2019).
In [185], the relation between the iDAScore system [183] and biological events during the pre-implantation period was explored. The study concluded that iDAScore was correlated with morphokinetics and morphological alterations of preimplantation embryos. The score is correlated with the embryonic development speed through fertilization up to blastulation and also related to the incidence of irregular first division, fragmentation at cleavage stage, blastomere exclusion and extrusion during the peri-compaction period, and blastocyst morphology. The dataset consisted of 925 cycles from individual patients from the Kato Ladies Clinic in Japan (2019–2020).
The work [186] studied how an embryo evaluation algorithm may perform across different IVF clinics, where age differences occur. The iDAScore v1.0 [183] was considered. A method for age-based standardization of the AUCs was proposed. After this standardization, the variance among clinics was reduced by 16%. The data included 4805 transferred embryos from 4086 treatments and were collected from four IVF clinics (2013–2022).
In [187], the authors studied whether the use of an annotation-free embryo scoring system using deep learning on time-lapse sequence images has correlation with live birth and neonatal outcomes. The model used was iDAScore, which uses a 3D CNN [183]. The authors concluded that iDAScore correlates with decreased miscarriage and increased live birth and has no correlation to neonatal outcomes. As the iDAScore does not require manual annotations, it can serve as an objective evaluation tool. The dataset consisted of 3010 patients undergoing autologous single vitrified/warmed blastocyst transfer cycles (2019–2020).
In [188], the iDAScore v2.0 model was studied for the evaluation of embryos after 2, 3, and 5 or more days of incubation. It was also observed that the prediction was correlated with morphokinetic parameters. The model consists of two separate components. The first part consists of two 3D CNN networks that take data from day 2/3 (20 to 84 HPI) and predict implantation potential and direct cleavages (from one to three cells and from two to five cells), followed by separate calibration models for each day. For embryos incubated for more than 84 hpi, the time lapse images from 20 to 148 hpi are fed into a 3D CNN model, followed by calibration. The model here consists of a CNN similar to [183]. The scores are estimates of pregnancy probabilities representative of the average patient population and range from 1 to 9.9. The models of iDAScore v2.0 and KIDScore D3 [189] performed on par, while iDAScore v2.0 performed better than KIDScore D5 v3 [184] and iDAScore v1. Of course, KIDScore D3 and KIDScore D5 v3 are manual. The dataset consisted of 181,428 embryos gathered from 22 IVF clinics worldwide.
In [190], pregnancy prediction was studied for frozen embryo transfers through the analysis of endometrial histology on hematoxylin and eosin. Endometrial biopsies were extracted from healthy individuals in natural menstrual cycles and from infertile ones in mock artificial cycles. The ResNet-18 model was considered [21], with a three-fold cross validation strategy. At first, a DL model was trained to differentiate between these two groups, and achieved 100% accuracy. The second group then underwent Frozen–Thawed Embryo Transfers (FETs), which resulted in a positive (pregnant) or negative (non-pregnant) outcome. The DL model achieved 77.8% for accuracy in predicting this outcome. The accuracy was also 75% for patients who had euploid embryo transfers. The dataset consisted of 61 endometrium biopsy (EB) samples.
In [191], a histogram-based gradient boosting decision tree model [60] was developed for predicting clinical pregnancy in IVF and ICSI cycles. The AUC achieved was 0.704. The variables with the most predictive importance were the women’s age, the number of two-pronuclear embryos (2PNs), the AMH level, the number of oocytes retrieved, and the endometrial thickness. Some additional interrelations between the variables were revealed. For example, women having a lower AMH may be better off following the short strategy. The dataset consisted of 37,062 cycles from the Women’s Hospital School of Medicine at Zhejiang University (2010–2020).
In [192], a model was developed to predict the pregnancy outcome and multiple pregnancy risk after embryo transfer. Six models were considered, logistic regression, random forest, SVM, light gradient boosting machine [60], XGBoost [154], and multilayer perceptron (MLP). The XGBoost model was the best, with 0.716 pregnancy prediction accuracy and 0.711 for multiple pregnancy prediction accuracy. The initial dataset consisted of 1507 embryo cycles from the Center for Reproductive Medicine, Chi Mei Medical Center, Tainan, Taiwan (2010–2019).
In [193], live birth prediction was studied. Several ML models are considered, including logistic regression, K-nearest neighbor, multilayer perceptron, decision tree, and a 1D DL model, consisting of nine dense layers and dropout layers. Training with and without feature selection is considered. The random forest algorithm yielded the best result, with a random forest model achieving the highest F1-score of 76.49% and a ROC Curve (ROC AUC) of 84.60%, following the training without feature selection. The authors suggest the usability of this tool, although, as with most DL tools, it should not be solely used for decision making, as it is limited by the training set. The authors also note the importance of including important metadata for future use, like alcohol consumption, smoking, caffeine consumption, hypertension, and other lifestyle factors. The data considered is obtained from the Human Fertilisation & Embryology Authority, for the years 2010–2016 [194]. It includes 495,630 patient records with 94 features on treatment cycles.
In [195], a ResNet CNN architecture was used to predict the outcome of live birth from time-lapse data. The architecture consisted of seven convolution modules and two fully connected layers, based on ResNet18 [21]. The model achieved an AUC of 0.968. The time-lapse embryo data used consisted of 15,434 embryos with positive and negative labels, depending on the embryo outcome, from the Reproductive Medicine Center of Tongji Hospital, Huazhong University of Science and Technology, Wuhan, China.
In [196], early pregnancy loss prediction was studied. Fetal heart rate was observed to be a strong feature for this prediction. The models considered were logistic regression, support vector machine, decision tree, back propagation neural network, XGBoost [154], and random forest. The random forest outperformed the rest, with an AUC of 0.97. Especially for longer embryo transfer day data, it achieved a 99% accuracy for prediction at 10 weeks after embryo transfer. The models were tested with and without the fetal heart rate measurement and observed an improvement in performance in the second case. The dataset, after some cleaning, included 31,030 cases at 6–10 weeks gestation, which included 19,929 ongoing pregnancy samples and 11,101 early pregnancy-loss samples from the Reproductive and Genetic Hospital of CITIC-Xiangya.
In [197], the model ranked static images of blastocysts, for predicting clinical pregnancy. The model architecture was an ensemble of three Resnet-18 models, which were trained at different image resolutions. This was motivated by the way embryologists evaluate embryos by looking at them in different magnifications. The AUC achieved was from 0.6 to 0.7 and outperformed manual grading. The authors also identified two sources of bias in the data, the first being the use of different microscopes and the second being the presence of an embryo-holding micropipette in some images. These sources of bias were also identified in an earlier work [198]. These were mitigated by applying appropriate balancing between the images in the dataset in the positive and negative classes. The dataset consisted of 5923 transferred blastocysts and 2614 non-transferred aneuploid blastocysts from 11 IVF clinics in the USA (2015 to 2020).
In [199], the problem of implantation outcome prediction was addressed using time-lapse images from day 3 and day 5. The architecture consists of two CNN models, one evaluates the day-3 embryos and the other the day-5 ones. Each model is trained separately. A Data Length Schedular is used to improve the training performance. This algorithm groups the training sequences in groups and sequentially feeds them to the training process after a set number of epochs. The prediction is made by averaging the output of each model. The accuracy reached was 76.9%, which outperformed the individual submodels, as well as the same model trained without the Data Length Schedular. It also outperformed two architectures proposed in [77]. The dataset included 130 time-lapse sequences of individual embryos, captured using EmbryoScope (Vitrolife). In total, there were 12,480 and 10,097 images used for day-3 and day-5 embryos.
In [200], pregnancy prediction after embryo transfer was studied. An AIR E prototype system is used to perform segmentation, feature extraction, feature selection, and classification. The models considered were probabilistic Bayesian, support vector machines, DNN, decision tree, and random forest. In the first dataset used, the SVM achieved an AUC of 0.77. In the second dataset, the random forest achieved an AUC of 0.75. The dataset consisted of micrographs of 221 blastocysts transferred as single embryos at two fertility centers in Mexico (2015–2019). The first dataset contained the ones captured using an Olympus IX71 and the second dataset the ones captured using an Olympus IX73.
In [201], the identification of the most important predictors for day-5 blastocyst utilization rate was considered. The key indicators considered were the number of metaphase II (MII) oocytes injected (intracytoplasmic sperm injection), the use of autologous/donated gametes, the maternal age at oocyte retrieval, the sperm concentration, the progressive sperm motility rate, and the fertilization rate. A neural network concluded that all six variables are of importance in predicting the day-5 blastocyst utilization rate, with the most prominent being the number of MII oocytes injected. The analysis showed a negative correlation between the number of MII oocytes injected and the utilization rate. The injection of up to six MII oocytes had an over 60% utilization rate. The dataset was collected from 885 patients from two IVF centers, Dogus IVF Centre, Cyprus, and Al Hadi Laboratory and Medical Centre, Lebanon (2021–2022).
The work [202] studied the relation between the aneuploidy risk score provided by a morphokinetic ploidy prediction model, termed Predicting Euploidy for Embryos in Reproductive Medicine (PREFER), from [151], and the outcomes of miscarriage and live birth. The PREFER model uses morphokinetic and clinical data. Another model was considered, PREFER-MK, using only morphokinetic data. The models rank embryos on three categories, high, medium, and low risk. The PREFER scores were indeed associated with live births and miscarriages. The model though can be affected heavily by age and clinical predictors to the point that it cannot properly rank embryos. The PREFER-MK can thus be a better choice. This model was associated with live birth but not with miscarriage. The dataset consisted of 3587 fresh single-embryo transfers collected from nine IVF clinics in the UK (2016–2019).
In [203], pregnancy prediction was studied using time-lapse data in combination with multi-centric clinical data. The model consisted of a 3D ConvNet using a ResNet backbone [21], pretrained on Kinetics [67], that analyzed the videos of embryonic development up to 2 or 3 days of development or up to 5 or 6 days of development. The output of this model was then combined with the clinical data and fed into a gradient boosting algorithm XGBoost that made the final prediction. This hybrid model outperformed the one using only the time-lapse data, with their AUC scores being 0.727 and 0.684. The model also outperformed a group of embryologists. In addition to the video score, the most important predictive features were oocyte age, total gonadotrophin dose intake, number of embryos generated, number of oocytes retrieved, and endometrium thickness. The dataset consisted of time-lapse videos and 31 clinical variables for 9986 embryos, and 447 more for testing, collected from 14 clinics in France and Spain (2016–2022).
In [204], a model was developed for blastocyst viability assessment of live birth and clinical pregnancy, termed Fertility Image Testing Through Embryo (FiTTE). The model was then appended into an ensemble model, which uses a ResNet-18 [18] architecture with a binary cross-entropy layer for embryo image processing (FiTTE) and a random forest classifier that combined the output of this architecture with clinical data to make the prediction. For predicting clinical pregnancy, the FiTTE model achieved an accuracy of 62.7%, while the ensemble model achieved 65.2%. For the ensemble model, along with the images, the most important clinical data for prediction were the age, pregnancy history, serum AMH, serum estradiol, and progesterone at the time of embryo transfer. Interestingly, a gradient-weighted class activation method was used to color-grade important parts in the images [110]. This generates color-coded blastocyst images, which helps in highlighting the important regions and aids in the interpretability of the model. The dataset consisted of 19,342 blastocyst images, with corresponding inspection history, from 9961 patients from the Hanabusa Women’s Clinic (2011–2019).
In [205], a live-birth prediction model combining blastocyst images and the couple’s clinical data was developed. The model consisted of a CNN, based on EfficientNetV2-S [179], to process the blastocyst images, and a multilayer perceptron to process the clinical data. The model achieved an AUC score of 0.77 for live-birth prediction and outperformed the simpler model that only used the blastocyst images. The most important clinical features for prediction were the maternal age, the day of blastocyst transfer, antral follicle count, retrieved oocyte number, and endometrium thickness measured before transfer. CNN heatmaps were also generated using XGradCAM [206], showing that the model focused on the ICM and TE regions for prediction. The dataset used consisted of 17,580 blastocysts with two blastocyst images and 16 clinical variables from the Reproductive and Genetic Hospital of CITIC-Xiangya (2016–2020).
In [207], PGT-A was evaluated for patients who underwent single thawed euploid embryo transfer. Subjective Next-Generation Sequencing (NGS) was compared to two AI algorithms, the PGTai, Technology Platform, CooperSurgical, Inc. (AI 1), and PGTai 2.0 (AI 2). The algorithms overall outperformed subjective NGS, showing increased euploidy rate and decreased simple mosaicism rate. The ongoing pregnancy and/or live-birth outcome was also higher in the AI 2 algorithm compared to the subjective NGS. The dataset consisted of 4,765 retrieval cycles, including 24,908 embryos that used PGT-A (2015–2020), and 1174 eulpoid embryos were transfered (2015–2020).
In [208], the problem of pregnancy prediction was considered for recurrent implantation failure patients. The dataset used, which had 44 patient features and the “Early Outcome” output, was divided into two groups, one group that had a double-embryo transfer (A) and one that had a single-embryo transfer (B). Then, four machine learning algorithms were considered for outcome prediction, random forest, AdaBoost, gradient boosted decision tree, and multilayer perceptron. For the first group, AdaBoost was the best model, and for the second group, gradient boosted decision tree was the best. Both of these models had good predictions for the transfer outcome. The use of these two models could be considered as a support tool to provide suggestion on whether single- or double-embryo transfer is the best option for future patients. The dataset included 45,921 patient records from the Recurrent Implantation Failure data from the Human Fertilisation & Embryology Authority (HFEA) database (2005–2016).
In [209], a model was developed to predict a single-embryo transfer pregnancy, a double-embryo transfer pregnancy, and a twin risk of double-embryo transfer. The developed model had a hierarchical structure, based on XGBoost [154]. In the first level, the characteristics of the patient and the embryo were considered to predict the implantation probability for single-embryo transfer. For double-embryo transfer, the implantation probability of two embryos (P1, P2) was calculated from the first level, which were used on the second level, along with patient features, to predict the probability of double-embryo transfer pregnancy and twin risk. The model achieved an AUC of 0.7945 for single-embryo transfer pregnancy, 0.8385 for double-embryo transfer pregnancy, and 0.7229 for double-embryo transfer twin risk. For these three tasks, important variables were age, attempts at IVF, estradiol level on hCG day, and endometrial thickness for the first task; the age, attempts at IVF, endometrial thickness, and the added probabilities P1 + P2 for the second one; and age, attempts at IVF, 2PN/MII, and P1*P2 for the third one. The dataset consisted of 9211 patients and 10,076 embryos (2016–2018), with 38 features, from the Tongji Hospital, Wuhan, China.
In [210], an algorithm was developed to predict clinical pregnancy and live-birth rates using time-lapse images in EmbryoScope. The models considered included logistic regression, XGBoost, decision tree, and random forest. The supervised random forest algorithm was the best for predicting clinical pregnancy outcome, which achieved an AUC of 0.91 in the training set and 0.69 in the test set, with similar results for live-birth outcomes. An additional study was conducted in which embryo growth morphokinetics was separated into five groups using unsupervised clustering. The clusters with the fastest morphokinetics were then observed to have 54% pregnancy rates, while the cluster with the slowest morphokinetics had a 71% pregnancy rate, although the differences did not have statistical significance. The dataset consisted of 367 embryos from the Texas Children’s Hospital Family Fertility Center (2014–2018).
In [211], a model was developed for live-birth prediction. The architecture considered is the Attention Branch Network (ABN) [212], using ResNet56 [21] pretrained on ImageNet [62]. The effect of the embryo confidence score cut-off level was also studied. The model can also visualize areas of importance by color-grading the embryo images. However, no common visual features were found that could predict a live or non-live birth. The dataset consisted of time-lapse images of 470 transferred embryos, with 141,444 time-lapse images overall, from the Nagoya City University Hospital and the Sawada Women’s Clinic (2014–2018).
In [213], a comparison was made between three different methods for predicting fetal heartbeat. iDAScore v1.0 [183], KIDScore D5 v3.0, and Gardner criteria were compared between different age groups. In general, the iDAScore achieved the highest AUC or equal to the highest AUC among all age groups. The dataset consisted of 3018 patients who had undergone their first single vitrified blastocyst transfer (2019–2020).
In [214], the impact of endometrial thickness on the continuing pregnancy rate was studied. For this, a random forest model algorithm and a logistic regression algorithm were considered for pregnancy prediction, of which the random forest was the best. Based on this model, cut-off values for the endometrial thickness were derived. The dataset consisted of 729 couples with unexplained infertility, some undergoing IUI and some undergoing IVF/ICSI treatments, gathered from two infertility centers.
4.10. Intrauterine Insemination (IUI)
The work [215] developed a model for predicting the time of ovulation, as well as the optimal fertilization window for intrauterine insemination or synchronized intercourse, using blood tests. Two algorithms were developed, one for predicting ovulation and one for treatment management. The first was an NGBoost model [154] that estimated the probability of ovulation occurring on each cycle day. The second algorithm decided whether the best day for insemination can be determined or an additional blood test must be performed first to determine which day. The model correctly determined the timing for IUI in 92.9% and intercourse in 92.4% of the cases. The dataset included 2467 cycles of frozen-embryo transfer during the natural cycle (2018–2022), from the Herzliya Medical Center, Israel. The importance of this study was also discussed in [216], noting that it could limit patient’s visits to the clinic, even potentially to the point that patients do not have to visit the clinic prior to insemination.
4.11. Sperm Analysis
Of course, AI also contributes significantly to male reproductive health. Sperm analysis is an important part of IVF in which AI contributes.
In [217], a model was developed to predict the sperm DNA fragmentation index, using the sperm chromatin dispersion (SCD) test. Azure’s custom vision was used [218] to build an ensemble model. Two configurations were considered. For one part of the dataset (8887 images), a binary classification was studied (halo/no halo). For the second part of the dataset (15,528 images), multiple classes were considered (big/medium/small halo/degraded (DEG)/dust). The accuracy on the binary and multi-class cases were 80.15% and 75.25%. The complete dataset consisted of 24,415 images from 30 patients after the SCD test, obtained with a phase-contrast microscope.
There are numerous other aspects of male fertility in which AI is used, for example, to predict the upgrade of sperm parameters after varicocele repair surgery [219] and to predict the fertility rate [220]. As this review is more focused on embryo-related studies, the interested reader may see the review [221].
4.12. Quality Assurance
Quality assurance is another important aspect of clinic functionality, as it is important to ensure regulatory compliance and consistency of performance for all personnel [222].
In [223], a comparison was made between AI predictions and real outcomes for MD and embryologists performing embryo transfer, embryo vitrification, embryo warming, and trophectoderm biopsy. This allowed monitoring staff performance and identifying the appearance of significant gaps between the predicted and actual performance rates. In fact, in some cases, there was a significant deviation between the predicted AI and actual performance in embryo transfer performed by an MD and an embryologist.
In [224], an algorithm was developed to improve the IVF workflow, reduce the number of visits, and level-load the embryology work. The model optimized several tasks. One was the prediction of the best day to monitor. Then, the model predicted a single day for triggering and predicted the number of oocytes for this day, one day prior, and one day after. Using again the IVF data and the observations made on the best day, the algorithm also predicted the total number of oocytes and mature oocytes. The model was a stacking ensemble that combined multiple models such as linear regression, random forest, extra-tree regression, K-nearest neighbor, and XGBoost. The precision in predicting the total number of oocytes was 0.76 when using only baseline information and 0.77 when also considering the data from the observation day. The dataset consisted of 1591 cycles over 4731 visits.
5. Open Challenges
Below, some of the limitations of AI in IVF and the challenges that lie ahead in the field are covered.
5.1. Selecting the Best Architecture
From the numerous works discussed above, in the various aspects of IVF, it is evident that many different ML and DL algorithms and architectures have been considered. This naturally opens up the discussion of which model is the most appropriate for each task. It is understandable though, as with other problems in ML, that there is not a single architecture that could collectively outperform the rest. The model’s choice should be made based on the task at hand and the available data. Nonetheless, it seems that a promising approach for moving forward in the future is the development of hybrid models that combine characteristics from different architectures to build combinations of them. This of course makes the design more intricate, as numerous different layers are connected with different types of interconnections. Such models are harder to develop and train, but the results can be better than individual architectures. Of course, for each model at hand, it is important to also take into account its computational requirements and the cost of integrating, maintaining, updating, and overall managing such a platform in a clinic. So implementation issues in real clinical environments should not be overlooked and should definitely be taken into account when considering suitable models.
5.2. Data Availability
Large databases are important for studies and statistical analyses in all medical fields and IVF [225]. Naturally, large image datasets are essential to effectively train very deep DL models for CV and clinical medical research [226]. For medical imaging, such as human-embryo analysis, the collection, processing, and publication of image datasets have several challenges and should follow certain standardized procedures, as discussed in [227]. This issue was also mentioned in [8], where five limitations of AI were identified, the lack of available data, the existence of bias in the data, the limited interpretability of the models, the inability to handle uncertainty, and ethical concerns. Similar issues, as well as legal liability, the definition of the "normal embryo", and the selection bias under the prism of sociocultural norms, were identified in [228,229,230,231].
The key elements of a properly collected and processed DL dataset for medical use are ample data volume, complete annotation, truth, and reusability [227]. In other words, it should be related to the Findability, Accessibility, Interoperability, and Reusability (FAIR) principles for managing data [232]. Medical image datasets must first describe the high-level attributes of the data. Metadata must also be provided.
A significant contribution to data availability for DL is the recent work [233], which constructed an annotated dataset of blastocyst images. It includes 2344 blastocysts from 837 patients. The authors also included metrics from expert embryologists to serve as a benchmark for DL models for grading blastocyst expansion (EXP), inner cell mass (ICM), and trophectoderm (TE). They also provided the performance of some baseline DL models, XCeption [20], Deit [234] transformer, and Swin [133] transformer.
Another important dataset is provided in [166]. It includes 704 time-lapse videos of developing embryos, in all seven focal planes. The dataset is fully annotated, with 16 different development phases. There are 2.4 million images overall. In addition, the work [76] provided a dataset of 211 Hoffman Modulation Contrast (HMC) blastocyst images.
5.3. Data Limitations
In addition to the above issues in data collection, the use of existing datasets for DL IV suffers from problems like bias in the data themselves, publishing incentives by the authors, incorrectly chosen baselines, and more, as discussed in [235]. For example, regarding data bias, the medical data from a hospital are a reflection of only a limited population size, and as that population may be nonuniform, this nonuniformity may create a bias in the data themselves. This means that the probability distribution of the training data does not reflect the whole set of actual cases. These issues that can appear in the gathered data can naturally lead to ethical concerns around the use of AI in IVF. This population imbalance that may appear in the data could lead to discriminatory decisions against underrepresented populations. Thus, it may be unclear as to what features may be reinforced over the rest [228]. All of these issues put significant limitations on a model’s generalizability, as the existence of dataset bias prohibits the model’s transferability from clinic to clinic.
Another ethical concern has to do with data confidentiality. Although all data that are used in a study are first de-identified, an open question is whether it would be possible to partially re-identify them, using techniques like data triangulation [236]. This confidentiality issue also has to do with data ownership. What happens in a scenario where a clinic that holds a specific dataset closes down, and where will these data end up? Or what if a private clinic or faculty is bought up by another organization, and then ownership of the dataset is also transferred?
Another concern is the use of benchmarks and the choice of evaluation metrics. There is an open discussion in the research community about the effectiveness of benchmarks and the validity of some standard metrics [235]. So to reliably perform DL-IVF, the metrics used for training and evaluation must be carefully examined. There are also other issues, especially when combining data from multiple clinics, like the use of different microscopes and the presence of embryo-holding micropipettes [197]. Data normalization is essential here to avoid bias due to image characteristics, like brightness, which are irrelevant to embryo quality. Another essential action is standardization, both for images and the clinical data gathered. For images, this would include actions like cropping, resizing, removing blank frames, and other such actions. For clinical data, standardization involves the way the data are reported, for example, the number and range of the bins used for categorical data like age.
As is evident from the above reviewed works, most research groups use their own datasets, which have been gathered from clinics they belong to or collaborate with. Most of these datasets are private and are not shared due to legal or other reasons. This creates a barrier in making advancements in the field, especially from smaller research groups and clinics that do not have the resources to collect their own large datasets and have to work with limited data [36]. So, due to the sensitive nature of the content, legal issues arise with respect to their use by other research groups, which can limit their accessibility; see the discussions in [236,237]. Regulatory bodies should work towards building a legal framework for the protection of patient privacy, as well as the fair use of medical data; see the discussions in [236,237,238], where the ethical and legal issues in the use of AI in healthcare in general are reviewed. Examples include consent and anonymity, transparency, liability, cybersecurity, and data ownership.
Naturally, the availability of large datasets for specific conditions facilitates further research on the topic [235]. However, this can lead to underdeveloped research topics due to the lack of available datasets for training. Similar problems may arise in DL-empowered IVF. So for the future progression of the field, it is important that regulatory authorities such as health ministries and hospitals assign a portion of their budgets to experienced personnel to properly gather and annotate data, to enrich existing collections; see, for example, the Human Fertilization & Embryology Authority archive [194], the works [76,166], and the PhysioNet archive [239].
Overall, data collection is in itself a difficult and time-consuming task, which is accompanied by several potential issues, like the demanding task of adding annotations, the existence of bias, sharing limitations due to legal and other issues, and more. So a collective effort must be made towards addressing these data issues to further improve their effective usage and also facilitate reproducibility of each study. For example, as mentioned in [36], the use of collaborative federated learning may be key for sharing data among clinics without compromising security and delicate patient information. It would also be important for future developments to establish a common benchmark dataset or even a synthetic data generation protocol.
5.4. Transfer Learning
Transfer learning is a widely used technique in DL. The core idea behind transfer learning is to use an architecture that has proven efficient for a given problem to solve another similar task. This is very common in CV problems. However, adjustments must always be made when an architecture is moved to a new problem to accommodate the new characteristics of the dataset. For example, some features extracted from the original dataset may be less relevant for the new one, so considering them may hinder performance. So, further tuning must be performed for the model to be properly trained. Moreover, the relation between the datasets of the solved problem and the new one should be carefully examined for similarities and differences to properly choose the correct features to be preserved or discarded.
To address these challenges, a more focused approach should be taken in transfer learning. As mentioned in the previous sections, many of the models considered were pre-trained in public datasets like ImageNet [62]. It would be interesting to study though whether models with weights initialized from training on relative medical data would converge more quickly and give a better performance. Examples include the datasets [166,233].
5.5. Model Interpretability
An important aspect which is essential in the integration of AI systems in decision making is the model’s interpretability, which has been identified in [8,36,228,230]. As DL architectures have many layers and often combine multiple sub-architectures and take as input embryo images and clinical data, they are often seen as a black-box structure. So, it is hard to unmask the causality between the input information and the output decision made by the algorithm. This foggy relation may hide biases, and until fully explained, it is hard to have full confidence in AI decision-making systems.
Thus, it is essential that future studies also devote time in interpreting the way that decisions are made by their algorithms, with many works considering this already; see [161,168,170]. As a starting point for addressing interpretability, many works list the most important features in their algorithm’s decision process; see, for example, [130,153,169,171,173,177,191,201,203,204,205,209,215,224]. Another notable example is the approach in [108,204], where the important parts of the embryo images were color-graded. Of course, as the DL architectures become larger, addressing interpretability will be a challenging task. This is of course a challenge that encompasses all applications of DL [240], not just IVF. For all future applications of artificial intelligence, being able to provide explainable decisions would be essential to establish reliable tools and gain the user’s trust. Addressing it will help DL systems gain the acceptance of embryologists and patients alike.
5.6. AI and Responsibility
A question that arises when using AI systems to support clinical descisions is who should take responsibility for the descision. As AI systems do not have consciousness, they cannot be held directly responsible for their suggestions, whether positive or erroneous. This is one of many reasons why the last link in the decision-making process should always be the clinical practitioner, and the AI system should have a secondary supporting role.
On the other hand, an open question that could be asked here is whether a chain of accountability may apply; that is, in case of erroneous suggestions by the AI system, does the accountability translate to its designer? Accountability in AI systems is of course a problem that is not limited to IVF but spans other fields of AI-assisted medical practice [241,242,243,244] and has implications for relevant topics, such as insurance claims. As the use of AI is becoming widespread, legal authorities must push for establishing the relevant regulations and legislation for its ethical use and address issues related to accountability.
5.7. The Role of Embryologists
The role of embryologists in the lab can shift more towards decision making as the degree of automation in the lab increases [245]. This will also increase the need for embryologists to develop critical thinking and scientific and technical skills to interact with engineers and bioinformaticians to develop new lines of diagnosis and treatment, as well as to improve existing AI architectures [245]. Future embryologists would also need to develop their statistics and database management skills to be able to evaluate and process large datasets, as well as explain and evaluate the decisions of AI automated systems [246].
Of course, since AI systems are far from being considered 100% reliable, they can only be used as a support tool for trained medical practitioners. The AI tools can certainly help in reducing inter- and intra-variability, especially in less experienced personnel, but the final say always falls on the practitioners. These are the people who will take into account all the existing parameters, make the final suggestion, and take responsibility for it. This is also paired with the fact that patients need to build trust in their doctors, accompanied by an emotional support during the IVF process, which cannot be provided by an AI tool. So, the role of the human practitioner may certainly change, but it will remain indispensable in the future. As the authors in [247] argue, human practitioners are not yet dispensable!
6. Conclusions
This work provided a review of recent advances in the field of AI-assisted IVF. Several topics were covered, with the aim of providing a roadmap for researchers in the field. In addition, several challenges for the future have been identified, such as the collection of large and unbiased datasets, the secure sharing of data, the efficient exploitation of transfer learning, the interpretability of the model, the accountability of AI, and the future role of embryologists. Certainly, with collective efforts by the research community and regulatory authorities, any of the aforementioned or unforeseen challenges can be successfully surpassed.
The use of AI spans numerous sub-fields of IVF, like strategy selection, embryo grading, ovarian stimulation, pregnancy prediction, quality assurance, and more. The field is rapidly expanding, so more and more problems in the fields of IVF and obsteretics, gynecology, and male fertility in general will be supported by the use of AI. It is certain that the use of AI will revolutionize IVF and the relevant fields. The development of artificial intelligence and its integration into IVF is based on collaboration between people from diverse backgrounds, such as engineers, physicists, biologists, and physicians. The joint expertise of these groups will bring about a new era of IVF with better prospects for patients than ever.
Author Contributions
Conceptualization, L.M., L.A.I., G.V. and S.K.G.; formal analysis, L.M. and L.A.I.; writing—original draft preparation, L.M., L.A.I., G.V. and S.K.G.; writing—review and editing, L.M. and S.K.G.; supervision, S.P.S., A.D.B., A.P., K.-I.D.K., M.A.M., P.S., I.S., V.A. and S.K.G.; All authors have read and agreed to the published version of the manuscript.
Funding
This research was carried out as part of the project “Classification and characterization of fetal images for assisted reproduction using artificial intelligence and computer vision” (Project code: KP6-0079459) under the framework of the Action “Investment Plans of Innovation” of the Operational Program “Central Macedonia 2014 2020”, which is co-funded by the European Regional Development Fund and Greece. This project has received funding from the European Union’s Horizon Europe research and innovation programme under grant agreement No. 101095435 (REALM).
Data Availability Statement
This work generated no data.
Acknowledgments
The authors are thankful to the anonymous reviewers for their helpful comments.
Conflicts of Interest
The authors declare no conflicts of interest.
Abbreviations
The following abbreviations are used in this manuscript:
| nD (1D,…) | n-dimensional (one-dimensional etc.) |
| AFC | Antral Follicle Count |
| AI | Artificial Intelligence |
| ART | Assisted Reproductive Technology |
| BC | Blastocoel |
| BG | Background |
| CNN | Convolutional Neural Network |
| CV | Computer Vision |
| DL | Deep Learning |
| ET | Embryo Transfer |
| FCDNN | Fully Connected Deep Neural Network |
| FSH | Follicle-Stimulating Hormone |
| GAN | Generative Adversarial Network |
| GLOM | Grey Level Co-Occurrence Matrix |
| hpi | Hours Post Insemination |
| ICM | Inner Cell Mass |
| ICSI | Intra-Cytoplasmic Sperm Injection |
| IVF | In Vitro Fertilization |
| LSTM | Long Short-Term Memory |
| MII | Metaphase II |
| ML | Machine Learning |
| NLP | Natural Language Processing |
| NN | Neural Network |
| PGT | Preimplantation Genetic Testing |
| ResNet | Residual Network |
| RGB | Red–Green–Blue |
| SVM | Support Vector Machine |
| TE | Trophectoderm |
| TLI | Time-Lapse Imaging |
| ViT | Vision Transformer |
| VGG | Visual Geometry Group |
| ZP | Zona Pellucida |
References
- CDC. What is Assisted Reproductive Technology? Available online: https://www.cdc.gov/art/about/ (accessed on 31 January 2024).
- Bormann, C.L. ART: Laboratory Aspects. In Clinical Reproductive Medicine and Surgery: A Practical Guide; Springer: Berlin/Heidelberg, Germany, 2022; pp. 393–408. [Google Scholar]
- CDC. ART Success Rates. Available online: https://www.cdc.gov/art/success-rates/?CDC_AAref_Val=https://www.cdc.gov/art/artdata/index.html (accessed on 31 January 2024).
- CDCC. 2020 National ART Summary. Available online: https://www.cdc.gov/art/php/national-summary/ (accessed on 31 January 2024).
- Miyagi, Y.; Miyake, T. Potential of artificial intelligence for estimating Japanese fetal weights. Acta Medica Okayama 2020, 74, 483–493. [Google Scholar] [PubMed]
- Looney, P.; Stevenson, G.N.; Nicolaides, K.H.; Plasencia, W.; Molloholli, M.; Natsis, S.; Collins, S.L. Fully automated, real-time 3D ultrasound segmentation to estimate first trimester placental volume using deep learning. JCI Insight 2018, 3, e120178. [Google Scholar] [CrossRef] [PubMed]
- Petrozziello, A.; Jordanov, I.; Papageorghiou, T.A.; Redman, W.C.; Georgieva, A. Deep learning for continuous electronic fetal monitoring in labor. In Proceedings of the 2018 40th Annual International Conference of the IEEE Engineering in Medicine and Biology Society (EMBC), Honolulu, HI, USA, 18–21 July 2018; IEEE: New York, NY, USA, 2018; pp. 5866–5869. [Google Scholar]
- Yazdani, A.; Costa, S.; Kroon, B. Artificial intelligence: Friend or foe? Aust. New Zealand J. Obstet. Gynaecol. 2023, 63, 127–130. [Google Scholar] [CrossRef]
- Curchoe, C.L. For whom the artificial intelligence bell tolls: Preimplantation genetic testing for aneuploidy, does it toll for thee? Fertil. Steril. 2022, 117, 536–538. [Google Scholar] [CrossRef]
- Gardner, D.K.; Sakkas, D. Making and selecting the best embryo in the laboratory. Fertil. Steril. 2023, 120, 457–466. [Google Scholar] [CrossRef]
- Charnpinyo, N.; Suthicharoenpanich, K.; Onthuam, K.; Engphaiboon, S.; Chaichaowarat, R.; Suebthawinkul, C.; Siricharoen, P. Embryo Selection for IVF using Machine Learning Techniques Based on Light Microscopic Images of Embryo and Additional Factors. In Proceedings of the 2023 45th Annual International Conference of the IEEE Engineering in Medicine & Biology Society (EMBC), Sydney, Australia, 24–27 July 2023; IEEE: New York, NY, USA, 2023; pp. 1–4. [Google Scholar]
- Basile, N.; Carbajosa, A.R.; Meseguer, M. Evaluation of embryo quality: Time-lapse imaging to assess embryo morphokinesis. In Textbook of Assisted Reproductive Techniques; CRC Press: Boca Raton, FL, USA, 2017; pp. 285–298. [Google Scholar]
- Fadon, P.; Gallegos, E.; Jalota, S.; Muriel, L.; Diaz-Garcia, C. Time-lapse systems: A comprehensive analysis on effectiveness. In Proceedings of the Seminars in Reproductive Medicine; Thieme Medical Publishers, Inc.: New York, NY, USA, 2021; Volume 39, pp. e12–e18. [Google Scholar]
- Yu, C.; Liu, J.; Nemati, S.; Yin, G. Reinforcement Learning in Healthcare: A Survey. ACM Comput. Surv. 2021, 55, 1–36. [Google Scholar] [CrossRef]
- Prayitno; Shyu, C.R.; Putra, K.T.; Chen, H.C.; Tsai, Y.Y.; Hossain, K.S.M.T.; Jiang, W.; Shae, Z.Y. A Systematic Review of Federated Learning in the Healthcare Area: From the Perspective of Data Properties and Applications. Appl. Sci. 2021, 11, 11191. [Google Scholar] [CrossRef]
- Krishnan, R.; Rajpurkar, P.; Topol, E. Self-supervised learning in medicine and healthcare. Nat. Biomed. Eng. 2022, 6, 1346–1352. [Google Scholar] [CrossRef]
- Goodfellow, I.; Bengio, Y.; Courville, A. Deep Learning; Adaptive Computation and Machine Learning; MIT Press: Cambridge, MA, USA, 2016; p. 800. [Google Scholar]
- Simonyan, K.; Zisserman, A. Very Deep Convolutional Networks for Large-Scale Image Recognition. In Proceedings of the 3rd International Conference on Learning Representations, ICLR 2015, San Diego, CA, USA, 7–9 May 2015. [Google Scholar]
- Szegedy, C.; Ioffe, S.; Vanhoucke, V.; Alemi, A.A. Inception-v4, Inception-ResNet and the Impact of Residual Connections on Learning. In Proceedings of the Thirty-First AAAI Conference on Artificial Intelligence, AAAI’17, San Francisco, CA, USA, 4–9 February 2017; AAAI Press: San Francisco, CA, USA, 2017; pp. 4278–4284. [Google Scholar]
- Chollet, F. Xception: Deep learning with depthwise separable convolutions. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Honolulu, HI, USA, 21–26 July 2017; pp. 1251–1258. [Google Scholar]
- He, K.; Zhang, X.; Ren, S.; Sun, J. Deep Residual Learning for Image Recognition. In Proceedings of the 2016 IEEE Conference on Computer Vision and Pattern Recognition (CVPR), Las Vegas, NV, USA, 27–30 June 2016; pp. 770–778. [Google Scholar] [CrossRef]
- Huang, G.; Liu, Z.; Van Der Maaten, L.; Weinberger, K.Q. Densely Connected Convolutional Networks. In Proceedings of the 2017 IEEE Conference on Computer Vision and Pattern Recognition (CVPR), Honolulu, HI, USA, 21–26 July 2017; pp. 2261–2269. [Google Scholar] [CrossRef]
- Vaswani, A.; Shazeer, N.; Parmar, N.; Uszkoreit, J.; Jones, L.; Gomez, A.N.; Lu, K.; Polosukhin, I. Attention is All you Need. In Proceedings of the Advances in Neural Information Processing Systems; Guyon, I., Luxburg, U.V., Bengio, S., Wallach, H., Fergus, R., Vishwanathan, S., Garnett, R., Eds.; Curran Associates, Inc.: Red Hook, NY, USA, 2017; Volume 30. [Google Scholar]
- Wolf, T.; Debut, L.; Sanh, V.; Chaumond, J.; Delangue, C.; Moi, A.; Cistac, P.; Rault, T.; Louf, R.; Funtowicz, M.; et al. Transformers: State-of-the-art natural language processing. In Proceedings of the 2020 Conference on Empirical Methods in Natural Language Processing: System Demonstrations, Online, 16–20 November 2020; pp. 38–45. [Google Scholar]
- Dosovitskiy, A.; Beyer, L.; Kolesnikov, A.; Weissenborn, D.; Zhai, X.; Unterthiner, T.; Dehghani, M.; Minderer, M.; Heigold, G.; Gelly, S.; et al. An Image is Worth 16 × 16 Words: Transformers for Image Recognition at Scale. In Proceedings of the International Conference on Learning Representations, Vienna, Austria, 4 May 2021. [Google Scholar]
- Goodfellow, I.; Pouget-Abadie, J.; Mirza, M.; Xu, B.; Warde-Farley, D.; Ozair, S.; Courville, A.; Bengio, Y. Generative adversarial nets. In Proceedings of the Advances in Neural Information Processing Systems, Montreal, QC, Canada, 8–13 December 2014; pp. 2672–2680. [Google Scholar]
- Moriya, T.; Roth, H.R.; Nakamura, S.; Oda, H.; Nagara, K.; Oda, M.; Mori, K. Unsupervised segmentation of 3D medical images based on clustering and deep representation learning. In Proceedings of the Medical Imaging, Houston, TX, USA, 11–13 February 2018. [Google Scholar]
- Hengstschläger, M. Artificial intelligence as a door opener for a new era of human reproduction. Hum. Reprod. Open 2023, 2023, hoad043. [Google Scholar] [CrossRef]
- Miloski, B. Opportunities for artificial intelligence in healthcare and in vitro fertilization. Fertil. Steril. 2023, 120, 3–7. [Google Scholar] [CrossRef]
- Gardner, D.K. The way to improve ART outcomes is to introduce more technologies in the laboratory. Reprod. Biomed. Online 2022, 44, 389–392. [Google Scholar] [CrossRef] [PubMed]
- Abdullah, K.A.L.; Atazhanova, T.; Chavez-Badiola, A.; Shivhare, S.B. Automation in ART: Paving the way for the future of infertility treatment. Reprod. Sci. 2023, 30, 1006–1016. [Google Scholar] [CrossRef] [PubMed]
- Louis, C.M.; Erwin, A.; Handayani, N.; Polim, A.A.; Boediono, A.; Sini, I. Review of computer vision application in in vitro fertilization: The application of deep learning-based computer vision technology in the world of IVF. J. Assist. Reprod. Genet. 2021, 38, 1627–1639. [Google Scholar] [CrossRef] [PubMed]
- Zaninovic, N.; Rosenwaks, Z. Artificial intelligence in human in vitro fertilization and embryology. Fertil. Steril. 2020, 114, 914–920. [Google Scholar] [CrossRef]
- Jiang, V.S. Artificial Intelligence in the IVF Laboratory: A Review of Advancements Over the Last Decade. Fertil. Steril. 2023, 120, S0015–S0282. [Google Scholar] [CrossRef]
- Letterie, G. Artificial Intelligence and assisted reproductive technologies: 2023. Ready for prime time? Or not. Fertil. Steril. 2023, 120, 32–37. [Google Scholar] [CrossRef]
- Luong, T.M.T.; Le, N.Q.K. Artificial intelligence in time-lapse system: Advances, applications, and future perspectives in reproductive medicine. J. Assist. Reprod. Genet. 2024, 41, 239–252. [Google Scholar] [CrossRef]
- Narmadha, K.; Varalakshmi, P. Federated Learning in Healthcare: A Privacy Preserving Approach. In Proceedings of the MIE, Nice, France, 27–30 May 2022; pp. 194–198. [Google Scholar]
- Hariton, E.; Pavlovic, Z.; Fanton, M.; Jiang, V.S. Applications of artificial intelligence in ovarian stimulation: A tool for improving efficiency and outcomes. Fertil. Steril. 2023, 120, 8–16. [Google Scholar] [CrossRef]
- Salih, M.; Austin, C.; Warty, R.; Tiktin, C.; Rolnik, D.; Momeni, M.; Rezatofighi, H.; Reddy, S.; Smith, V.; Vollenhoven, B.; et al. Embryo selection through artificial intelligence versus embryologists: A systematic review. Hum. Reprod. Open 2023, 2023, hoad031. [Google Scholar] [CrossRef]
- Wang, J.; Guo, Y.; Zhang, N.; Li, T. Research progress of time-lapse imaging technology and embryonic development potential: A review. Medicine 2023, 102, e35203. [Google Scholar] [CrossRef]
- Kim, J.; Lee, J.; Jun, J.H. Non-invasive evaluation of embryo quality for the selection of transferable embryos in human in vitro fertilization-embryo transfer. Clin. Exp. Reprod. Med. 2022, 49, 225. [Google Scholar] [CrossRef] [PubMed]
- Berman, A.; Anteby, R.; Efros, O.; Klang, E.; Soffer, S. Deep Learning for Embryo Evaluation Using Time-Lapse: A Systematic Review of Diagnostic Test Accuracy. Am. J. Obstet. Gynecol. 2023, 229, 490–501. [Google Scholar] [CrossRef] [PubMed]
- Isa, I.S.; Yusof, U.K.; Mohd Zain, M. Image Processing Approach for Grading IVF Blastocyst: A State-of-the-Art Review and Future Perspective of Deep Learning-Based Models. Appl. Sci. 2023, 13, 1195. [Google Scholar] [CrossRef]
- Jiang, V.S.; Bormann, C.L. Non-invasive genetic screening: Current advances in artificial intelligence for embryo ploidy prediction. Fertil. Steril. 2023, 120, 228–234. [Google Scholar] [CrossRef]
- Shobha, R.B.; Bharathi, S.; Pareek, P.K. Deep Learning Methods to Automate Embryo Classification and Evaluation. In Proceedings of the International Conference on Applied Machine Learning and Data Analytics, Reynosa, Mexico, 22–23 December 2022; Springer: Berlin/Heidelberg, Germany, 2022; pp. 1–12. [Google Scholar]
- Glatstein, I.; Chavez-Badiola, A.; Curchoe, C.L. New frontiers in embryo selection. J. Assist. Reprod. Genet. 2023, 40, 223–234. [Google Scholar] [CrossRef]
- Cimadomo, D.; Fernandez, L.S.; Soscia, D.; Fabozzi, G.; Benini, F.; Cesana, A.; Dal Canto, M.B.; Maggiulli, R.; Muzzì, S.; Scarica, C.; et al. Inter-centre reliability in embryo grading across several IVF clinics is limited: Implications for embryo selection. Reprod. BioMed. Online 2022, 44, 39–48. [Google Scholar] [CrossRef]
- Giménez-Rodríguez, C.; Meseguer, M. The patient or the blastocyst; which leads to the perfect outcome prediction? Fertil. Steril. 2023, 120, 811–812. [Google Scholar] [CrossRef]
- Sfakianoudis, K.; Maziotis, E.; Grigoriadis, S.; Pantou, A.; Kokkini, G.; Trypidi, A.; Giannelou, P.; Zikopoulos, A.; Angeli, I.; Vaxevanoglou, T.; et al. Reporting on the value of artificial intelligence in predicting the optimal embryo for transfer: A systematic review including data synthesis. Biomedicines 2022, 10, 697. [Google Scholar] [CrossRef]
- Dimitriadis, I.; Zaninovic, N.; Badiola, A.C.; Bormann, C.L. Artificial intelligence in the embryology laboratory: A review. Reprod. BioMed. Online 2022, 44, 435–448. [Google Scholar] [CrossRef]
- Brayboy, L.M.; Quaas, A.M. The DIY IVF cycle—harnessing the power of deeptech to bring ART to the masses. J. Assist. Reprod. Genet. 2023, 40, 259–263. [Google Scholar] [CrossRef]
- Cherouveim, P.; Velmahos, C.; Bormann, C.L. Artificial Intelligence (AI) for Sperm Selection–a Systematic Review. Fertil. Steril. 2023, 120, 24–31. [Google Scholar] [CrossRef] [PubMed]
- Voliotis, M.; Hanassab, S.; Abbara, A.; Heinis, T.; Dhillo, W.S.; Tsaneva-Atanasova, K. Quantitative approaches in clinical reproductive endocrinology. Curr. Opin. Endocr. Metab. Res. 2022, 27, 100421. [Google Scholar] [CrossRef] [PubMed]
- Guo, X.; Zhan, H.; Zhang, X.; Pang, Y.; Xu, H.; Zhang, B.; Lao, K.; Ding, P.; Wang, Y.; Han, L. Predictive models for starting dose of gonadotropin in controlled ovarian hyperstimulation: Review and progress update. Hum. Fertil. 2023, 26, 1609–1616. [Google Scholar] [CrossRef] [PubMed]
- Chen, Z.; Wang, Z.; Du, M.; Liu, Z. Artificial intelligence in the assessment of female reproductive function using ultrasound: A review. J. Ultrasound Med. 2022, 41, 1343–1353. [Google Scholar] [CrossRef]
- Rolfes, V.; Bittner, U.; Gerhards, H.; Krüssel, J.S.; Fehm, T.; Ranisch, R.; Fangerau, H. Artificial intelligence in reproductive medicine—An ethical perspective. Geburtshilfe Frauenheilkd. 2023, 83, 106–115. [Google Scholar] [CrossRef]
- Go, K.J.; Hudson, C. Deep technology for the optimization of cryostorage. J. Assist. Reprod. Genet. 2023, 40, 1829–1834. [Google Scholar] [CrossRef]
- Wang, R.; Pan, W.; Yu, L.; Zhang, X.; Pan, W.; Hu, C.; Wen, L.; Jin, L.; Liao, S. AI-Based Optimal Treatment Strategy Selection for Female Infertility for First and Subsequent IVF-ET Cycles. J. Med. Syst. 2023, 47, 87. [Google Scholar] [CrossRef]
- Bauer, E.; Kohavi, R. An empirical comparison of voting classification algorithms: Bagging, boosting, and variants. Mach. Learn. 1999, 36, 105–139. [Google Scholar] [CrossRef]
- Ke, G.; Meng, Q.; Finley, T.; Wang, T.; Chen, W.; Ma, W.; Ye, Q.; Liu, T.Y. Lightgbm: A highly efficient gradient boosting decision tree. Adv. Neural Inf. Process. Syst. 2017, 30, 3149–3157. [Google Scholar]
- Liu, Z.; Huang, B.; Cui, Y.; Xu, Y.; Zhang, B.; Zhu, L.; Wang, Y.; Jin, L.; Wu, D. Multi-task deep learning with dynamic programming for embryo early development stage classification from time-lapse videos. IEEE Access 2019, 7, 122153–122163. [Google Scholar] [CrossRef]
- Deng, J.; Dong, W.; Socher, R.; Li, L.J.; Li, K.; Fei-Fei, L. Imagenet: A large-scale hierarchical image database. In Proceedings of the 2009 IEEE Conference on Computer Vision and Pattern Recognition, Miami, FL, USA, 20–25 June 2009; IEEE: New York, NY, USA; pp. 248–255. [Google Scholar]
- Fjeldstad, J.; Qi, W.; Mercuri, N.; Siddique, N.; Meriano, J.; Krivoi, A.; Nayot, D. An artificial intelligence tool predicts blastocyst development from static images of fresh mature oocytes. Reprod. BioMed. Online 2024, 48, 103842. [Google Scholar] [CrossRef] [PubMed]
- Tan, M.; Le, Q. Efficientnet: Rethinking model scaling for convolutional neural networks. In Proceedings of the International Conference on Machine Learning, PMLR, Long Beach, CA, USA, 10–15 June 2019; pp. 6105–6114. [Google Scholar]
- Lee, C.I.; Su, Y.R.; Chen, C.H.; Chang, T.A.; Kuo, E.E.S.; Zheng, W.L.; Hsieh, W.T.; Huang, C.C.; Lee, M.S.; Liu, M. End-to-end deep learning for recognition of ploidy status using time-lapse videos. J. Assist. Reprod. Genet. 2021, 38, 1655–1663. [Google Scholar] [CrossRef] [PubMed]
- Carreira, J.; Zisserman, A. Quo vadis, action recognition? Anew model and the kinetics dataset. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Honolulu, HI, USA, 21–26 July 2017; pp. 6299–6308. [Google Scholar]
- Kay, W.; Carreira, J.; Simonyan, K.; Zhang, B.; Hillier, C.; Vijayanarasimhan, S.; Viola, F.; Green, T.; Back, T.; Natsev, P.; et al. The kinetics human action video dataset. arXiv 2017, arXiv:1705.06950. [Google Scholar]
- Targosz, A.; Myszor, D.; Mrugacz, G. Human oocytes image classification method based on deep neural networks. BioMed. Eng. Online 2023, 22, 92. [Google Scholar] [CrossRef]
- Chen, L.C.; Zhu, Y.; Papandreou, G.; Schroff, F.; Adam, H. Encoder-decoder with atrous separable convolution for semantic image segmentation. In Proceedings of the European Conference on Computer Vision (ECCV), Munich, Germany, 8–14 September 2018; pp. 801–818. [Google Scholar]
- Iandola, F.N.; Han, S.; Moskewicz, M.W.; Ashraf, K.; Dally, W.J.; Keutzer, K. SqueezeNet: AlexNet-level accuracy with 50x fewer parameters and <0.5 MB model size. arXiv 2016, arXiv:1602.07360. [Google Scholar]
- Danardono, G.B.; Erwin, A.; Purnama, J.; Handayani, N.; Polim, A.A.; Boediono, A.; Sini, I. A Homogeneous Ensemble of Robust Pre-defined Neural Network Enables Automated Annotation of Human Embryo Morphokinetics. J. Reprod. Infertil. 2022, 23, 250. [Google Scholar] [CrossRef]
- Einy, S.; Sen, E.; Saygin, H.; Hivehchi, H.; Dorostkar Navaei, Y. Local Binary Convolutional Neural Networks’ Long Short-Term Memory Model for Human Embryos’ Anomaly Detection. Sci. Program. 2023, 2023. [Google Scholar] [CrossRef]
- Juefei-Xu, F.; Naresh Boddeti, V.; Savvides, M. Local binary convolutional neural networks. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Honolulu, HI, USA, 21–26 July 2017; pp. 19–28. [Google Scholar]
- Jiang, V.S.; Kartik, D.; Thirumalaraju, P.; Kandula, H.; Kanakasabapathy, M.K.; Souter, I.; Dimitriadis, I.; Bormann, C.L.; Shafiee, H. Advancements in the future of automating micromanipulation techniques in the IVF laboratory using deep convolutional neural networks. J. Assist. Reprod. Genet. 2023, 40, 251–257. [Google Scholar] [CrossRef]
- Aguirre-Espericueta, G.; Mendizabal-Ruiz, G. CNNs for ISCI Stage Recognition on Video Sequences. In Proceedings of the XLV Mexican Conference on Biomedical Engineering; Springer: Berlin/Heidelberg, Germany, 2022; pp. 111–118. [Google Scholar]
- Saeedi, P.; Yee, D.; Au, J.; Havelock, J. Automatic identification of human blastocyst components via texture. IEEE Trans. Biomed. Eng. 2017, 64, 2968–2978. [Google Scholar]
- Rad, R.M.; Saeedi, P.; Au, J.; Havelock, J. Predicting human embryos’ implantation outcome from a single blastocyst image. In Proceedings of the 2019 41st Annual International Conference of the IEEE Engineering in Medicine and Biology Society (EMBC), Berlin, Germany, 23–27 July 2019; IEEE: New York, NY, USA, 2019; pp. 920–924. [Google Scholar]
- Shi, W.; Caballero, J.; Huszár, F.; Totz, J.; Aitken, A.P.; Bishop, R.; Rueckert, D.; Wang, Z. Real-time single image and video super-resolution using an efficient sub-pixel convolutional neural network. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Las Vegas, NV, USA, 27–30 June 2016; pp. 1874–1883. [Google Scholar]
- Chen, L.C.; Papandreou, G.; Schroff, F.; Adam, H. Rethinking atrous convolution for semantic image segmentation. arXiv 2017, arXiv:1706.05587. [Google Scholar]
- Szegedy, C.; Vanhoucke, V.; Ioffe, S.; Shlens, J.; Wojna, Z. Rethinking the inception architecture for computer vision. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Las Vegas, NV, USA, 27–30 June 2016; pp. 2818–2826. [Google Scholar]
- Arsalan, M.; Haider, A.; Choi, J.; Park, K.R. Detecting blastocyst components by artificial intelligence for human embryological analysis to improve success rate of in vitro fertilization. J. Pers. Med. 2022, 12, 124. [Google Scholar] [CrossRef] [PubMed]
- Ronneberger, O.; Fischer, P.; Brox, T. U-net: Convolutional networks for biomedical image segmentation. In Proceedings of the Medical Image Computing and Computer-Assisted Intervention–MICCAI 2015: 18th International Conference, Munich, Germany, 5–9 October 2015; Proceedings, Part III 18. Springer: Berlin/Heidelberg, Germany, 2015; pp. 234–241. [Google Scholar]
- Iglovikov, V.; Shvets, A. Ternausnet: U-net with vgg11 encoder pre-trained on imagenet for image segmentation. arXiv 2018, arXiv:1801.05746. [Google Scholar]
- Zhao, H.; Shi, J.; Qi, X.; Wang, X.; Jia, J. Pyramid scene parsing network. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Honolulu, HI, USA, 21–26 July 2017; pp. 2881–2890. [Google Scholar]
- Rad, R.M.; Saeedi, P.; Au, J.; Havelock, J. BLAST-NET: Semantic segmentation of human blastocyst components via cascaded atrous pyramid and dense progressive upsampling. In Proceedings of the 2019 IEEE International Conference on Image Processing (ICIP), Taipei, Taiwan, 22–25 September 2019; IEEE: New York, NY, USA, 2019; pp. 1865–1869. [Google Scholar]
- Mushtaq, A.; Mumtaz, M.; Raza, A.; Salem, N.; Yasir, M.N. Artificial Intelligence-Based Detection of Human Embryo Components for Assisted Reproduction by In Vitro Fertilization. Sensors 2022, 22, 7418. [Google Scholar] [CrossRef] [PubMed]
- Ishaq, M.; Raza, S.; Rehar, H.; Abadeen, S.E.Z.U.; Hussain, D.; Naqvi, R.A.; Lee, S.W. Assisting the Human Embryo Viability Assessment by Deep Learning for In Vitro Fertilization. Mathematics 2023, 11, 2023. [Google Scholar] [CrossRef]
- Jamal, A.; Dharmawan, A.P.; Septiandri, A.A.; Iffanolida, P.A.; Riayati, O.; Wiweko, B. Densely U-Net Models for Human Embryo Segmentation. In Proceedings of the 2023 4th International Conference on Artificial Intelligence and Data Sciences (AiDAS), Ipoh, Malaysia, 6–7 September 2023; IEEE: New York, NY, USA, 2023; pp. 17–22. [Google Scholar]
- Septiandri, A.A.; Jamal, A.; Iffanolida, P.A.; Riayati, O.; Wiweko, B. Human blastocyst classification after in vitro fertilization using deep learning. In Proceedings of the 2020 7th International Conference on Advance Informatics: Concepts, Theory and Applications (ICAICTA), Online, 8–9 September 2020; IEEE: New York, NY, USA, 2020; pp. 1–4. [Google Scholar]
- Khder, S.M.; Mohamed, E.A.; Yassine, I.A. A Clustering-Based Fusion System for Blastomere Localization. Biomed. Eng. Appl. Basis Commun. 2022, 34, 2250021. [Google Scholar] [CrossRef]
- Targosz, A.; Przystałka, P.; Wiaderkiewicz, R.; Mrugacz, G. Semantic segmentation of human oocyte images using deep neural networks. BioMed. Eng. OnLine 2021, 20, 40. [Google Scholar] [CrossRef]
- Badrinarayanan, V.; Kendall, A.; Cipolla, R. SegNet: A deep convolutional encoder-decoder architecture for image segmentation. IEEE Trans. Pattern Anal. Mach. Intell. 2017, 39, 2481–2495. [Google Scholar] [CrossRef]
- Bori, L.; Meseguer, F.; Valera, M.A.; Galan, A.; Remohi, J.; Meseguer, M. The higher the score, the better the clinical outcome: Retrospective evaluation of automatic embryo grading as a support tool for embryo selection in IVF laboratories. Hum. Reprod. 2022, 37, 1148–1160. [Google Scholar] [CrossRef]
- Pierson, H.E.; Invik, J.; Meriano, J.; Pierson, R.A. A novel system for rapid conversion of Gardner embryo grades to linear scale numeric variables. Reprod. BioMed. Online 2023, 46, 808–818. [Google Scholar] [CrossRef]
- Alkindy, F.K.; Yusof, U.K.; Zain, M.M. An Automated Day 3 Embryo Grading Based On Morphological Characteristics Using CNN with Transfer Learning Techniques. In Proceedings of the 2023 IEEE 13th International Conference on Control System, Computing and Engineering (ICCSCE), Penang, Malaysia, 25–26 August 2023; IEEE: New York, NY, USA, 2023; pp. 214–219. [Google Scholar]
- Mohamed, Y.A.; Yusof, U.K.; Isa, I.S.; Zain, M.M. An Automated Blastocyst Grading System Using Convolutional Neural Network and Transfer Learning. In Proceedings of the 2023 IEEE 13th International Conference on Control System, Computing and Engineering (ICCSCE), Penang, Malaysia, 25–26 August 2023; IEEE: New York, NY, USA, 2023; pp. 202–207. [Google Scholar]
- Garg, K.; Dev, A.; Bansal, P.; Mittal, H. An Efficient Deep Learning Model for Embryo Classification. In Proceedings of the 2024 14th International Conference on Cloud Computing, Data Science & Engineering (Confluence), Noida, India, 18–19 January 2024; IEEE: New York, NY, USA, 2024; pp. 358–363. [Google Scholar]
- Thirumalaraju, P.; Kanakasabapathy, M.K.; Bormann, C.L.; Gupta, R.; Pooniwala, R.; Kandula, H.; Souter, I.; Dimitriadis, I.; Shafiee, H. Evaluation of deep convolutional neural networks in classifying human embryo images based on their morphological quality. Heliyon 2021, 7, e06298. [Google Scholar] [CrossRef]
- Liao, Q.; Zhang, Q.; Feng, X.; Huang, H.; Xu, H.; Tian, B.; Liu, J.; Yu, Q.; Guo, N.; Liu, Q.; et al. Development of deep learning algorithms for predicting blastocyst formation and quality by time-lapse monitoring. Commun. Biol. 2021, 4, 415. [Google Scholar] [CrossRef] [PubMed]
- Hung Vuong Hospital, Ho Chi Minh City. Embryo Quality Classification Dataset. Available online: https://www.kaggle.com/competitions/world-championship-2023-embryo-classification/data (accessed on 21 April 2024).
- Wu, C.; Fu, L.; Tian, Z.; Liu, J.; Song, J.; Guo, W.; Zhao, Y.; Zheng, D.; Jin, Y.; Yi, D.; et al. LWMA-Net: Light-weighted morphology attention learning for human embryo grading. Comput. Biol. Med. 2022, 151, 106242. [Google Scholar] [CrossRef] [PubMed]
- Cho, J.; Brumar, C.; Maeder-York, P.; Barash, O.; Malmsten, J.; Zaninovic, N.; Sakkas, D.; Miller, K.; Levy, M.; VerMilyea, M.; et al. P-171 Sensitivity analysis of an embryo grading artificial intelligence model to different focal planes. Hum. Reprod. 2022, 37, deac107–deac166. [Google Scholar] [CrossRef]
- Kragh, M.F.; Rimestad, J.; Berntsen, J.; Karstoft, H. Automatic grading of human blastocysts from time-lapse imaging. Comput. Biol. Med. 2019, 115, 103494. [Google Scholar] [CrossRef]
- Chen, T.J.; Zheng, W.L.; Liu, C.H.; Huang, I.; Lai, H.H.; Liu, M. Using deep learning with large dataset of microscope images to develop an automated embryo grading system. Fertil. Reprod. 2019, 1, 51–56. [Google Scholar] [CrossRef]
- SFC. Stork Fertility Center. Available online: https://e-stork.com.tw/ (accessed on 31 January 2024).
- Vaidya, G.; Chandrasekhar, S.; Gajjar, R.; Gajjar, N.; Patel, D.; Banker, M. Time series prediction of viable embryo and automatic grading in IVF using deep learning. Open Biomed. Eng. J. 2021, 15, 190–203. [Google Scholar] [CrossRef]
- Nova IVF Fertility. Available online: www.novaivffertility.com/ivf-centre/ahmedabad/fertility-clinic-ahmedabad (accessed on 1 February 2024).
- Wang, S.; Zhou, C.; Zhang, D.; Chen, L.; Sun, H. A deep learning framework design for automatic blastocyst evaluation with multifocal images. IEEE Access 2021, 9, 18927–18934. [Google Scholar] [CrossRef]
- Buiu, C.; Dănăilă, V.R.; Răduţă, C.N. MobileNetV2 ensemble for cervical precancerous lesions classification. Processes 2020, 8, 595. [Google Scholar] [CrossRef]
- Selvaraju, R.R.; Cogswell, M.; Das, A.; Vedantam, R.; Parikh, D.; Batra, D. Grad-cam: Visual explanations from deep networks via gradient-based localization. In Proceedings of the IEEE International Conference on Computer Vision, Venice, Italy, 22–29 October 2017; pp. 618–626. [Google Scholar]
- Bormann, C.L.; Thirumalaraju, P.; Kanakasabapathy, M.K.; Kandula, H.; Souter, I.; Dimitriadis, I.; Gupta, R.; Pooniwala, R.; Shafiee, H. Consistency and objectivity of automated embryo assessments using deep neural networks. Fertil. Steril. 2020, 113, 781–787. [Google Scholar] [CrossRef]
- Zeman, A.; Maerten, A.S.; Mengels, A.; Sharon, L.F.; Spiessens, C.; de Beeck, H.O. Deep learning for human embryo classification at the cleavage stage (Day 3). In Proceedings of the Pattern Recognition. ICPR International Workshops and Challenges, Virtual, 10–15 January 2021; Proceedings, Part I. Springer: Berlin/Heidelberg, Germany, 2021; pp. 278–292. [Google Scholar]
- Zoph, B.; Vasudevan, V.; Shlens, J.; Le, Q.V. Learning transferable architectures for scalable image recognition. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Salt Lake City, UT, USA, 18–23 June 2018; pp. 8697–8710. [Google Scholar]
- Xie, S.; Girshick, R.; Dollár, P.; Tu, Z.; He, K. Aggregated residual transformations for deep neural networks. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Honolulu, HI, USA, 21–26 July 2017; pp. 1492–1500. [Google Scholar]
- Bormann, C.L.; Kanakasabapathy, M.K.; Thirumalaraju, P.; Gupta, R.; Pooniwala, R.; Kandula, H.; Hariton, E.; Souter, I.; Dimitriadis, I.; Ramirez, L.B.; et al. Performance of a deep learning based neural network in the selection of human blastocysts for implantation. Elife 2020, 9, e55301. [Google Scholar] [CrossRef]
- Fitz, V.; Kanakasabapathy, M.; Thirumalaraju, P.; Kandula, H.; Ramirez, L.; Boehnlein, L.; Swain, J.; Curchoe, C.; James, K.; Dimitriadis, I.; et al. Should there be an “AI” in TEAM? Embryologists selection of high implantation potential embryos improves with the aid of an artificial intelligence algorithm. J. Assist. Reprod. Genet. 2021, 38, 2663–2670. [Google Scholar] [CrossRef] [PubMed]
- Loewke, K.E.; Cho, J.H.; Maeder-York, P.; Barash, O.O.; Meseguer, M.; Zaninovic, N.; Miller, K.A.; Sakkas, D.; Levy, M.; VerMilyea, M.D. A Generalizable Model for Ranking Blastocyst Stage Embryos Using Deep Learning. Fertil. Steril. 2021, 116, e152. [Google Scholar] [CrossRef]
- Pamungkasari, P.D.; Uchida, K.; Saito, S.; Juwono, F.H.; Hanoum, I.F.; Shirakawa, S. Embryo Grade Prediction for In-Vitro Fertilization. In Deep Learning for Biomedical Applications; CRC Press: Boca Raton, FL, USA, 2021; pp. 21–40. [Google Scholar]
- Huang, G.; Liu, Z.; Pleiss, G.; Van Der Maaten, L.; Weinberger, K.Q. Convolutional networks with dense connectivity. IEEE Trans. Pattern Anal. Mach. Intell. 2019, 44, 8704–8716. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.; Li, D.; Dai, C.; Shan, G.; Zhang, Z.; Zhuang, S.; Lee, C.W.; Wong, A.; Yue, C.; Huang, Z.; et al. Automated Morphological Grading of Human Blastocysts From Multi-Focus Images. IEEE Trans. Autom. Sci. Eng. 2023, 21, 2584–2592. [Google Scholar] [CrossRef]
- Huang, B.; Tan, W.; Li, Z.; Jin, L. An artificial intelligence model (euploid prediction algorithm) can predict embryo ploidy status based on time-lapse data. Reprod. Biol. Endocrinol. 2021, 19, 1–10. [Google Scholar] [CrossRef]
- Hara, K.; Kataoka, H.; Satoh, Y. Can spatiotemporal 3D CNNs retrace the history of 2D CNNs and ImageNet? In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Salt Lake City, UT, USA, 18–23 June 2018; pp. 6546–6555. [Google Scholar]
- Khosravi, P.; Kazemi, E.; Zhan, Q.; Malmsten, J.E.; Toschi, M.; Zisimopoulos, P.; Sigaras, A.; Lavery, S.; Cooper, L.A.; Hickman, C.; et al. Deep learning enables robust assessment and selection of human blastocysts after in vitro fertilization. npj Digit. Med. 2019, 2, 21. [Google Scholar] [CrossRef]
- Szegedy, C.; Liu, W.; Jia, Y.; Sermanet, P.; Reed, S.; Anguelov, D.; Erhan, D.; Vanhoucke, V.; Rabinovich, A. Going deeper with convolutions. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Boston, MA, USA, 7–12 June 2015; pp. 1–9. [Google Scholar]
- Barnes, J.; Brendel, M.; Gao, V.R.; Rajendran, S.; Kim, J.; Li, Q.; Malmsten, J.E.; Sierra, J.T.; Zisimopoulos, P.; Sigaras, A.; et al. A non-invasive artificial intelligence approach for the prediction of human blastocyst ploidy: A retrospective model development and validation study. Lancet Digit. Health 2023, 5, e28–e40. [Google Scholar] [CrossRef]
- Diakiw, S.; Hall, J.; VerMilyea, M.; Amin, J.; Aizpurua, J.; Giardini, L.; Briones, Y.; Lim, A.; Dakka, M.; Nguyen, T.; et al. Development of an artificial intelligence model for predicting the likelihood of human embryo euploidy based on blastocyst images from multiple imaging systems during IVF. Hum. Reprod. 2022, 37, 1746–1759. [Google Scholar] [CrossRef]
- VerMilyea, M.; Hall, J.; Diakiw, S.; Johnston, A.; Nguyen, T.; Perugini, D.; Miller, A.; Picou, A.; Murphy, A.; Perugini, M. Development of an artificial intelligence-based assessment model for prediction of embryo viability using static images captured by optical light microscopy during IVF. Hum. Reprod. 2020, 35, 770–784. [Google Scholar] [CrossRef]
- Dakka, M.; Nguyen, T.; Hall, J.; Diakiw, S.; VerMilyea, M.; Linke, R.; Perugini, M.; Perugini, D. Automated detection of poor-quality data: Case studies in healthcare. Sci. Rep. 2021, 11, 18005. [Google Scholar] [CrossRef]
- Hinton, G.; Vinyals, O.; Dean, J. Distilling the knowledge in a neural network. arXiv 2015, arXiv:1503.02531. [Google Scholar]
- Ortiz, J.A.; Morales, R.; Lledo, B.; Vicente, J.A.; Gonzalez, J.; García-Hernandez, E.M.; Cascales, A.; Ten, J.; Bernabeu, A.; Bernabeu, R. Application of machine learning to predict aneuploidy and mosaicism in embryos from in vitro fertilization cycles. AJOG Glob. Rep. 2022, 2, 100103. [Google Scholar] [CrossRef] [PubMed]
- Ou, Z.; Zhang, Q.; Li, Y.; Meng, X.; Wang, Y.; Ouyang, Y.; Li, Z.; Zeng, K. Classification of human embryos by using deep learning. In Proceedings of the International Conference on Cloud Computing, Performance Computing, and Deep Learning (CCPCDL 2023), Huzhou, China, 17–19 February 2023; SPIE: Bellingham, DC, USA, 2023; Volume 12712, pp. 342–347. [Google Scholar]
- Xie, Q.; Luong, M.T.; Hovy, E.; Le, Q.V. Self-training with noisy student improves imagenet classification. In Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition, Seattle, WA, USA, 13–19 June 2020; pp. 10687–10698. [Google Scholar]
- Liu, Z.; Lin, Y.; Cao, Y.; Hu, H.; Wei, Y.; Zhang, Z.; Lin, S.; Guo, B. Swin transformer: Hierarchical vision transformer using shifted windows. In Proceedings of the IEEE/CVF International Conference on Computer Vision, Montreal, QC, Canada, 10–17 October 2021; pp. 10012–10022. [Google Scholar]
- Krizhevsky, A.; Sutskever, I.; Hinton, G.E. Imagenet classification with deep convolutional neural networks. Adv. Neural Inf. Process. Syst. 2012, 25, 84–90. [Google Scholar] [CrossRef]
- Marsh, P.; Radif, D.; Rajpurkar, P.; Wang, Z.; Hariton, E.; Ribeiro, S.; Simbulan, R.; Kaing, A.; Lin, W.; Rajah, A.; et al. A proof of concept for a deep learning system that can aid embryologists in predicting blastocyst survival after thaw. Sci. Rep. 2022, 12, 21119. [Google Scholar] [CrossRef]
- Chavez-Badiola, A.; Flores-Saiffe-Farías, A.; Mendizabal-Ruiz, G.; Drakeley, A.J.; Cohen, J. Embryo Ranking Intelligent Classification Algorithm (ERICA): Artificial intelligence clinical assistant predicting embryo ploidy and implantation. Reprod. BioMed. Online 2020, 41, 585–593. [Google Scholar] [CrossRef]
- Darus, R.; Yusuf, U.K.; Yu, S.J.; Isa, I.S.; Zain, M.M.; Fauzi, N.A. EmbryoSys: An Intelligence-Web-based In Vitro Fertilization (IVF) Embryo Tracking & Grading System. In Proceedings of the 2023 IEEE Industrial Electronics and Applications Conference (IEACon), Penang, Malaysia, 6–7 November 2023; IEEE: New York, NY, USA, 2023; pp. 243–248. [Google Scholar]
- Lockhart, L.; Saeedi, P.; Au, J.; Havelock, J. Multi-Label Classification for Automatic Human Blastocyst Grading with Severely Imbalanced Data. In Proceedings of the 2019 IEEE 21st International Workshop on Multimedia Signal Processing (MMSP), Kuala Lumpur, Malaysia, 27–29 September 2019; IEEE: New York, NY, USA, 2019; pp. 1–6. [Google Scholar]
- Cimadomo, D.; Chiappetta, V.; Innocenti, F.; Saturno, G.; Taggi, M.; Marconetto, A.; Casciani, V.; Albricci, L.; Maggiulli, R.; Coticchio, G.; et al. Towards Automation in IVF: Pre-Clinical Validation of a Deep Learning-Based Embryo Grading System during PGT-A Cycles. J. Clin. Med. 2023, 12, 1806. [Google Scholar] [CrossRef]
- Wu, C.; Yan, W.; Li, H.; Li, J.; Wang, H.; Chang, S.; Yu, T.; Jin, Y.; Ma, C.; Luo, Y.; et al. A classification system of day 3 human embryos using deep learning. Biomed. Signal Process. Control 2021, 70, 102943. [Google Scholar] [CrossRef]
- Liang, S.; Zhang, R.; Liang, D.; Song, T.; Ai, T.; Xia, C.; Xia, L.; Wang, Y. Multimodal 3D DenseNet for IDH genotype prediction in gliomas. Genes 2018, 9, 382. [Google Scholar] [CrossRef]
- Mednikov, Y.; Nehemia, S.; Zheng, B.; Benzaquen, O.; Lederman, D. Transfer representation learning using Inception-V3 for the detection of masses in mammography. In Proceedings of the 2018 40th Annual International Conference of the IEEE Engineering in Medicine and Biology Society (EMBC), Honolulu, HI, USA, 18–21 July 2018; IEEE: New York, NY, USA, 2018; pp. 2587–2590. [Google Scholar]
- Brito, C.; Machado, A.; Sousa, A.L. Electrocardiogram Beat-Classification Based on a ResNet Network. In Proceedings of the MedInfo, Lyon, France, 25–30 August 2019; pp. 55–59. [Google Scholar]
- Cheng, P.M.; Malhi, H.S. Transfer learning with convolutional neural networks for classification of abdominal ultrasound images. J. Digit. Imaging 2017, 30, 234–243. [Google Scholar] [CrossRef]
- Veeck, L.L. An Atlas of Human Gametes and Conceptuses: An Illustrated Reference for Assisted Reproductive Technology; CRC Press: Boca Raton, FL, USA, 1999. [Google Scholar]
- Sandler, M.; Howard, A.; Zhu, M.; Zhmoginov, A.; Chen, L.C. Mobilenetv2: Inverted residuals and linear bottlenecks. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Salt Lake City, UT, USA, 18–23 June 2018; pp. 4510–4520. [Google Scholar]
- Howard, J.; Gugger, S. Fastai: A layered API for deep learning. Information 2020, 11, 108. [Google Scholar] [CrossRef]
- Kanakasabapathy, M.K.; Thirumalaraju, P.; Bormann, C.L.; Kandula, H.; Dimitriadis, I.; Souter, I.; Yogesh, V.; Pavan, S.K.S.; Yarravarapu, D.; Gupta, R.; et al. Development and evaluation of inexpensive automated deep learning-based imaging systems for embryology. Lab A Chip 2019, 19, 4139–4145. [Google Scholar] [CrossRef] [PubMed]
- Bormann, C.L.; Curchoe, C.L.; Thirumalaraju, P.; Kanakasabapathy, M.K.; Gupta, R.; Pooniwala, R.; Kandula, H.; Souter, I.; Dimitriadis, I.; Shafiee, H. Deep learning early warning system for embryo culture conditions and embryologist performance in the ART laboratory. J. Assist. Reprod. Genet. 2021, 38, 1641–1646. [Google Scholar] [CrossRef] [PubMed]
- Yuan, Z.; Yuan, M.; Song, X.; Huang, X.; Yan, W. Development of an artificial intelligence based model for predicting the euploidy of blastocysts in PGT-A treatments. Sci. Rep. 2023, 13, 2322. [Google Scholar] [CrossRef] [PubMed]
- Bamford, T.; Easter, C.; Montgomery, S.; Smith, R.; Dhillon-Smith, R.K.; Barrie, A.; Campbell, A.; Coomarasamy, A. A comparison of 12 machine learning models developed to predict ploidy, using a morphokinetic meta-dataset of 8147 embryos. Hum. Reprod. 2023, 38, 569–581. [Google Scholar] [CrossRef]
- Danardono, G.B.; Handayani, N.; Louis, C.M.; Polim, A.A.; Sirait, B.; Periastiningrum, G.; Afadlal, S.; Boediono, A.; Sini, I. Embryo ploidy status classification through computer-assisted morphology assessment. AJOG Glob. Rep. 2023, 3, 100209. [Google Scholar] [CrossRef]
- Houri, O.; Gil, Y.; Danieli-Gruber, S.; Shufaro, Y.; Sapir, O.; Hochberg, A.; Ben-Haroush, A.; Wertheimer, A. Prediction of oocyte maturation rate in the GnRH antagonist flexible IVF protocol using a novel machine learning algorithm–A retrospective study. Eur. J. Obstet. Gynecol. Reprod. Biol. 2023, 284, 100–104. [Google Scholar] [CrossRef]
- Chen, T.; Guestrin, C. Xgboost: A scalable tree boosting system. In Proceedings of the 22nd ACM Sigkdd International Conference on Knowledge Discovery and Data Mining, San Francisco, CA, USA, 13–17 August 2016; pp. 785–794. [Google Scholar]
- Vergos, G.; Iliadis, L.A.; Kritopoulou, P.; Papatheodorou, A.; Boursianis, A.D.; Kokkinidis, K.I.D.; Papadopoulou, M.S.; Goudos, S.K. Ensemble Learning Technique for Artificial Intelligence Assisted IVF Applications. In Proceedings of the 2023 12th International Conference on Modern Circuits and Systems Technologies (MOCAST), Athens, Greece, 28–30 June 2023; IEEE: New York, NY, USA, 2023; pp. 1–4. [Google Scholar]
- Toporcerová, S.; Špaková, I.; Šoltys, K.; Klepcová, Z.; Kl’oc, M.; Bohošová, J.; Trachtová, K.; Peterová, L.; Mičková, H.; Urdzík, P.; et al. Small Non-Coding RNAs as New Biomarkers to Evaluate the Quality of the Embryo in the IVF Process. Biomolecules 2022, 12, 1687. [Google Scholar] [CrossRef]
- Hammer, K.C.; Jiang, V.S.; Kanakasabapathy, M.K.; Thirumalaraju, P.; Kandula, H.; Dimitriadis, I.; Souter, I.; Bormann, C.L.; Shafiee, H. Using artificial intelligence to avoid human error in identifying embryos: A retrospective cohort study. J. Assist. Reprod. Genet. 2022, 39, 2343–2348. [Google Scholar] [CrossRef]
- Kanakasabapathy, M.K.; Thirumalaraju, P.; Bormann, C.L.; Gupta, R.; Pooniwala, R.; Kandula, H.; Souter, I.; Dimitriadis, I.; Shafiee, H. Deep learning mediated single time-point image-based prediction of embryo developmental outcome at the cleavage stage. arXiv 2020, arXiv:2006.08346. [Google Scholar]
- Chen, L.; Li, W.; Liu, Y.; Peng, Z.; Cai, L.; Zhang, N.; Xu, J.; Wang, L.; Teng, X.; Yao, Y.; et al. Non-invasive embryo selection strategy for clinical IVF to avoid wastage of potentially competent embryos. Reprod. BioMed. Online 2022, 45, 26–34. [Google Scholar] [CrossRef]
- Ibrahim, H.A.; Thamilvanan, M.N.; Zaian, A.; Supriyanto, E. Fertility Assessment Model For Embryo Grading Using Convolutional Neural Network (CNN). In Proceedings of the 2022 International Conference on Healthcare Engineering (ICHE), Johor, Malaysia, 23–25 September 2022; IEEE: New York, NY, USA, 2022; pp. 1–4. [Google Scholar]
- Yang, H.Y.; Leahy, B.D.; Jang, W.D.; Wei, D.; Kalma, Y.; Rahav, R.; Carmon, A.; Kopel, R.; Azem, F.; Venturas, M.; et al. BlastAssist: A deep learning pipeline to measure interpretable features of human embryos. Hum. Reprod. 2024, 39, 698–708. [Google Scholar] [CrossRef] [PubMed]
- Jang, W.D.; Wei, D.; Zhang, X.; Leahy, B.; Yang, H.; Tompkin, J.; Ben-Yosef, D.; Needleman, D.; Pfister, H. Learning vector quantized shape code for amodal blastomere instance segmentation. In Proceedings of the 2023 IEEE 20th International Symposium on Biomedical Imaging (ISBI), Cartagena, Colombia, 18–21 April 2023; IEEE: New York, NY, USA, 2023; pp. 1–5. [Google Scholar]
- Leahy, B.D.; Jang, W.D.; Yang, H.Y.; Struyven, R.; Wei, D.; Sun, Z.; Lee, K.R.; Royston, C.; Cam, L.; Kalma, Y.; et al. Automated measurements of key morphological features of human embryos for IVF. In Proceedings of the Medical Image Computing and Computer Assisted Intervention–MICCAI 2020: 23rd International Conference, Lima, Peru, 4–8 October 2020; Proceedings, Part V 23. Springer: Berlin/Heidelberg, Germany, 2020; pp. 25–35. [Google Scholar]
- Lukyanenko, S.; Jang, W.D.; Wei, D.; Struyven, R.; Kim, Y.; Leahy, B.; Yang, H.; Rush, A.; Ben-Yosef, D.; Needleman, D.; et al. Developmental stage classification of embryos using two-stream neural network with linear-chain conditional random field. In Proceedings of the Medical Image Computing and Computer Assisted Intervention–MICCAI 2021: 24th International Conference, Strasbourg, France, 27 September–1 October 2021; Proceedings, Part VIII 24. Springer: Berlin/Heidelberg, Germany, 2021; pp. 363–372. [Google Scholar]
- Celard, P.; Seara Vieira, A.; Sorribes-Fdez, J.M.; Iglesias, E.L.; Borrajo, L. Improving Generation and Evaluation of Long Image Sequences for Embryo Development Prediction. Electronics 2024, 13, 476. [Google Scholar] [CrossRef]
- Gomez, T.; Feyeux, M.; Boulant, J.; Normand, N.; David, L.; Paul-Gilloteaux, P.; Fréour, T.; Mouchère, H. A time-lapse embryo dataset for morphokinetic parameter prediction. Data Brief 2022, 42, 108258. [Google Scholar] [CrossRef] [PubMed]
- Letterie, G.; Mac Donald, A. Artificial intelligence in in vitro fertilization: A computer decision support system for day-to-day management of ovarian stimulation during in vitro fertilization. Fertil. Steril. 2020, 114, 1026–1031. [Google Scholar] [CrossRef]
- Fanton, M.; Nutting, V.; Rothman, A.; Maeder-York, P.; Hariton, E.; Barash, O.; Weckstein, L.; Sakkas, D.; Copperman, A.B.; Loewke, K. An interpretable machine learning model for individualized gonadotrophin starting dose selection during ovarian stimulation. Reprod. BioMed. Online 2022, 45, 1152–1159. [Google Scholar] [CrossRef]
- Hariton, E.; Chi, E.A.; Chi, G.; Morris, J.R.; Braatz, J.; Rajpurkar, P.; Rosen, M. A machine learning algorithm can optimize the day of trigger to improve in vitro fertilization outcomes. Fertil. Steril. 2021, 116, 1227–1235. [Google Scholar] [CrossRef]
- Fanton, M.; Nutting, V.; Solano, F.; Maeder-York, P.; Hariton, E.; Barash, O.; Weckstein, L.; Sakkas, D.; Copperman, A.B.; Loewke, K. An interpretable machine learning model for predicting the optimal day of trigger during ovarian stimulation. Fertil. Steril. 2022, 118, 101–108. [Google Scholar] [CrossRef]
- Reuvenny, S.; Youngster, M.; Luz, A.; Hourvitz, R.; Maman, E.; Baum, M.; Hourvitz, A. An artificial intelligence-based approach for selecting the optimal day for triggering in antagonist protocol cycles. Reprod. BioMed. Online 2024, 48, 103423. [Google Scholar] [CrossRef]
- Correa, N.; Cerquides, J.; Arcos, J.L.; Vassena, R. Supporting first FSH dosage for ovarian stimulation with machine learning. Reprod. BioMed. Online 2022, 45, 1039–1045. [Google Scholar] [CrossRef]
- Ferrand, T.; Boulant, J.; He, C.; Chambost, J.; Jacques, C.; Pena, C.A.; Hickman, C.; Reignier, A.; Fréour, T. Predicting the number of oocytes retrieved from controlled ovarian hyperstimulation with machine learning. Hum. Reprod. 2023, 38, 1918–1926. [Google Scholar] [CrossRef]
- Galtier, M.N.; Marini, C. Substra: A framework for privacy-preserving, traceable and collaborative machine learning. arXiv 2019, arXiv:1910.11567. [Google Scholar]
- Liang, X.; Liang, J.; Zeng, F.; Lin, Y.; Li, Y.; Cai, K.; Ni, D.; Chen, Z. Evaluation of oocyte maturity using artificial intelligence quantification of follicle volume biomarker by three-dimensional ultrasound. Reprod. BioMed. Online 2022, 45, 1197–1206. [Google Scholar] [CrossRef] [PubMed]
- Yang, X.; Li, H.; Wang, Y.; Liang, X.; Chen, C.; Zhou, X.; Zeng, F.; Fang, J.; Frangi, A.; Chen, Z.; et al. Contrastive rendering with semi-supervised learning for ovary and follicle segmentation from 3D ultrasound. Med. Image Anal. 2021, 73, 102134. [Google Scholar] [CrossRef] [PubMed]
- Liu, L.; Shen, F.; Liang, H.; Yang, Z.; Yang, J.; Chen, J. Machine learning-based modeling of ovarian response and the quantitative evaluation of comprehensive impact features. Diagnostics 2022, 12, 492. [Google Scholar] [CrossRef]
- Fordham, D.E.; Rosentraub, D.; Polsky, A.L.; Aviram, T.; Wolf, Y.; Perl, O.; Devir, A.; Rosentraub, S.; Silver, D.H.; Gold Zamir, Y.; et al. Embryologist agreement when assessing blastocyst implantation probability: Is data-driven prediction the solution to embryo assessment subjectivity? Hum. Reprod. 2022, 37, 2275–2290. [Google Scholar] [CrossRef]
- Tan, M.; Le, Q. Efficientnetv2: Smaller models and faster training. In Proceedings of the International Conference on Machine Learning, PMLR, Online, 18–24 July 2021; pp. 10096–10106. [Google Scholar]
- Alpha Scientists in Reproductive Medicine; ESHRE Special Interest Group of Embryology. The Istanbul consensus workshop on embryo assessment: Proceedings of an expert meeting. Hum. Reprod. 2011, 26, 1270–1283. [Google Scholar] [CrossRef]
- Dai, Z.; Liu, H.; Le, Q.V.; Tan, M. Coatnet: Marrying convolution and attention for all data sizes. Adv. Neural Inf. Process. Syst. 2021, 34, 3965–3977. [Google Scholar]
- Diakiw, S.M.; Hall, J.M.; VerMilyea, M.; Lim, A.Y.; Quangkananurug, W.; Chanchamroen, S.; Bankowski, B.; Stones, R.; Storr, A.; Miller, A.; et al. An artificial intelligence model correlated with morphological and genetic features of blastocyst quality improves ranking of viable embryos. Reprod. BioMed. Online 2022, 45, 1105–1117. [Google Scholar] [CrossRef]
- Berntsen, J.; Rimestad, J.; Lassen, J.T.; Tran, D.; Kragh, M.F. Robust and generalizable embryo selection based on artificial intelligence and time-lapse image sequences. PLoS ONE 2022, 17, e0262661. [Google Scholar] [CrossRef]
- Vitrolife. KIDScore D5 Decision Support Tool. 2019. Available online: https://www.vitrolife.com/support/support-material/ (accessed on 31 January 2024).
- Ezoe, K.; Shimazaki, K.; Miki, T.; Takahashi, T.; Tanimura, Y.; Amagai, A.; Sawado, A.; Akaike, H.; Mogi, M.; Kaneko, S.; et al. Association between a deep learning-based scoring system with morphokinetics and morphological alterations in human embryos. Reprod. BioMed. Online 2022, 45, 1124–1132. [Google Scholar] [CrossRef]
- Johansen, M.N.; Parner, E.T.; Kragh, M.F.; Kato, K.; Ueno, S.; Palm, S.; Kernbach, M.; Balaban, B.; Keleş, İ.; Gabrielsen, A.V.; et al. Comparing performance between clinics of an embryo evaluation algorithm based on time-lapse images and machine learning. J. Assist. Reprod. Genet. 2023, 40, 2129–2137. [Google Scholar] [CrossRef] [PubMed]
- Ueno, S.; Berntsen, J.; Ito, M.; Okimura, T.; Kato, K. Correlation between an annotation-free embryo scoring system based on deep learning and live birth/neonatal outcomes after single vitrified-warmed blastocyst transfer: A single-centre, large-cohort retrospective study. J. Assist. Reprod. Genet. 2022, 39, 2089–2099. [Google Scholar] [CrossRef] [PubMed]
- Theilgaard Lassen, J.; Fly Kragh, M.; Rimestad, J.; Nygård Johansen, M.; Berntsen, J. Development and validation of deep learning based embryo selection across multiple days of transfer. Sci. Rep. 2023, 13, 4235. [Google Scholar] [CrossRef] [PubMed]
- Petersen, B.M.; Boel, M.; Montag, M.; Gardner, D.K. Development of a generally applicable morphokinetic algorithm capable of predicting the implantation potential of embryos transferred on Day 3. Hum. Reprod. 2016, 31, 2231–2244. [Google Scholar] [CrossRef]
- Li, T.; Liao, R.; Chan, C.; Greenblatt, E.M. Deep learning analysis of endometrial histology as a promising tool to predict the chance of pregnancy after frozen embryo transfers. J. Assist. Reprod. Genet. 2023, 40, 1–10. [Google Scholar] [CrossRef]
- Fu, K.; Li, Y.; Lv, H.; Wu, W.; Song, J.; Xu, J. Development of a Model Predicting the Outcome of In Vitro Fertilization Cycles by a Robust Decision Tree Method. Front. Endocrinol. 2022, 13, 877518. [Google Scholar] [CrossRef]
- Wen, J.Y.; Liu, C.F.; Chung, M.T.; Tsai, Y.C. Artificial intelligence model to predict pregnancy and multiple pregnancy risk following in vitro fertilization-embryo transfer (IVF-ET). Taiwan J. Obstet. Gynecol. 2022, 61, 837–846. [Google Scholar] [CrossRef]
- Goyal, A.; Kuchana, M.; Ayyagari, K.P.R. Machine learning predicts live-birth occurrence before in-vitro fertilization treatment. Sci. Rep. 2020, 10, 20925. [Google Scholar] [CrossRef]
- HFEA. Human Fertilisation & Embryology Authority Dataset. Available online: https://www.hfea.gov.uk/media/2667/ar-2015-2016-xlsb.xlsb (accessed on 31 January 2024).
- Huang, B.; Zheng, S.; Ma, B.; Yang, Y.; Zhang, S.; Jin, L. Using deep learning to predict the outcome of live birth from more than 10,000 embryo data. BMC Pregnancy Childbirth 2022, 22, 36. [Google Scholar] [CrossRef]
- Liu, L.; Jiao, Y.; Li, X.; Ouyang, Y.; Shi, D. Machine learning algorithms to predict early pregnancy loss after in vitro fertilization-embryo transfer with fetal heart rate as a strong predictor. Comput. Methods Programs Biomed. 2020, 196, 105624. [Google Scholar] [CrossRef]
- Loewke, K.; Cho, J.H.; Brumar, C.D.; Maeder-York, P.; Barash, O.; Malmsten, J.E.; Zaninovic, N.; Sakkas, D.; Miller, K.A.; Levy, M.; et al. Characterization of an artificial intelligence model for ranking static images of blastocyst stage embryos. Fertil. Steril. 2022, 117, 528–535. [Google Scholar] [CrossRef] [PubMed]
- Loewke, K.E.; Cho, J.H.; Maeder-York, P.; Barash, O.O.; Meseguer, M.; Malmsten, J.; Miller, K.A.; Sakkas, D.; Levy, M.; VerMilyea, M.D. Identifying Potential Sources of Bias in Deep Learning Models for Embryo Assessment. Fertil. Steril. 2021, 116, e158–e159. [Google Scholar] [CrossRef]
- Abbasi, M.; Saeedi, P.; Au, J.; Havelock, J. A deep learning approach for prediction of IVF implantation outcome from day 3 and day 5 time-lapse human embryo image sequences. In Proceedings of the 2021 IEEE International Conference on Image Processing (ICIP), Anchorage, AL, USA, 19–22 September 2021; IEEE: New York, NY, USA, 2021; pp. 289–293. [Google Scholar]
- Chavez-Badiola, A.; Flores-Saiffe Farias, A.; Mendizabal-Ruiz, G.; Garcia-Sanchez, R.; Drakeley, A.J.; Garcia-Sandoval, J.P. Predicting pregnancy test results after embryo transfer by image feature extraction and analysis using machine learning. Sci. Rep. 2020, 10, 4394. [Google Scholar] [CrossRef] [PubMed]
- Serdarogullari, M.; Raad, G.; Yarkiner, Z.; Bazzi, M.; Mourad, Y.; Alpturk, S.; Fakih, F.; Fakih, C.; Liperis, G. Identifying predictors of Day 5 blastocyst utilization rate using an artificial neural network. Reprod. BioMed. Online 2023, 47, 103399. [Google Scholar] [CrossRef]
- Bamford, T.; Smith, R.; Easter, C.; Dhillon-Smith, R.; Barrie, A.; Montgomery, S.; Campbell, A.; Coomarasamy, A. Association between a morphokinetic ploidy prediction model risk score and miscarriage and live birth: A multicentre cohort study. Fertil. Steril. 2023, 120, 834–843. [Google Scholar] [CrossRef]
- Duval, A.; Nogueira, D.; Dissler, N.; Maskani Filali, M.; Delestro Matos, F.; Chansel-Debordeaux, L.; Ferrer-Buitrago, M.; Ferrer, E.; Antequera, V.; Ruiz-Jorro, M.; et al. A hybrid artificial intelligence model leverages multi-centric clinical data to improve fetal heart rate pregnancy prediction across time-lapse systems. Hum. Reprod. 2023, 38, 596–608. [Google Scholar] [CrossRef]
- Enatsu, N.; Miyatsuka, I.; An, L.M.; Inubushi, M.; Enatsu, K.; Otsuki, J.; Iwasaki, T.; Kokeguchi, S.; Shiotani, M. A novel system based on artificial intelligence for predicting blastocyst viability and visualizing the explanation. Reprod. Med. Biol. 2022, 21, e12443. [Google Scholar] [CrossRef]
- Liu, H.; Zhang, Z.; Gu, Y.; Dai, C.; Shan, G.; Song, H.; Li, D.; Chen, W.; Lin, G.; Sun, Y. Development and evaluation of a live birth prediction model for evaluating human blastocysts from a retrospective study. eLife 2023, 12, e83662. [Google Scholar] [CrossRef]
- Fu, R.; Hu, Q.; Dong, X.; Guo, Y.; Gao, Y.; Li, B. Axiom-based grad-cam: Towards accurate visualization and explanation of cnns. arXiv 2020, arXiv:2008.02312. [Google Scholar]
- Buldo-Licciardi, J.; Large, M.J.; McCulloh, D.H.; McCaffrey, C.; Grifo, J.A. Utilization of standardized preimplantation genetic testing for aneuploidy (PGT-A) via artificial intelligence (AI) technology is correlated with improved pregnancy outcomes in single thawed euploid embryo transfer (STEET) cycles. J. Assist. Reprod. Genet. 2023, 40, 289–299. [Google Scholar] [CrossRef]
- Shen, L.; Zhang, Y.; Chen, W.; Yin, X. The application of artificial intelligence in predicting embryo transfer outcome of recurrent implantation failure. Front. Physiol. 2022, 13, 885661. [Google Scholar] [CrossRef] [PubMed]
- Xi, Q.; Yang, Q.; Wang, M.; Huang, B.; Zhang, B.; Li, Z.; Liu, S.; Yang, L.; Zhu, L.; Jin, L. Individualized embryo selection strategy developed by stacking machine learning model for better in vitro fertilization outcomes: An application study. Reprod. Biol. Endocrinol. 2021, 19, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Yang, L.; Peavey, M.; Kaskar, K.; Chappell, N.; Zhu, L.; Devlin, D.; Valdes, C.; Schutt, A.; Woodard, T.; Zarutskie, P.; et al. Development of a dynamic machine learning algorithm to predict clinical pregnancy and live birth rate with embryo morphokinetics. F&S Rep. 2022, 3, 116–123. [Google Scholar]
- Sawada, Y.; Sato, T.; Nagaya, M.; Saito, C.; Yoshihara, H.; Banno, C.; Matsumoto, Y.; Matsuda, Y.; Yoshikai, K.; Sawada, T.; et al. Evaluation of artificial intelligence using time-lapse images of IVF embryos to predict live birth. Reprod. BioMed. Online 2021, 43, 843–852. [Google Scholar] [CrossRef]
- Fukui, H.; Hirakawa, T.; Yamashita, T.; Fujiyoshi, H. Attention branch network: Learning of attention mechanism for visual explanation. In Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition, Long Beach, CA, USA, 15–20 June 2019; pp. 10705–10714. [Google Scholar]
- Ueno, S.; Berntsen, J.; Ito, M.; Uchiyama, K.; Okimura, T.; Yabuuchi, A.; Kato, K. Pregnancy prediction performance of an annotation-free embryo scoring system on the basis of deep learning after single vitrified-warmed blastocyst transfer: A single-center large cohort retrospective study. Fertil. Steril. 2021, 116, 1172–1180. [Google Scholar] [CrossRef]
- Mehrjerd, A.; Rezaei, H.; Eslami, S.; Ghaebi, N.K. Determination of Cut Off for Endometrial Thickness in Couples with Unexplained Infertility: Trustable AI. In Proceedings of the MIE, Nice, France, 27–30 May 2022; pp. 264–268. [Google Scholar]
- Youngster, M.; Luz, A.; Baum, M.; Hourvitz, R.; Reuvenny, S.; Maman, E.; Hourvitz, A. Artificial intelligence in the service of intrauterine insemination and timed intercourse in spontaneous cycles. Fertil. Steril. 2023, 120, 1004–1012. [Google Scholar] [CrossRef]
- Hariton, E.; Andrusier, M.A.; Khorshid, A. Timing intrauterine inseminations: Do we need an ultrasound, or can artificial intelligence do the trick? Fertil. Steril. 2023, 120, 985–986. [Google Scholar] [CrossRef]
- Kumar, R.S.; Sharma, S.; Halder, A.; Gupta, V. Deep learning-based robust automated system for predicting human sperm DNA fragmentation index. J. Hum. Reprod. Sci. 2023, 16, 16–21. [Google Scholar] [CrossRef]
- Salvaris, M.; Dean, D.; Tok, W.H.; Salvaris, M.; Dean, D.; Tok, W.H. Cognitive services and custom vision. In Deep Learning with Azure: Building and Deploying Artificial Intelligence Solutions on the Microsoft AI Platform; Apress: New York, NY, USA, 2018; pp. 99–128. [Google Scholar]
- Ory, J.; Tradewell, M.B.; Blankstein, U.; Lima, T.F.; Nackeeran, S.; Gonzalez, D.C.; Nwefo, E.; Moryousef, J.; Madhusoodanan, V.; Lau, S.; et al. Artificial intelligence based machine learning models predict sperm parameter upgrading after varicocele repair: A multi-institutional analysis. World J. Men’s Health 2022, 40, 618. [Google Scholar] [CrossRef]
- Naseem, S.; Mahmood, T.; Saba, T.; Alamri, F.S.; Bahaj, S.A.; Ateeq, H.; Farooq, U. DeepFert: An Intelligent Fertility Rate Prediction Approach for Men Based on Deep Learning Neural Networks. IEEE Access 2023, 11, 75006–75022. [Google Scholar] [CrossRef]
- Abou Ghayda, R.; Cannarella, R.; Calogero, A.E.; Shah, R.; Rambhatla, A.; Zohdy, W.; Kavoussi, P.; Avidor-Reiss, T.; Boitrelle, F.; Mostafa, T.; et al. Artificial intelligence in andrology: From semen analysis to image diagnostics. World J. Men’s Health 2024, 42, 39. [Google Scholar] [CrossRef] [PubMed]
- Curchoe, C.L.; Bormann, C.; Hammond, E.; Salter, S.; Timlin, C.; Williams, L.B.; Gilboa, D.; Seidman, D.; Campbell, A.; Morbeck, D. Assuring quality in assisted reproduction laboratories: Assessing the performance of ART Compass—A digital art staff management platform. J. Assist. Reprod. Genet. 2023, 40, 265–278. [Google Scholar] [CrossRef] [PubMed]
- Cherouveim, P.; Jiang, V.S.; Kanakasabapathy, M.K.; Thirumalaraju, P.; Souter, I.; Dimitriadis, I.; Bormann, C.L.; Shafiee, H. Quality assurance (QA) for monitoring the performance of assisted reproductive technology (ART) staff using artificial intelligence (AI). J. Assist. Reprod. Genet. 2023, 40, 241–249. [Google Scholar] [CrossRef] [PubMed]
- Letterie, G.; MacDonald, A.; Shi, Z. An artificial intelligence platform to optimize workflow during ovarian stimulation and IVF: Process improvement and outcome-based predictions. Reprod. BioMed. Online 2022, 44, 254–260. [Google Scholar] [CrossRef]
- Curchoe, C.L.; Tarafdar, O.; Aquilina, M.C.; Seifer, D.B. SART CORS IVF registry: Looking to the past to shape future perspectives. J. Assist. Reprod. Genet. 2022, 39, 2607–2616. [Google Scholar] [CrossRef]
- Liu, K.; Zhang, Y.; Martin, C.; Ma, X.; Shen, B. Translational bioinformatics for human reproductive biology research: Examples, opportunities and challenges for a future reproductive medicine. Int. J. Mol. Sci. 2022, 24, 4. [Google Scholar] [CrossRef]
- Kohli, M.D.; Summers, R.M.; Geis, J.R. Medical Image Data and Datasets in the Era of Machine Learning—Whitepaper from the 2016 C-MIMI Meeting Dataset Session. J. Digit. Imaging 2017, 30, 392–399. [Google Scholar] [CrossRef]
- Muthu, S.; Nabi, F.; Nabi, J. AIM in Obstetrics and Gynecology. In Artificial Intelligence in Medicine; Springer: Berlin/Heidelberg, Germany, 2021; pp. 1–4. [Google Scholar]
- Shafiee, H.; Kanakasabapathy, M.K.; Bormann, C.L.; Topol, E.J. Digitising the human embryo. Lancet 2022, 400, 1577. [Google Scholar] [CrossRef]
- Bhaskar, D.; Chang, T.A.; Wang, S. Current trends in artificial intelligence in reproductive endocrinology. Curr. Opin. Obstet. Gynecol. 2022, 34, 159–163. [Google Scholar] [CrossRef]
- Sadeghi, M.R. Will Artificial Intelligence Change the Future of IVF? J. Reprod. Infertil. 2022, 23, 139. [Google Scholar] [CrossRef]
- Wilkinson, M.D.; Dumontier, M.; Aalbersberg, I.J.; Appleton, G.; Axton, M.; Baak, A.; Blomberg, N.; Boiten, J.W.; da Silva Santos, L.B.; Bourne, P.E.; et al. The FAIR Guiding Principles for scientific data management and stewardship. Sci. Data 2016, 3, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Kromp, F.; Wagner, R.; Balaban, B.; Cottin, V.; Cuevas-Saiz, I.; Schachner, C.; Fancsovits, P.; Fawzy, M.; Fischer, L.; Findikli, N.; et al. An annotated human blastocyst dataset to benchmark deep learning architectures for in vitro fertilization. Sci. Data 2023, 10, 271. [Google Scholar] [CrossRef] [PubMed]
- Touvron, H.; Cord, M.; Douze, M.; Massa, F.; Sablayrolles, A.; Jégou, H. Training data-efficient image transformers & distillation through attention. In Proceedings of the International Conference on Machine Learning, PMLR, Online, 18–24 July 2021; pp. 10347–10357. [Google Scholar]
- Varoquaux, G.; Cheplygina, V. Machine learning for medical imaging: Methodological failures and recommendations for the future. Npj Digit. Med. 2022, 5, 48. [Google Scholar] [CrossRef] [PubMed]
- Price, W.N.; Cohen, I.G. Privacy in the age of medical big data. Nat. Med. 2019, 25, 37–43. [Google Scholar] [CrossRef]
- Gerke, S.; Minssen, T.; Cohen, G. Ethical and legal challenges of artificial intelligence-driven healthcare. In Artificial Intelligence in Healthcare; Elsevier: Amsterdam, The Netherlands, 2020; pp. 295–336. [Google Scholar]
- Mostert, M.; Bredenoord, A.L.; Biesaart, M.C.; Van Delden, J.J. Big Data in medical research and EU data protection law: Challenges to the consent or anonymise approach. Eur. J. Hum. Genet. 2016, 24, 956–960. [Google Scholar] [CrossRef]
- Goldberger, A.L.; Amaral, L.A.; Glass, L.; Hausdorff, J.M.; Ivanov, P.C.; Mark, R.G.; Mietus, J.E.; Moody, G.B.; Peng, C.K.; Stanley, H.E. PhysioBank, PhysioToolkit, and PhysioNet: Components of a new research resource for complex physiologic signals. Circulation 2000, 101, e215–e220. [Google Scholar] [CrossRef]
- Li, X.; Xiong, H.; Li, X.; Wu, X.; Zhang, X.; Liu, J.; Bian, J.; Dou, D. Interpretable deep learning: Interpretation, interpretability, trustworthiness, and beyond. Knowl. Inf. Syst. 2022, 64, 3197–3234. [Google Scholar] [CrossRef]
- Smith, H.; Fotheringham, K. Artificial intelligence in clinical decision-making: Rethinking liability. Med. Law Int. 2020, 20, 131–154. [Google Scholar] [CrossRef]
- Maliha, G.; Gerke, S.; Cohen, I.G.; Parikh, R.B. Artificial Intelligence and Liability in Medicine. Milbank Q. 2021, 99, 629–647. [Google Scholar] [CrossRef]
- Price, W.N.; Gerke, S.; Cohen, I.G. Potential liability for physicians using artificial intelligence. JAMA 2019, 322, 1765–1766. [Google Scholar] [CrossRef]
- Cestonaro, C.; Delicati, A.; Marcante, B.; Caenazzo, L.; Tozzo, P. Defining medical liability when artificial intelligence is applied on diagnostic algorithms: A systematic review. Front. Med. 2023, 10, 1305756. [Google Scholar] [CrossRef] [PubMed]
- Serdarogullari, M.; Ammar, O.F.; Sharma, K.; Kohlhepp, F.; Montjean, D.; Meseguer, M.; Fraire-Zamora, J.J. #ESHREjc report: Seeing is believing! How time lapse imaging can improve IVF practice and take it to the future clinic. Hum. Reprod. 2022, 37, 1370–1372. [Google Scholar] [PubMed]
- Bori, L.; Meseguer, M. Will the introduction of automated ART laboratory systems render the majority of embryologists redundant? Reprod. BioMedicine Online 2021, 43, 979–981. [Google Scholar] [CrossRef] [PubMed]
- Allahbadia, G.N.; Allahbadia, S.G.; Gupta, A. In Contemporary Reproductive Medicine Human Beings are Not Yet Dispensable. J. Obstet. Gynecol. India 2023, 73, 295–300. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).