Abstract
Cardiovascular diseases stand as the leading cause of mortality worldwide, underscoring the urgent need for effective tools that enable early detection and monitoring of at-risk patients. This study combines Artificial Intelligence (AI) techniques—specifically the k-means clustering algorithm—alongside dimensionality reduction methods like Principal Component Analysis (PCA) and Uniform Manifold Approximation and Projection (UMAP) to identify patient groups with varying levels of heart attack risk. We used a publicly available clinical dataset with 1319 patient records, which included variables such as age, gender, blood pressure, glucose levels, CK-MB Creatine Kinase MB (KCM), and troponin levels. We normalized and prepared the data, then we employed PCA and UMAP to reduce dimensionality and facilitate visualization. Using the k-means algorithm, we segmented the patients into distinct groups based on their clinical features. Our analysis revealed two distinct patient groups. Group 2 exhibited significantly higher levels of troponin (mean 0.4761 ng/mL), KCM (18.65 ng/mL), and glucose (mean 148.19 mg/dL) and was predominantly composed of men (97%). These factors indicate an increased risk of cardiac events compared to Group 1, which had lower levels of these biomarkers and a slightly higher average age. Interestingly, no significant differences in blood pressure were observed between the groups. This study demonstrates the effectiveness of combining Machine Learning (ML) techniques with dimensionality reduction methods to enhance risk stratification accuracy in cardiology. By enabling more targeted interventions for high-risk patients, our unsupervised segmentation approach focuses on intrinsic data patterns rather than predefined diagnostic labels, serves as a powerful complement to traditional risk assessment tools.
1. Introduction
Cardiovascular diseases (CVDs) represent one of the most significant public health challenges of the 21st century. According to the World Health Organization (WHO), CVDs are the leading cause of death globally, accounting for approximately 31% of all deaths worldwide [1]. These conditions encompass a wide range of disorders affecting the heart and blood vessels, including coronary heart disease, cerebrovascular disease, rheumatic heart disease, and other conditions. The high prevalence and mortality associated with CVDs underscore the urgent need to improve strategies for prevention, diagnosis, and treatment.
The impact of CVDs extends beyond mortality; they also affect patients’ quality of life and impose a considerable economic burden on healthcare systems. Risk factors such as high blood pressure, elevated blood glucose, high cholesterol levels, smoking, obesity, and physical inactivity significantly contribute to the development and progression of these diseases [2]. Moreover, the complex interplay between genetic, environmental, and lifestyle factors makes early identification of at-risk individuals challenging.
In this context, early detection and effective monitoring of patients at risk of heart attacks are crucial to reducing the incidence and mortality associated with CVDs. Preventive interventions and timely treatments can substantially improve clinical outcomes and alleviate the strain on healthcare systems. However, traditional risk assessment methods often fail to capture the full complexity of the factors involved, limiting their effectiveness.
Artificial Intelligence (AI) and Machine Learning (ML) emerge as promising tools to address these challenges. AI enables the analysis of large volumes of data and the discovery of hidden patterns that may go unnoticed using conventional statistical methods. ML, as a subfield of AI, focuses on developing algorithms capable of learning from data and improving their accuracy over time without being explicitly programmed for each task [3]. These techniques have proven effective in various medical applications, such as computer-assisted diagnosis, prediction of clinical outcomes, and treatment personalization [4,5].
Specifically, the use of unsupervised learning algorithms, such as clustering, allows for the identification of patient groups with similar characteristics without the need for predefined labels. The k-means algorithm is one of the most widely used methods in this field due to its simplicity and efficiency in handling large datasets [6]. By combining it with dimensionality reduction techniques like Principal Component Analysis (PCA) and Uniform Manifold Approximation and Projection (UMAP), it is possible to simplify high-dimensional data and better visualize relationships between variables [7,8].
The present study aims to apply AI techniques—specifically the k-means clustering algorithm along with PCA and UMAP—to identify groups of patients with different levels of risk for heart attacks. By analyzing clinical and laboratory variables such as age, gender, blood pressure, glucose levels, and troponin levels, we seek to detect patterns and groupings that may not be evident through traditional analyses. This approach can contribute to improving risk stratification and developing more effective prevention strategies in cardiology. Particularly, our unsupervised segmentation approach, by focusing on these intrinsic data patterns rather than being constrained by predefined diagnostic labels, can serve as a powerful complement to traditional risk assessment tools [9,10].
The main contributions of this study are as follows:
- ➢
- Effective Dimensionality Reduction Using UMAP: We successfully applied the nonlinear dimensionality reduction technique UMAP to the clinical dataset, reducing complexity while preserving essential data structures. This allowed for clearer visualization and better handling of nonlinear relationships between medical variables.
- ➢
- Identification of Patient Groups with Varying Cardiac Risk Levels: By utilizing the k-means clustering algorithm on the reduced dataset, we identified two distinct groups of patients with different levels of risk for heart attacks. This highlights the potential of unsupervised ML methods in uncovering hidden patterns in medical data.
- ➢
- Insights into Critical Biomarkers for Heart Attack Risk: We identified troponin, CK-MB Creatine Kinase MB (KCM), and glucose levels, along with gender, as significant factors in stratifying cardiovascular risk among patients. This finding can aid clinicians in focusing on key biomarkers for early detection and intervention.
- ➢
- Contribution to Personalized Medicine and Preventive Cardiology: Our approach demonstrates how AI and machine learning techniques can enhance risk stratification accuracy, leading to more targeted interventions for high-risk patients and improved prevention strategies in cardiology.
These results can benefit healthcare professionals by providing advanced tools for patient risk assessment and supporting the integration of AI in clinical decision-making processes.
The rest of this article is organized as follows: Section 2 details the methodology employed, including a description of the dataset and the analytical techniques used. Section 3 presents the results obtained and their interpretation. In Section 4, we discuss the clinical implications of our findings. Finally, Section 5 concludes by highlighting the main contributions of the study and suggesting directions for future research.
2. Materials and Methods
In this section, we detail the process we followed to evaluate the effectiveness of the clustering model and identify significant patterns in the medical variables of each patient. To achieve the study’s objectives and validate our hypothesis, we developed a methodology that integrates several techniques, including Exploratory Data Analysis (EDA), dimensionality reduction, and machine learning algorithms. The focus of the study was to identify hidden patterns and natural groupings in a clinical dataset, aiming to classify patients into different risk groups for heart attacks.
Figure 1 presents a block diagram illustrating the workflow of the methodology used in this study. This diagram allows us to visualize the interrelationships between each stage of the process, facilitating replication of the methodology by other researchers. The implementation was entirely coded in Python 3.11.12 using open-source libraries (e.g., scikit-learn for UMAP and k-means, Matplotlib/Seaborn for visualizations).
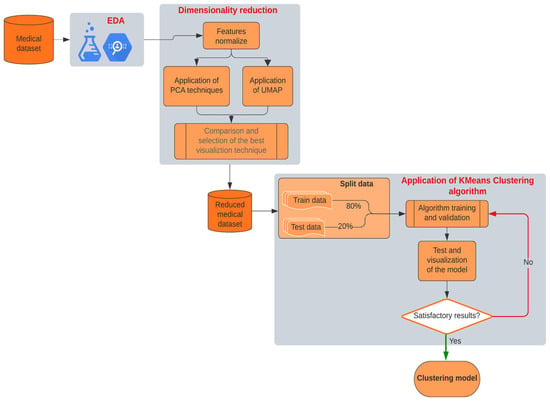
Figure 1.
Block diagram of our proposal methodology.
We explain each of the phases in detail below, highlighting the key contributions and aspects of each process.
- Exploratory Data Analysis (EDA): We began the study with a thorough analysis of the medical data, identifying the most relevant features for subsequent analysis. During this phase, we evaluated the distribution of variables, checked for missing data, and explored possible relationships between variables. Our goal in the EDA was to gain a clear understanding of the data and prepare the dataset for the next phases of analysis. Detecting outliers and normalizing variables were critical steps to ensure data homogeneity and readiness for modeling. By thoroughly understanding the data’s characteristics, we aimed to minimize biases and enhance the accuracy of our modeling efforts.
- Dimensionality Reduction: The original dataset contained multiple medical features that could have nonlinear relationships. To facilitate clustering and pattern visualization, we implemented dimensionality reduction techniques. We applied the Uniform Manifold Approximation and Projection (UMAP), a nonlinear dimensionality reduction method ideal for preserving both local and global data structures. We initially applied Principal Component Analysis (PCA) for dimensionality reduction, but observed overlapping clusters due to its linearity assumption, which failed to capture critical nonlinear interactions (e.g., troponin’s exponential relationship with glucose levels in high-risk patients). Switching to UMAP, which models local and global structures, improved cluster separation and revealed two distinct risk phenotypes. This phase reduced the dataset’s complexity to a two-dimensional space, simplifying the subsequent clustering stage and enhancing pattern discernibility.
- K-means Clustering Algorithm: After reducing the data to two dimensions using UMAP, we applied the k-means clustering algorithm to identify natural groupings among the patients. K-means partitions the data into a predefined number of clusters, aiming to minimize intra-cluster distances and maximize separation between clusters. In this study, we chose to establish two clusters after evaluating several options and determining that this number provided the most meaningful segmentation of patients based on critical biomarkers like troponin, KCM, and glucose levels. This decision was guided by methods such as the elbow method and silhouette analysis, ensuring that our clustering approach was both data-driven and clinically relevant.
- Validation and Visualization: After applying k-means, we validated the model using internal validation metrics and visual inspection of the clusters. We adjusted based on the cluster cohesion and separation observed in the visualization. Once satisfied with the cluster quality, we proceeded to interpret the clusters clinically, identifying relevant patterns and differences between patient groups that could inform risk stratification and intervention strategies.
This methodological approach enabled us to achieve a meaningful classification of patients based on their cardiac risk, highlighting the utility of AI techniques in the early identification of risk factors in the clinical setting. By combining UMAP and k-means, we effectively segmented the patient population and achieved a clear visualization of the results, providing valuable insights that could enhance clinical decision-making and patient outcomes.
Although PCA and UMAP are established techniques for dimensionality reduction, their application to explore linear patterns and nonlinear relationships has seen growing interest in recent years, with emerging studies applying these techniques to explore clinical data. Prior studies have focused on linear methods (e.g., PCA + k-means) for clinical data visualization [11], but these approaches fail to capture nonlinear interactions between biomarkers, which are critical for identifying subclinical risk patterns. Our work addresses this gap by proposing a workflow that prioritizes UMAP over PCA when data exhibit nonlinear relationships, demonstrating its superiority in cohorts with complex metabolic profiles. Table A1 in Appendix A provides a comparative analysis of related works, highlighting this study’s main contribution and the research gap addressed by our novel integration.
2.1. Medical Dataset and EDA
This work uses the clinical dataset Heart Attack Analysis & Prediction Dataset, provided by Rashik Rahman Pritom, available on Kaggle under the CC BY 4.0 license. The dataset contains 1319 patient records, each containing eight relevant variables selected based on their significance in cardiovascular health assessment. These variables are [12,13]:
- Age: Patient’s age in years.
- Gender: Male or Female (represented as 0 and 1, respectively).
- Pulse Rate: Heart rate measured in beats per minute.
- High Blood Pressure (Systolic Pressure): Maximum arterial pressure during heart contraction.
- Low Blood Pressure (Diastolic Pressure): Minimum arterial pressure between heartbeats.
- Glucose Level: Blood glucose concentration in mg/dL.
- CK-MB (Creatine Kinase MB) (KCM): an enzyme primarily found in the heart and, to a lesser extent, in skeletal muscles.
- Troponin Level: Blood troponin concentration in ng/mL, a specific biomarker for myocardial damage.
The dataset’s dimensions (1319 samples × 8 features) provide a robust foundation for applying ML techniques. The data were retrospectively collected from electronic medical records of local hospitals, ensuring diversity and representativeness within the sample population.
We conducted an observational, descriptive, and cross-sectional study. No direct interventions were made with the patients; instead, we analyzed existing data to identify groupings based on similarities in clinical and laboratory variables.
Although the dataset is publicly available, its application in this context (stratification based on troponin/glucose levels) is novel. Previous studies using these data have focused on supervised models [14], while our unsupervised approach uncovers hidden patterns without the bias of prior labeling. An excerpt of the first ten rows of the dataset is presented in Table A2 of Appendix A, illustrating the structure and type of data used in this study.
Before proceeding with the modeling phase, we performed an extensive EDA to understand the underlying patterns and distributions within the dataset. This step was crucial for identifying data quality issues, uncovering relationships between variables, and informing subsequent analytical choices.
We conducted an EDA to understand the distribution and relationships among the variables. Using histograms, box-and-whisker plots, and correlation matrices, we identified patterns and potential outliers in the data.
Since the variables were on different scales, we applied normalization using the StandardScaler method. This process allowed us to standardize the data, ensuring that each variable contributed equally to the analysis and enhancing the effectiveness of the clustering algorithms.
We identified and analyzed outliers to determine whether they should be excluded or if they provided relevant information. Outliers can offer valuable insights into extreme or unusual cases that might significantly influence the study’s results (see Figure A1 in Appendix B). We decided to retain these outliers because they could represent patients with higher risk and are essential for the integrity of the analysis.
To identify linear relationships between variables, we calculated the Pearson correlation matrix. We observed a moderate correlation between systolic and diastolic blood pressure (r ≈ 0.59), suggesting that patients with high systolic pressure tend to have high diastolic pressure. The other variables showed low correlations, indicating that each contributes unique information to the dataset and is valuable for the clustering process.
By thoroughly understanding and preprocessing the data, we ensured that the dataset was suitable for dimensionality reduction and clustering. The insights gained from the EDA guided our methodological choices and helped us interpret the results within a clinical context.
2.2. Dimensionality Reduction
In clinical datasets with multiple variables, it is common for some variables to correlate with each other, leading to redundant information that can complicate analysis. Dimensionality reduction techniques help simplify high-dimensional data while preserving as much relevant information as possible. This simplification facilitates data processing, enhances the performance of machine learning algorithms, and makes data visualization more manageable [15].
Although the dataset contains only 8 variables, dimensionality reduction was essential to visualize nonlinear patterns in an interpretable 2D space. This allowed groups of patients to be identified who, although not highly linearly correlated, share similar risk profiles when complex multivariate interactions are considered.
Principal Component Analysis (PCA) is a widely used statistical method that transforms a set of possibly correlated variables into a set of uncorrelated variables known as principal components [16]. These components are linear combinations of the original variables and are ordered so that the first principal component captures the maximum possible variance in the data, the second component captures the next highest variance, and so on. We selected the initial components that explained at least 95% of the total variance, aiming to reduce dimensionality while retaining most of the information [17].
Uniform Manifold Approximation and Projection (UMAP) is a nonlinear dimensionality reduction technique that constructs a high-dimensional graph based on probabilistic distances between data points and optimizes a low-dimensional representation preserving both local and global data structures [8]. Mathematically, UMAP minimizes the cross-entropy cost function between two fuzzy sets, one representing high-dimensional similarities, , and the other low-dimensional similarities, :
where is the probability of association between points i and j in high-dimensional space, and is the corresponding probability in the reduced space [8]. This formulation, derived from fuzzy set theory and Riemannian geometry, allows UMAP to capture nonlinear relationships between clinical variables (e.g., troponin–glucose interactions), which often exhibit non-additive relationships in cardiovascular risk assessment [18].
Compared to linear methods like PCA [15], UMAP better preserves both local and global structures in biomedical datasets, a critical advantage for visualizing natural patient groupings [19]. We used UMAP to visualize the data in a two-dimensional space, facilitating the identification of natural groupings among patients.
2.3. Application of K-Means
The k-means algorithm clusters data into k groups by iteratively minimizing the sum of squared distances between data points and the centroid of their assigned cluster [20,21]. This method aims to partition the dataset into distinct, non-overlapping subsets where each data point belongs to the cluster with the nearest mean value. Formally, given a dataset X containing n x-dimensional data points , where i = 1, 2, …, n, X is partitioned into ‘k’ clusters , where j = 1, 2, …, k such that:
The primary goal is to minimize the total within-cluster variance (J), ensuring that data points within each cluster are as homogeneous as possible while maintaining separation between clusters. That is, minimize Equation (3):
where denotes the centroid (mean) of cluster . The algorithm proceeds as follows [21]:
- Initialization: Randomly select k initial centroids from the dataset.
- Assignment: For each data point, compute its Euclidean distance to all centroids and assign it to the nearest cluster.
- Update: Recalculate each centroid as the arithmetic mean of all points assigned to its cluster.
- Iteration: Repeat steps 2–3 until cluster assignments stabilize (i.e., centroids no longer change significantly between iterations).
To identify the optimal number of clusters or groups (k), we employed the elbow method and the silhouette coefficient:
- Elbow Method: This technique involves running k-means clustering on the dataset for a range of k values and computing the within-cluster sum of squares (WCSS). By plotting WCSS against the number of clusters, we look for an “elbow” point where the rate of decrease sharply changes, indicating diminishing returns with additional clusters [22]. In our analysis, the elbow point suggested that k = 2 was optimal.
- Silhouette Coefficient: This metric measures how well each data point fits within its assigned cluster compared to other clusters. It ranges from −1 to 1, where a higher value indicates better clustering quality [23]. We calculated the silhouette scores for different k values and found that the highest average silhouette score occurred at k = 2, reinforcing the result from the elbow method.
By combining these two methods, we confidently determined that dividing the patients into two clusters was the most meaningful approach for our dataset. This allowed us to effectively group patients based on critical biomarkers such as troponin, KCM, and glucose levels, which are significant indicators of cardiac risk.
3. Simulation Results
In this section, we explain the steps taken to execute the entire process outlined in the block diagram of Figure 1. The process is divided into three main parts: EDA, dimensionality reduction, and application of ML algorithms. Each subsection provides a detailed account of the methodologies employed and the results obtained, offering insights into how each step contributes to identifying patient groups with varying levels of cardiac risk.
We begin with the EDA to understand the dataset’s characteristics and prepare the data for analysis. Next, we apply dimensionality reduction techniques to simplify the dataset while preserving essential information. Finally, we utilize machine learning algorithms to cluster the patients and interpret the results within a clinical context.
3.1. EDA Application
In this subsection, we present the correlation matrix between the features in the dataset. Also, we show a statistical summary of all variables in the dataset and the outlier analysis.
We plotted the correlation matrix to understand how closely related the variables are to each other. In a correlation matrix, values closer to 1 indicate a stronger positive correlation between variables, meaning they tend to increase together. Correlation matrices are fundamental tools in exploratory data analysis, helping to identify relationships between variables and detect potential multicollinearity issues [24].
From the correlation matrix (see Figure 2), we observed that the variables pressurehigh (systolic blood pressure) and pressurelow (diastolic blood pressure) have a correlation coefficient of approximately 0.59. Although this correlation is not extremely high, it indicates a moderate relationship between the two variables. This makes clinical sense, as patients with higher systolic pressure also have higher diastolic pressure [25]. This correlation reflects the physiological relationship between the two measures of blood pressure, influenced by factors such as arterial stiffness and vascular resistance.
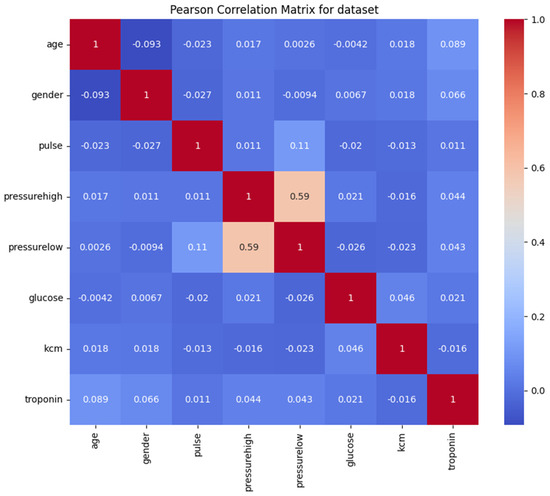
Figure 2.
Correlation matrix of the features.
Depending on the analytical approach, one might consider whether both variables are necessary or if one could suffice. However, since the correlation is moderate rather than strong, both variables may still provide unique information and contribute valuable insights to the analysis.
The other variables did not show significant correlations with each other (see Figure 2), suggesting that there is no strong collinearity in the dataset. This lack of high correlation among most variables is advantageous for clustering purposes because it indicates that each variable contributes distinct information. When variables are not highly correlated, clustering algorithms like k-means can more effectively utilize the unique characteristics of each variable to segment patients into meaningful groups.
Understanding the correlations between variables helped us ensure that the dataset was suitable for clustering without the need to remove or combine variables due to redundancy [26]. It also provided confidence that each variable could potentially influence the formation of clusters, aiding in the identification of patient groups with different risk profiles.
To further understand the dataset, we performed a statistical analysis of all variables, focusing on measures such as mean, standard deviation (std), minimum and maximum values, and quartiles. Table 1 below presents a comprehensive summary of these statistical metrics, offering deeper insights into the distribution and variability within the data.

Table 1.
Statistical summary of dataset variables.
From the statistical data, we notice that the pulse variable exhibits a high standard deviation (51.63) relative to its mean (78.34). Additionally, the maximum value of pulse is 1111, which is significantly greater than the third quartile (85). This disparity suggests the presence of outliers that could be affecting the distribution of this variable.
Similarly, the variables glucose, KCM, and troponin show maximum values considerably higher than their respective third quartile values. This indicates the presence of outliers that might influence the interpretation of these variables.
By carefully analyzing these statistical summaries, we gained valuable insights into the dataset’s characteristics, which guided our subsequent steps in the analysis. Recognizing the presence of outliers and understanding their potential significance helped us make informed decisions about data preprocessing and ensured that our clustering results would be meaningful in a clinical context. Appendix B shows all the outliers presented in the features of this dataset (see Figure A1).
Given that our objective is to identify groups of patients, we decided to retain these outliers because they could represent a specific subgroup within the dataset. These extreme values might correspond to patients with higher risk profiles, and excluding them could lead to a loss of critical information.
By maintaining the outliers, we aim to ensure that the clustering algorithm captures the full spectrum of patient data, potentially revealing important patterns associated with elevated cardiac risk.
3.2. Dimensionality Reduction Applications
In this section, we normalized all the data to the same scale. This step is essential because the algorithms need to interpret and differentiate the meaning of each numerical variable, given that they were originally on different scales. By standardizing the data using Standardscaler normalization, we ensured that each variable contributed equally to the analysis, preventing variables with larger scales from dominating the results.
With all data on the same scale, we proceeded to visualize the variance distributed among each principal component. PCA operates by transforming the original variables into a new set of uncorrelated variables called principal components, ordered by the amount of variance they capture from the data [16]. In other words, the more variance a component captures, the more it contributes to the segmentation and separation of the data.
In our study, since we have eight variables, we can obtain a maximum of eight principal components. However, the human eye can only visualize up to three dimensions effectively, making it challenging to interpret more than three components visually. Therefore, we focused on the first few components that capture the most variance.
Ideally, the first two or three principal components should capture a substantial portion of the total variance to allow for meaningful visualization and analysis [7]. In Figure 3, we present the variance of each principal component individually and how they accumulate to reach 100% of the total variance.
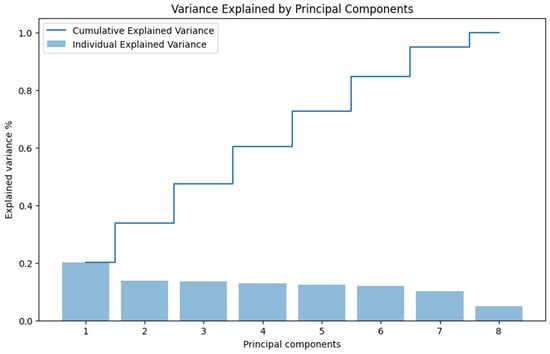
Figure 3.
Variance explained by each principal component.
Observing the figure, we notice that the sum of the first two components barely reaches 40% of the accumulated variance. This low percentage indicates that the data are not well represented in just two dimensions using PCA, as a significant amount of information (variance) remains in the higher components.
The limited variance captured by the first two principal components suggests that PCA may not be the most effective dimensionality reduction technique for our dataset, especially if nonlinear relationships exist between variables [27]. This finding led us to consider alternative methods better suited for preserving complex data structures.
In Figure 4, we present the plot of the data distributed using the first two principal components, which account for almost 40% of the total variance. The figure shows a high concentration of data points clustered together, with a few points scattered away from this dense area. The points are quite dispersed overall, and there do not appear to be evident clusters indicating a natural separation between groups in the data.
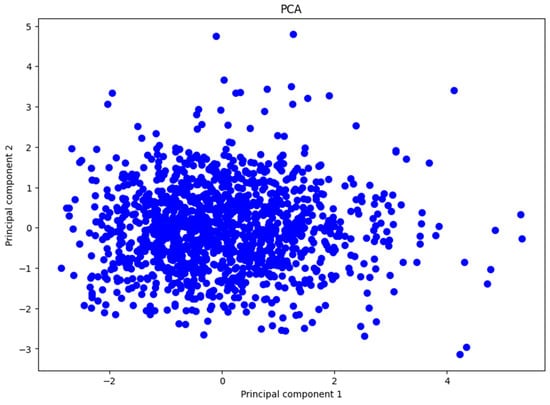
Figure 4.
Representation of the first two principal components.
This observation is consistent with our earlier findings, where we noted that the first two principal components capture a relatively small portion of the total variance. Because these components do not explain a significant amount of variance, important information may be lost when projecting the data onto this lower-dimensional space. This loss of information makes it difficult to distinguish distinct groups or patterns within the dataset using PCA.
Based on these insights, we conclude that PCA is not effective for reducing the dimensionality of our dataset in this specific problem. The ineffectiveness of PCA may be attributed to the data not exhibiting linear relationships among variables. Since PCA is a linear dimensionality reduction technique, it struggles to capture the complex, nonlinear structures that may exist in the data [16,27]. This limitation suggests the need for alternative methods that can handle nonlinear relationships more effectively.
UMAP
We decided to apply the UMAP technique, which proved to be more effective for our problem due to two main reasons [8,28]:
- Preservation of Local and Global Structure: UMAP is specifically designed to preserve both the local and global structures of the data. This means it attempts to maintain close relationships between similar data points as well as the broader relationships among groups of points in the high-dimensional space. By doing so, UMAP provides a more faithful representation of the data’s intrinsic geometry in a lower-dimensional space. This characteristic is crucial when dealing with complex datasets where important patterns may exist at different scales.
- Manifold Approximation: UMAP operates under the assumption that the data lie on a low-dimensional manifold within the high-dimensional space. It seeks to find a representation of this manifold in a lower-dimensional space. This approach can result in a clearer separation of clusters or patterns, making it easier to identify distinct groups within the data. UMAP’s ability to capture nonlinear relationships enhances the visualization and interpretability of data.
After applying UMAP to the normalized data, we obtained Figure 5. From this figure, we can clearly observe two distinct groups of patients, one on the left and one on the right. This outcome indicates that UMAP successfully reduced the dimensionality of the dataset while preserving meaningful structures relevant for clustering.
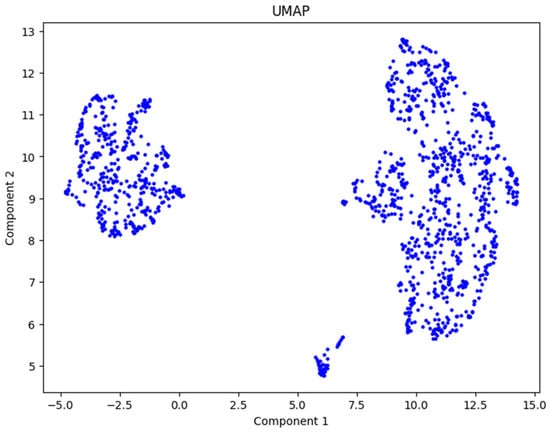
Figure 5.
UMAP representation by two components.
Using UMAP, we achieved our objective of dimensionality reduction for this dataset. The technique effectively unveiled the underlying structure of the data, showing the presence of two natural groupings among the patients. This result sets a strong foundation for the next step, which involves applying the k-means algorithm to divide the patients into clusters and interpret the findings.
3.3. K-Means Application
Although Figure 5 suggests the presence of two distinct patient groups, we verified this observation using the elbow method and the silhouette score, as explained in the methodology section. These methods helped us determine the optimal number of clusters (k) for our dataset.
After calculating and plotting these indicators, as shown in Figure 6, we found that both methods indicated that k = 2 is the optimal number of clusters. This finding aligns with our initial hypothesis based on the UMAP visualization.
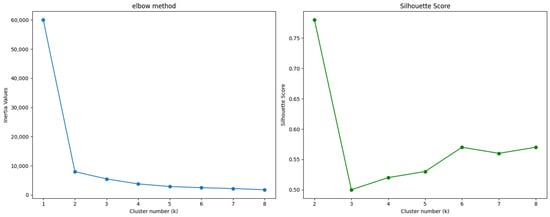
Figure 6.
Elbow method and Silhouette score.
With the optimal number of clusters determined, we applied the k-means algorithm to divide the patients into two groups. By confirming the optimal number of clusters through these methods, we ensured that our clustering approach was robust and data-driven. The resulting clusters provided a foundation for analyzing the characteristics of each group and interpreting their clinical significance. The clustering results are presented in Figure 7.
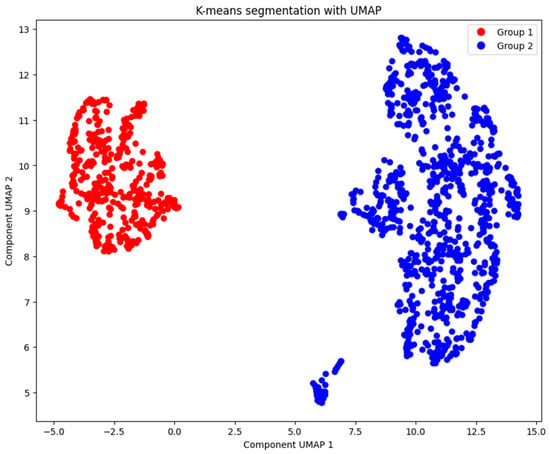
Figure 7.
Patients’ segmentation with k-means (k = 2).
Our unsupervised approach offers key advantages over traditional supervised models. First, it allows us to uncover hidden patterns that escape label-based methods: we identify high-risk patients with moderate elevation of troponin combined with severe hyperglycemia, a profile that supervised models might ignore by focusing only on binary diagnoses of infarction. Second, by not relying on predefined clinical labels, this approach is ideal for analyzing retrospective data with incomplete or biased diagnoses, common in hospital registries. This allows emerging risks to be detected in subclinical populations, offering opportunities for preventive interventions before major cardiovascular events occur.
The k-means algorithm has two key limitations in the context of clinical data. First, it assumes that the clusters are spherical and homogeneously sized (convex clusters), a simplification that may not reflect the complexity of biomedical patterns with irregular shapes or asymmetric distributions. Second, although we base the choice of k = 2 on the elbow method and silhouette coefficient, this decision could underestimate the heterogeneity of subpopulations in large-scale studies, where a higher k value could reveal more granular risk stratifications. These limitations were partially mitigated by clinical validation of the identified groups, prioritizing medical relevance over purely statistical criteria.
4. Discussion
In this section, we present a summarized comparison of the two patient groups identified and discuss the main differences observed. It is important to preface this discussion by noting that our primary aim was not to replicate a supervised classification of a single outcome (such as the presence or absence of a past heart attack), but rather to leverage UMAP and k-means to uncover potentially more nuanced patient segments that reflect the risk for a broader spectrum of cardiovascular risk profiles based on their comprehensive data signatures. This aligns with a growing body of research demonstrating the power of unsupervised learning to identify novel signatures of health and disease from complex biomedical data, even without explicit diagnostic labels [9], and to delineate cardiovascular risk patterns in populations [29].
The dataset’s ‘positive/negative’ labels, referring to the occurrence of a heart attack, represent a crucial clinical endpoint; however, cardiovascular health encompasses a wider array of conditions and risk gradations, and AI, including unsupervised methods, is increasingly recognized for its potential to enhance clinical value across this entire spectrum [30,31]. Our methodology sought to identify these underlying data-driven patient groupings, which may offer insights beyond the binary classification of a single outcome. Table 2 shows the average values of key variables for each group.

Table 2.
Average Values of Key Variables in the Segmented Groups.
The results obtained using UMAP and k-means clustering suggest that patients in Group 2 are at higher risk due to elevated levels of troponin, KCM, and glucose, three significant indicators for cardiac issues. Although Group 1 has a slightly higher average age, it does not exhibit the same elevated levels of these critical biomarkers.
The stark difference in gender distribution between the two groups is noteworthy; Group 2 consists of 97% male patients, whereas only 3% of Group 1 are male. This disparity could be clinically significant and may relate to gender-specific risk factors for cardiovascular diseases. Existing literature indicates that men have a higher risk of certain cardiac events compared to women, potentially due to differences in hormonal profiles, lifestyle factors, and prevalence of risk behaviors [32]. Exploring this aspect further would be valuable in future studies.
Interestingly, both groups have similar average blood pressure readings, implying that blood pressure may not be the distinguishing factor between these clusters. Instead, biomarkers like troponin and glucose appear to play a more pivotal role in differentiating patient risk profiles in this dataset.
Applying UMAP as a dimensionality reduction technique was crucial for clearly identifying the differences between the groups. By preserving both local and global data structures, UMAP facilitated the effective use of the k-means algorithm for patient segmentation. The combination of these AI techniques allowed us to uncover patterns that might not have been apparent using traditional linear methods like PCA.
4.1. Quantitative Alignment of Data-Driven Clusters with Heart Attack Labels
This data-driven segmentation, free from the constraints of predefined outcome labels for a single condition, allows for the discovery of emergent patient profiles that may represent different points along the cardiovascular risk continuum or distinct etiological pathways. Indeed, unsupervised learning has proven effective in identifying clinically relevant subgroups in conditions like coronary artery disease [33] and in discovering groups in patients with subclinical cardiac dysfunction before evident clinical progression [34], highlighting its utility for personalized prevention and early risk stratification.
To address the need for quantitative validation against available ground truth, we evaluated how our two k-means clusters align with the dataset’s binary ‘positive’/’negative’ labels for historical heart attack. A contingency analysis (Table 3) revealed the following distribution: Cluster 2 contained 308 ‘negative’ and 586 ‘positive’ patients, while Cluster 1 comprised 201 ‘negative’ and 224 ‘positive’ patients.

Table 3.
Contingency analysis of clusters and labels.
However, when we compared these clusters against the original clinical labels (positive/negative for cardiac event), we observed a purity of 60%, indicating substantial overlap rather than a clean separation (see Figure 8). The high-risk aligned Cluster 2 demonstrated a sensitivity (recall) of 72.3% in capturing patients with a ‘positive’ heart attack label, and a precision (positive predictive value) of 65.5%.
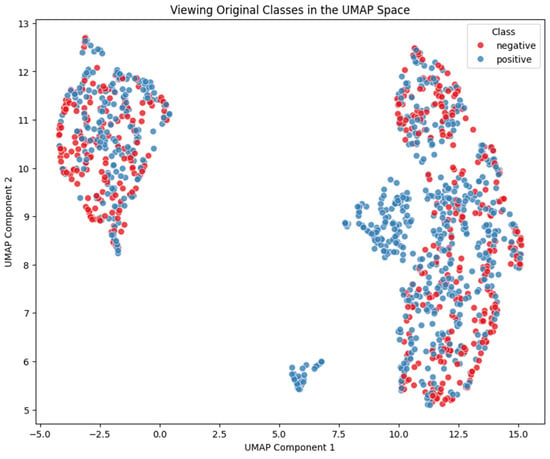
Figure 8.
Mapping of the classes into the clusters.
It is crucial to emphasize that k-means in this context is not intended as a supervised classifier for predicting cardiac events, but rather as an exploratory tool to reveal latent structure in the biomarker space reduced by UMAP. Thus, a 60% purity and a precision of 65.5% (which is only marginally above the baseline prevalence of ‘positive’ cases) should not be viewed as a 40% “error rate”, but as a measure of concordance between unsupervised groupings and predefined clinical categories [35].
This divergence highlights that the UMAP + k-means clusters capture a different, potentially orthogonal, structure from the binary labels. The fact that both clusters contain a majority of ‘positive’ cases yet are clearly distinguished by the k-means algorithm based on UMAP-transformed biomarker (as shown in Figure 7 and Table 2), strongly suggests that our unsupervised approach is identifying distinct patient phenotypes even within a population with a high prevalence of the ‘positive’ historical heart attack label.
In cardiovascular medicine, disease phenotypes are often heterogeneous and multifactorial. Patients labelled “negative” may share subclinical biomarker profiles with those labelled “positive”, reflecting intermediate or early-stage pathophysiology not encoded in a simple dichotomy. The observed overlap and 60% purity, therefore, suggest the existence of intermediate subpopulations or a continuous risk spectrum beyond the scope of the labels used [30].
Critically, the clinical utility of our clusters is further underscored by their distinct biomarker profiles (Table 2). For example, Cluster 2 not only captured 72.3% of patients with a prior heart attack label but also contained 308 patients labeled ‘negative’. These individuals, despite lacking a documented past event, share a high-risk biomarker signature (elevated troponin, KCM, glucose) with many ‘positive’ patients. This is precisely the type of subclinical risk profile that our unsupervised approach aims to uncover, potentially identifying individuals who could benefit from proactive monitoring or preventive strategies before an overt event occurs. Therefore, the quantitative alignment with the binary ‘heart attack’ label, while providing one dimension of evaluation, must be interpreted alongside the rich clinical and biomarker distinctions that define the clusters and point towards a broader cardiovascular risk assessment.
4.2. Clinical Implications and Pathway to Personalized Cardiology and Improved Outcomes
The findings of this study, stemming from an unsupervised, biomarker-driven patient segmentation, offer several compelling clinical implications and pave the way for impactful future research directions. The ability of our AI-powered approach to discern distinct patient groups, particularly Group 2, characterized by its high-risk biomarker profile (elevated troponin, KCM, and glucose), holds a significant and actionable promise for transforming proactive cardiovascular risk management. This approach aligns with the principles of personalized medicine, where interventions are customized to individual patient profiles [36].
As a possible clinical implication, the statistical and clinical analysis of key biomarkers may offer clinical significance, such as troponin is a highly specific biomarker of myocardial injury and is widely used in the diagnosis and prognosis of acute coronary syndromes. In this study, Group 2 patients show an average troponin level of 0.4761 ng/mL, significantly higher than the 0.1186 ng/mL observed in Group 1. This finding suggests possible underlying ischemia or latent myocardial damage in these patients, which could correlate with a higher risk of coronary events in the short to medium term [37].
The clinical and prognostic implications: The elevated troponin levels in Group 2 support the hypothesis of a high cardiovascular risk profile. Recent studies indicate that even moderately elevated troponin levels are associated with increased all-cause and cardiovascular mortality [38]. Thus, patients in this group may benefit from close monitoring and intensive preventive interventions, such as angiotensin-converting enzyme inhibitors (ACEIs) or beta-blockers, to mitigate the risk of cardiac events.
On cross-check data, glucose levels and metabolic risk: Hyperglycemia is a key marker of insulin resistance and metabolic dysfunction, factors closely linked to cardiovascular risk [39]. In this study, Group 2 shows an average glucose level of 150 mg/dL, compared to 143 mg/dL in Group 1. Although both groups present hyperglycemia, the elevated levels in Group 2 suggest a possible coexistence of prediabetes or type 2 diabetes, conditions that exacerbate the risk of atherosclerosis and other adverse cardiovascular events [40].
Implications for metabolic health and cardiovascular risk: Sustained hyperglycemia, as observed in both groups, is a risk factor for microvascular and macrovascular complications. Studies indicate that dysglycemia promotes a chronic inflammatory state and increased oxidative stress, pathological processes that contribute to endothelial damage and atherosclerosis [41]. The combination of elevated troponin and glucose levels in Group 2 may indicate an increased risk of metabolic syndrome and coronary artery disease.
To continue the comparison of metabolic repercussions between groups, Group 1 can be defined as having a lower risk profile. Clinical and risk profile: Patients in Group 1 exhibit moderately elevated glucose levels but relatively low troponin levels, suggesting a lower cardiovascular risk burden. Moderate hyperglycemia, in the absence of myocardial injury markers, may indicate an early phase of metabolic dysfunction without significant cardiovascular involvement. Intervention strategies: Recommendations for these patients may focus on primary prevention strategies, such as lifestyle modifications (healthy diet, regular exercise) and the use of metformin to improve insulin sensitivity and reduce the risk of progression to type 2 diabetes and cardiovascular events [42].
Group 2 can be defined as having a high-risk profile. Implications of high cardiovascular and metabolic risk: The co-elevation of troponin and glucose in Group 2 suggests more severe metabolic dysregulation and a substantially increased cardiovascular risk. This clinical profile is consistent with metabolic syndrome and a state of low-grade chronic inflammation, conditions that increase the risk of atherothrombosis and cardiovascular events [43]. Priority clinical interventions: For patients in this group, intensive management is essential, which may include optimizing glycemic control through insulin therapy or newer antidiabetics (such as SGLT2 inhibitors or GLP-1 agonists) and the use of statins to reduce cardiovascular risk. Moreover, troponin monitoring could provide valuable information on the response to interventions and the risk of short-term cardiac events [44].
5. Conclusions
This study demonstrates the feasibility of using machine learning techniques to identify groups of patients with different levels of relevant cardiovascular risk profiles specific to this cohort of 1319 patients. While this study introduces a novel, data-driven segmentation of individuals based on real-time biomarkers such as troponin, without incorporating labeled clinical outcomes like documented cardiovascular events.
In evaluating the alignment of these data-driven segments with the dataset’s ‘heart attack (positive/negative)’ labels, we observed a quantitative correspondence accuracy of 60% when mapping clusters to these binary outcomes. Notably, both identified clusters contained a majority of patients with a ‘positive’ heart attack label, yet were distinctly separated by our UMAP + k-means approach based on their comprehensive biomarker profiles. This underscores that while the clusters capture a significant portion of patients with a documented past event (e.g., the higher-risk biomarker cluster encompassed 72.3% of ‘positive’ cases), their primary utility lies in identifying nuanced patient phenotypes including individuals labeled ‘negative’ but exhibiting high-risk biomarker signatures that reflect a broader spectrum of cardiovascular risk beyond a single historical event.
Our unsupervised segmentation approach, by focusing on these intrinsic data patterns rather than being constrained by predefined diagnostic labels, can serve as a powerful complement to traditional risk assessment tools [9,10]. It can reveal “hidden risk” in patients whose biomarker profiles suggest underlying myocardial distress or metabolic derangement not yet clinically apparent or fully weighted in standard risk calculators.
In a clinical context, these insights can guide proactive screening and personalized monitoring strategies for patients who, despite being labeled “negative”, may share a biomarker signature with higher-risk individuals, thereby enabling truly precision-driven interventions beyond conventional diagnostics [30,35].
Group 2, characterized by high troponin (0.4761 ng/mL), KCM (18.65 ng/mL), glucose levels (150 mg/dL), and a predominance of male patients (97%), requires prioritized medical attention and specific intervention strategies such as Sodium-Glucose Transport Protein 2 (SGLT2) inhibitors [45,46] or troponin-guided monitoring [47,48]. In contrast, patients in Group 1, although older (mean age 58 vs. 55 years), appear to have a lower risk based on these biomarkers, suggesting reduced acute risk despite advanced age.
The extreme gender disparity in Group 2 (97% male) highlights potential sex-specific risk factors, such as hormonal or behavioral differences, warranting tailored prevention strategies. Notably, blood pressure, a conventional risk metric, showed no significant variation between groups (127/73 vs. 127/72 mmHg), emphasizing the primacy of biomarkers like troponin and glucose in risk stratification.
Applying the k-means clustering algorithm alongside dimensionality reduction techniques like PCA and UMAP allowed us to effectively group patients and clearly visualize the data. These findings can assist healthcare professionals in clinical decision-making and in designing prevention and treatment programs.
While PCA, UMAP, and k-means are individually established techniques, our work uniquely integrates them into a clinical decision-support pipeline that (1) prioritizes UMAP to model nonlinear biomarker interactions critical for risk stratification, and (2) delivers actionable, patient-specific interventions (e.g., SGLT2 inhibitors for high-risk groups). This two-tiered approach—bridging nonlinear ML and therapeutic targeting—represents a critical advance over purely algorithmic applications, translating unsupervised learning into real-world cardiology practice.
Segmenting patients using AI techniques offers a promising avenue for improving the diagnosis and prevention of cardiovascular diseases. It provides significant advantages:
- Early Risk Identification: Enables the detection of patterns and risk factors that might go unnoticed in traditional analyses.
- Personalized Treatments: Facilitates patient stratification, potentially leading to more personalized and effective interventions.
- Impact on Precision Medicine: Incorporating biomarker analysis into risk assessment offers an opportunity to implement targeted and evidence-based interventions. For instance, patients in Group 2 could benefit from comprehensive management programs to reduce cardiovascular risk and improve long-term clinical outcomes, aligning with the principles of personalized medicine [5].
- Optimization of Healthcare Resources: Helps prioritize medical care toward patients at higher risk, enhancing efficiency in resource allocation.
However, this study has some limitations. This study focused on unsupervised methods to uncover subclinical patterns without relying on diagnostic labels. While a supervised model could predict infarctions with high accuracy, it would not identify subgroups of patients with emerging risk profiles, which are critical for proactive preventive strategies. As such, future work will focus on a supervised validation phase, where clinical outcomes will be integrated to assess the prospective value of these subgroups in predicting cardiovascular events. Additionally, longitudinal data will be crucial to evaluate the temporal stability and progression of these phenotypic clusters, potentially guiding personalized preventive interventions.
We recommend expanding this study by incorporating more clinical variables and utilizing different clustering algorithms to validate and enrich the results. Additionally, future work should consider variables related to lifestyle habits, such as smoking, diet, physical activity, and cholesterol levels, among others.
Author Contributions
Conceptualization, J.D.G.-F., R.R.-R. and J.E.L.-R.; Methodology, J.D.G.-F., S.V.-R. and A.G.-M.; Software, J.D.G.-F., J.E.G.-T. and J.E.L.-R.; Validation, S.V.-R., A.-F.L.-N. and E.A.I.-F.; writing—original draft preparation, J.D.G.-F., R.R.-R. and J.E.L.-R.; writing—review and editing, R.R.-R., J.E.L.-R., A.G.-M., A.-F.L.-N., J.L.-A. and E.A.I.-F.; funding acquisition, R.R.-R.; Investigation, J.E.G.-T. and E.A.I.-F.; Formal Analysis, E.A.I.-F. All authors have read and agreed to the published version of the manuscript.
Funding
This research received no external funding.
Data Availability Statement
The data that support the findings of this study are available from the corresponding author upon reasonable request.
Acknowledgments
The authors would like to thank the CICESE Research Center and the Secretariat of Science, Humanities, Technology and Innovation (Secihti) for their support.
Conflicts of Interest
The authors declare no conflicts of interest.
Appendix A

Table A1.
Related works.
Table A1.
Related works.
| Previous Work | Contributions | Opportunity |
|---|---|---|
| Unsupervised Learning for Heart Disease Prediction: Clustering-Based Approach [10] | Comparison of unsupervised models (k-means, DBSCAN, etc.) for patient stratification. Web interface for clinical use. | Lack of integration with lifestyle variables. It is mainly based on age and gender. |
| Detecting cardiovascular diseases using unsupervised machine learning clustering based on electronic medical records [11] | Robust model with EMR (155k patients) using k-means and PCA. High accuracy (~86%) in CVD detection. | It does not include advanced biomarkers (e.g., troponin). Linear PCA: Reduction to 2D mixes nonlinear clusters, limiting visual/clinical separation. |
| Machine learning-based prediction models for cardiovascular disease risk using electronic health records data: systematic review and meta-analysis [49] | Meta-analysis demonstrates superiority of ML (AUC 0.86) over traditional models. Identifies methodological heterogeneity as a clinical barrier. | Lack of standardization in metrics and external validation. |
| Our study | Combines k-means with UMAP (not just PCA) for better visualization/clustering. Focus on specific biomarkers (troponin, CK-MB) for accurate stratification. Identifies at-risk groups with significant differences in biomarkers (not just demographic variables). | Could be scaled with temporal data to predict dynamic risk. It could include more variables related to lifestyle habits, such as smoking, diet, physical activity, and cholesterol levels. |

Table A2.
Fragment of ten entries of the medical dataset.
Table A2.
Fragment of ten entries of the medical dataset.
| Age | Gender | Pulse | Pressurehigh | Pressurelow | Glucose | KCM | Troponin |
|---|---|---|---|---|---|---|---|
| 64 | 1 | 66 | 160 | 83 | 160 | 1.80 | 0.012 |
| 21 | 1 | 94 | 98 | 46 | 296 | 6.75 | 1.060 |
| 55 | 1 | 64 | 160 | 77 | 270 | 1.99 | 0.003 |
| 64 | 1 | 70 | 120 | 55 | 270 | 13.87 | 0.122 |
| 55 | 1 | 64 | 112 | 65 | 300 | 1.08 | 0.003 |
| 58 | 0 | 61 | 112 | 58 | 87 | 1.83 | 0.004 |
| 32 | 0 | 40 | 179 | 68 | 102 | 0.71 | 0.003 |
| 63 | 1 | 60 | 214 | 82 | 87 | 300 | 2.370 |
| 44 | 0 | 60 | 154 | 81 | 135 | 2.35 | 0.004 |
| 67 | 1 | 61 | 160 | 95 | 100 | 2.84 | 0.011 |
Appendix B
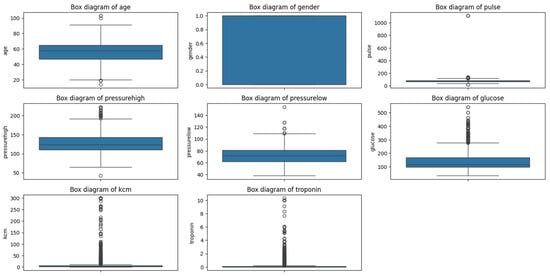
Figure A1.
Outliers in the medical dataset.
References
- World Health Organization (WHO). Cardiovascular Diseases (CVDs). Available online: https://www.who.int/news-room/fact-sheets/detail/cardiovascular-diseases-(cvds) (accessed on 23 October 2024).
- Chen, Y.; Tang, M.; Yuan, S.; Fu, S.; Li, Y.; Li, Y.; Wang, Q.; Cao, Y.; Liu, L.; Zhang, Q. Rhodiola rosea: A Therapeutic Candidate on Cardiovascular Diseases. Oxidative Med. Cell Longev. 2022, 2022, 1348795. [Google Scholar] [CrossRef] [PubMed]
- Heaton, J. Ian Goodfellow, Yoshua Bengio, and Aaron Courville: Deep learning. Genet. Program. Evolvable Mach. 2018, 19, 305–307. [Google Scholar] [CrossRef]
- Esteva, A.; Robicquet, A.; Ramsundar, B.; Kuleshov, V.; DePristo, M.; Chou, K.; Cui, C.; Corrado, G.; Thrun, S.; Dean, J. A guide to deep learning in healthcare. Nat. Med. 2019, 25, 24–29. [Google Scholar] [CrossRef]
- Topol, E.J. High-performance medicine: The convergence of human and artificial intelligence. Nat. Med. 2019, 25, 44–56. [Google Scholar] [CrossRef]
- Capó, M.; Pérez, A.; Lozano, J.A. An efficient approximation to the K-means clustering for massive data. Knowledge-Based Syst. 2017, 117, 56–69. [Google Scholar] [CrossRef]
- Jolliffe, I.T.; Cadima, J. Principal component analysis: A review and recent developments. Philos. Trans. R. Soc. A Math. Phys. Eng. Sci. 2016, 374, 20150202. [Google Scholar] [CrossRef]
- McInnes, L.; Healy, J.; Melville, J. UMAP: Uniform Manifold Approximation and Projection for Dimension Reduction. J. Mach. Learn. Res. 2020. [Google Scholar] [CrossRef]
- Shomorony, I.; Cirulli, E.T.; Huang, L.; Napier, L.A.; Heister, R.R.; Hicks, M.; Cohen, I.V.; Yu, H.-C.; Swisher, C.L.; Schenker-Ahmed, N.M.; et al. An unsupervised learning approach to identify novel signatures of health and disease from multimodal data. Genome Med. 2020, 12, 7. [Google Scholar] [CrossRef] [PubMed]
- Jetty, J.; Sk, S.S.; Polepalle, R.B.; Parusu, V. Unsupervised Learning for Heart Disease Prediction: Clustering-Based Approach. ITM Web Conf. 2025, 74, 01005. [Google Scholar] [CrossRef]
- Hu, Y.; Yan, H.; Liu, M.; Gao, J.; Xie, L.; Zhang, C.; Wei, L.; Ding, Y.; Jiang, H. Detecting cardiovascular diseases using unsupervised machine learning clustering based on electronic medical records. BMC Med. Res. Methodol. 2024, 24, 309. [Google Scholar] [CrossRef]
- Apple, F.S.; Sandoval, Y.; Jaffe, A.S.; Ordonez-Llanos, J. Cardiac Troponin Assays: Guide to Understanding Analytical Characteristics and Their Impact on Clinical Care. Clin. Chem. 2017, 63, 73–81. [Google Scholar] [CrossRef] [PubMed]
- Shah, A.S.V.; Griffiths, M.; Lee, K.K.; A McAllister, D.; Hunter, A.L.; Ferry, A.V.; Cruikshank, A.; Reid, A.; Stoddart, M.; Strachan, F.; et al. High sensitivity cardiac troponin and the under-diagnosis of myocardial infarction in women: Prospective cohort study. BMJ 2015, 350, g7873. [Google Scholar] [CrossRef] [PubMed]
- Ayushi; Sethi, S.; Jyoti. Heart Disease Prediction Integrating UMAP and XGBoost. Int. J. Recent Technol. Eng. 2020, 9, 2449–2457. [Google Scholar] [CrossRef]
- Cunningham, J.P.; Ghahramani, Z. Linear Dimensionality Reduction: Survey, Insights, and Generalizations. J. Ma-Chine Learn. Research 2015, 16, 2859–2900. [Google Scholar]
- Abdi, H.; Williams, L.J. Principal component analysis. WIREs Comput. Stat. 2010, 2, 433–459. [Google Scholar] [CrossRef]
- Gonzalez-Franco, J.D.; Preciado-Velasco, J.E.; Lozano-Rizk, J.E.; Rivera-Rodriguez, R.; Torres-Rodriguez, J.; Alonso-Arevalo, M.A. Comparison of Supervised Learning Algorithms on a 5G Dataset Reduced via Principal Component Analysis (PCA). Futur. Internet 2023, 15, 335. [Google Scholar] [CrossRef]
- Gale, C.P.; Kashinath, C.; Brooksby, P. The association between hyperglycaemia and elevated troponin levels on mortality in acute coronary syndromes. Diabetes Vasc. Dis. Res. 2006, 3, 80–83. [Google Scholar] [CrossRef] [PubMed]
- Becht, E.; McInnes, L.; Healy, J.; Dutertre, C.-A.; Kwok, I.W.H.; Ng, L.G.; Ginhoux, F.; Newell, E.W. Dimensionality reduction for visualizing single-cell data using UMAP. Nat. Biotechnol. 2019, 37, 38–44. [Google Scholar] [CrossRef] [PubMed]
- Xu, D.; Tian, Y. A Comprehensive Survey of Clustering Algorithms. Ann. Data Sci. 2015, 2, 165–193. [Google Scholar] [CrossRef]
- Ikotun, A.M.; Ezugwu, A.E.; Abualigah, L.; Abuhaija, B.; Heming, J. K-means clustering algorithms: A comprehensive review, variants analysis, and advances in the era of big data. Inf. Sci. 2023, 622, 178–210. [Google Scholar] [CrossRef]
- Bholowalia, P.; Kumar, A. EBK-Means: A Clustering Technique Based on Elbow Method and K-Means in WSN. Int. J. Comput. Appl. 2014, 105, 17–24. Available online: https://citeseerx.ist.psu.edu/document?repid=rep1&type=pdf&doi=5771aa21b2e151f3d93ba0a5f12d023a0bfcf28b (accessed on 20 March 2025).
- Arbelaitz, O.; Gurrutxaga, I.; Muguerza, J.; Pérez, J.M.; Perona, I. An extensive comparative study of cluster validity indices. Pattern Recognit. 2013, 46, 243–256. [Google Scholar] [CrossRef]
- Schober, P.; Boer, C.; Schwarte, L.A. Correlation Coefficients: Appropriate Use and Interpretation. Anesth. Analg. 2018, 126, 1763–1768. [Google Scholar] [CrossRef]
- Whelton, P.K.; Carey, R.M.; Aronow, W.S.; Casey, D.E.; Collins, K.J.; Dennison Himmelfarb, C.; DePalma, S.M.; Gidding, S.; Jamerson, K.A.; Jones, D.W.; et al. ACC/AHA/AAPA/ABC/ACPM/AGS/APhA/ASH/ASPC/NMA/PCNA Guideline for the Prevention, Detection, Evaluation, and Management of High Blood Pressure in Adults. J. Am. Coll. Cardiol. 2018, 71, e127–e248. [Google Scholar] [CrossRef]
- Everitt, B.S.; Landau, S.; Leese, M.; Stahl, D. Cluster Analysis, 5th ed.; Wiley Series in Probability and Statistics; John Wiley & Sons, Inc.: Chichester, UK, 2011. [Google Scholar] [CrossRef]
- Van Der Maaten, L.; Hinton, G. Visualizing Data Using t-SNE. Netherlands, Aug 2008. Available online: https://jmlr.csail.mit.edu/papers/volume9/vandermaaten08a/vandermaaten08a.pdf (accessed on 25 October 2024).
- McInnes, L.; Healy, J.; Saul, N.; Großberger, L. UMAP: Uniform Manifold Approximation and Projection. J. Open Source Softw. 2018, 3, 861. [Google Scholar] [CrossRef]
- Segar, M.W.; Rao, S.; Navar, A.M.; Michos, E.D.; Lewis, A.; Correa, A.; Sims, M.; Khera, A.; Hughes, A.E.; Pandey, A. County-level phenomapping to identify disparities in cardiovascular outcomes: An unsupervised clustering analysis. Am. J. Prev. Cardiol. 2020, 4, 100118. [Google Scholar] [CrossRef]
- Saba, P.S.; Al Kindi, S.; Nasir, K. Redefining Cardiovascular Risk Assessment as a Spectrum. Circ. 2024, 83, 574–576. [Google Scholar] [CrossRef]
- Gill, S.K.; Karwath, A.; Uh, H.-W.; Cardoso, V.R.; Gu, Z.; Barsky, A.; Slater, L.; Acharjee, A.; Duan, J.; Dall’Olio, L.; et al. Artificial intelligence to enhance clinical value across the spectrum of cardiovascular healthcare. Eur. Heart J. 2023, 44, 713–725. [Google Scholar] [CrossRef]
- Regitz-Zagrosek, V.; Kararigas, G. Mechanistic Pathways of Sex Differences in Cardiovascular Disease. Physiol. Rev. 2017, 97, 1–37. [Google Scholar] [CrossRef]
- Flores, A.M.; Schuler, A.; Eberhard, A.V.; Olin, J.W.; Cooke, J.P.; Leeper, N.J.; Shah, N.H.; Ross, E.G. Unsupervised Learning for Automated Detection of Coronary Artery Disease Subgroups. J. Am. Heart Assoc. 2021, 10, e021976. [Google Scholar] [CrossRef]
- Kaptein, Y.E.; Karagodin, I.; Zuo, H.; Lu, Y.; Zhang, J.; Kaptein, J.S.; Strande, J.L. Identifying Phenogroups in patients with subclinical diastolic dysfunction using unsupervised statistical learning. BMC Cardiovasc. Disord. 2020, 20, 367. [Google Scholar] [CrossRef] [PubMed]
- Gordon, M.M.; Moser, A.M.; Rubin, E. Unsupervised analysis of classical biomedical markers: Robustness and medical relevance of patient clustering using bioinformatics tools. PLoS ONE 2012, 7, e29578. [Google Scholar] [CrossRef]
- Ashley, E.A. The Precision Medicine Initiative. JAMA 2015, 313, 2119–2120. [Google Scholar] [CrossRef]
- Thygesen, K.; Alpert, J.S.; Jaffe, A.S.; Chaitman, B.R.; Bax, J.J.; Morrow, D.A.; White, H.D. Fourth Universal Definition of Myocardial Infarction (2018). Circulation 2018, 138, e618–e651. [Google Scholar] [CrossRef]
- Daccord, N.; Celton, J.-M.; Linsmith, G.; Becker, C.; Choisne, N.; Schijlen, E.; van de Geest, H.; Bianco, L.; Micheletti, D.; Velasco, R.; et al. High-quality de novo assembly of the apple genome and methylome dynamics of early fruit development. Nat. Genet. 2017, 49, 1099–1106. [Google Scholar] [CrossRef] [PubMed]
- American Diabetes Association. Standards of Medical Care in Diabetes—2021 Abridged for Primary Care Providers. Clin. Diabetes 2021, 39, 14–43. [Google Scholar] [CrossRef]
- Swinburn, B.A.; Kraak, V.I.; Allender, S.; Atkins, V.J.; Baker, P.I.; Bogard, J.R.; Brinsden, H.; Calvillo, A.; De Schutter, O.; Devarajan, R.; et al. The Global Syndemic of Obesity, Undernutrition, and Climate Change: The Lancet Commission report. Lancet 2019, 393, 791–846. [Google Scholar] [CrossRef]
- Brownlee, M. The Pathobiology of Diabetic Complications. Diabetes 2005, 54, 1615–1625. [Google Scholar] [CrossRef]
- Knowler, W.C.; Barrett-Connor, E.; Fowler, S.E.; Hamman, R.F.; Lachin, J.M.; Walker, E.A.; Nathan, D.M.; Diabetes Prevention Program Research Group. Reduction in the incidence of type 2 diabetes with lifestyle intervention or metformin. N. Engl. J. Med. 2002, 346, 393–403. [Google Scholar] [CrossRef]
- Grundy, S.M.; Cleeman, J.I.; Daniels, S.R.; Donato, K.A.; Eckel, R.H.; Franklin, B.A.; Gordon, D.J.; Krauss, R.M.; Savage, P.J.; Smith, S.C., Jr.; et al. Diagnosis and management of the metabolic syndrome: An American Heart Association/National Heart, Lung, and Blood Institute Scientific Statement. Circulation 2005, 112, 2735–2752. [Google Scholar] [CrossRef]
- Kumar, V.; Thakur, J.K.; Prasad, M. Histone acetylation dynamics regulating plant development and stress responses. Cell Mol. Life Sci. 2021, 78, 4467–4486. [Google Scholar] [CrossRef] [PubMed]
- Zannad, F.; Ferreira, J.P.; Pocock, S.J.; Anker, S.D.; Butler, J.; Filippatos, G.; Brueckmann, M.; Ofstad, A.P.; Pfarr, E.; Jamal, W.; et al. SGLT2 inhibitors in patients with heart failure with reduced ejection fraction: A meta-analysis of the EMPEROR-Reduced and DAPA-HF trials. Lancet 2020, 396, 819–829. [Google Scholar] [CrossRef] [PubMed]
- McMurray, J.J.V.; Solomon, S.D.; Inzucchi, S.E.; Køber, L.; Kosiborod, M.N.; Martinez, F.A.; Ponikowski, P.; Sabatine, M.S.; Anand, I.S.; Bělohlávek, J.; et al. Dapagliflozin in Patients with Heart Failure and Reduced Ejection Fraction. N. Engl. J. Med. 2019, 381, 1995–2008. [Google Scholar] [CrossRef] [PubMed]
- Allen, B.R.; Christenson, R.H.; Cohen, S.A.; Nowak, R.; Wilkerson, R.G.; Mumma, B.; Madsen, T.; McCord, J.; Veld, M.H.I.; Massoomi, M.; et al. Diagnostic Performance of High-Sensitivity Cardiac Troponin T Strategies and Clinical Variables in a Multisite US Cohort. Circulation 2021, 143, 1659–1672. [Google Scholar] [CrossRef]
- A Byrne, R.; Rossello, X.; Coughlan, J.J.; Barbato, E.; Berry, C.; Chieffo, A.; Claeys, M.J.; Dan, G.-A.; Dweck, M.R.; Galbraith, M.; et al. 2023 ESC Guidelines for the management of acute coronary syndromes. Eur. Heart J. 2023, 44, 3720–3826. [Google Scholar] [CrossRef]
- Liu, T.; Krentz, A.; Lu, L.; Curcin, V. Machine learning based prediction models for cardiovascular disease risk using electronic health records data: Systematic review and meta-analysis. Eur. Heart J. Digit. Health 2025, 6, 7–22. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).