1. Introduction
Potatoes are an important food crop around the world and is grown extensively to meet the basic nutritional needs of billions of people. The potato is the fourth most widely cultivated food crop in the world, following rice, wheat, and maize. It is grown in more than 150 countries, spanning a range of climatic zones from temperate regions to tropical highlands. The high nutritional value of the potato has led to its designation as a staple food in numerous countries, providing essential carbohydrates, vitamins, and minerals such as potassium. Moreover, the relatively short growth cycle and high yield potential of the potato make it a pivotal crop for addressing population growth and food shortages, which is of paramount importance for global food security [
1]. However, potato production is severely threatened by a variety of diseases, including those caused by viruses, bacteria, fungi, insect pests, and nematodes, as shown in
Figure 1. These diseases not only reduce the yield and quality of potato crops, but also result in significant economic losses for growers. Therefore, the early detection and effective management of these diseases are critical to protecting potato crops and increasing agricultural productivity. In addition, research focused on potato diseases is essential for improving crop yields and ensuring the stability of the global food supply [
2].
Traditional methods for identifying plant diseases depend on manual inspections requiring specialized knowledge to accurately diagnose the type of disease. This approach requires considerable computational effort and incurs significant costs. In addition, the failure to detect diseases in a timely manner can lead to their uncontrolled spread among plants, causing significant damage. The rapid development of deep learning technologies, particularly convolutional neural networks (CNNs), has led to significant advances in plant disease diagnosis [
3,
4,
5,
6]. In addition, CNNs are also effective in detecting abnormalities in plant leaves, such as spots, damage, and color changes. In recent years, an increasing number of studies have used these technologies for automated plant disease detection. The application of deep learning models has the potential to significantly improve the accuracy of potato leaf disease diagnosis while reducing the time required for diagnosis. This will allow growers to take timely action and effectively minimize the losses caused by these diseases. Numerous studies have focused on the detection of leaf diseases in crops such as potatoes [
7,
8,
9,
10], peanuts [
11], jasmine [
12], citrus, cucumber, grape, and tomato [
13,
14]. Consequently, the use of deep learning for plant leaf disease detection has become an important development direction in agricultural technology and can help address critical challenges in global agricultural production.
The use of a tri-CNN architecture, consisting of three deep convolutional neural networks (DenseNet169, Inception, and Xception), was proposed for disease identification [
11]. The model was initially pre-trained on the ImageNet dataset and later trained and tested on the Soa comprehensive dataset, achieving an accuracy of 98.46%. However, the high computational complexity of the tri-CNN architecture may limit its deployment on resource-constrained devices.
A novel MobileNetV3-based classifier specifically designed for classifying jasmine plant leaf spot disease was proposed [
12]. The classifier incorporates a data enhancement technique using a conditional generative adversarial network (cGAN). Evaluation using the jasmine plant leaf spot disease dataset demonstrated strong performance, with a training accuracy of 97%, highlighting its effectiveness. Real-world testing under varying conditions, such as extreme camera angles and different lighting conditions, showed the model’s flexibility, with its test accuracies ranging from 94% to 96%. However, since the model was developed exclusively for jasmine, further testing is required to assess its applicability to other crops.
Sharma et al. [
15] introduced a novel deep learning architecture called the Deeper Lightweight Convolutional Neural Network (DLMC-Net) for detecting leaf diseases across a wide variety of crops. Unlike many traditional models that are limited to specific crops, DLMC-Net features an optimized architecture capable of generalizing across multiple crops, including citrus, cucumber, grape, and tomato. The model incorporates integrated block and channel layers, which enhance feature propagation and reuse while addressing the gradient vanishing problem. It achieved accuracies of 93.56% on citrus, 92.34% on cucumber, 99.50% on grape, and 96.56% on tomato. While DLMC-Net offers advantages in terms of its lightweight design and high accuracy, there is still potential for improvement. Future research should focus on testing the model’s robustness under different environmental conditions to ensure its effectiveness in real-world agricultural scenarios.
Sholihati et al. [
16] employed the deep learning model called the Visual Geometry Group 16-layer network (VGG-16) in conjunction with data augmentation techniques to detect potato leaf diseases using the PlantVillage dataset [
17], which encompasses five categories of diseases. The VGG-16 model leverages the stacking of multiple convolutional layers to effectively extract complex image features. The model ultimately achieved a classification accuracy of 91% on 1220 test images from the PlantVillage dataset. The VGG16 model is known for its high computational intensity due to its deep layers, making it unsuitable for real-time applications or deployment on resource-constrained edge devices. As more efficient models become available, it would be beneficial for future studies to explore alternatives that can deliver comparable or superior accuracy while reducing the computational load.
Pandiri et al. [
18] introduced an optimized deep convolutional neural network (ODNet) specifically designed for the identification of potato leaf diseases. The performance of ODNet was enhanced through the adjustment of hyperparameters using the slime mold optimization (SMO) algorithm. The model was tested on the Potato Leaf Disease dataset [
19], which includes three categories: healthy, early blight, and late blight. The ODNet model achieved a classification accuracy of 96.67%, demonstrating its effectiveness in accurately identifying these diseases. However, the tests were limited to only three disease types, which restricts the model’s ability to generalize to a broader dataset. Additionally, the study did not evaluate the effectiveness of ODNet in real-world conditions. Disease behavior in actual farm fields is likely to be more diverse and complex than in controlled datasets.
Shaheed et al. [
20] introduced a composite model, EfficientRMT-Net, which integrates EfficientNet, vision transformer (ViT), depth-wise convolution (DWC), and ResNet architectures for the detection of potato leaf diseases using images from the PlantVillage dataset. To address the issue of class imbalance within the dataset, they applied data augmentation techniques. Additionally, transfer learning was employed to enhance the classification efficiency of the proposed approach. In their experiments, the individual architectures—EfficientNet, ViT, DWC, and ResNet—achieved accuracies of 76%, 84.1%, 84.6%, and 83.5%, respectively. The proposed integrated EfficientRMT-Net architecture, however, achieved a remarkable recognition accuracy of 99.12% on the PlantVillage dataset. Although the model demonstrated impressive accuracy (99.12%), the integration of multiple architectures makes it overly complex and computationally expensive, which could pose significant challenges in real-world applications, particularly in resource-constrained agricultural settings. Moreover, the study did not validate the model’s performance in a realistic agricultural environment.
Mahum et al. [
21] employed the DenseNet model to classify potato leaf diseases using data primarily collected from the PlantVillage dataset. In addition to the existing categories of early blight (EB) and late blight (LB) within the PlantVillage dataset, the authors manually collected additional data for categories such as potato leafroll virus (PLR), potato verticillium wilt (PVW), and healthy potato (PH). The classification of potato leaf diseases was conducted using a pre-trained efficient DenseNet model. To address the issue of data imbalance within the training set, the authors utilized a reweighted cross-entropy loss function to stabilize the training process. The model achieved a classification accuracy of 97.2% on the customized PlantVillage dataset. Nevertheless, the model’s efficacy on real-world data remains uncertain, especially considering the expansion and manual adjustment of the dataset. While the reweighted cross-entropy loss function addresses data imbalances, it is unclear whether the model will perform equally well on more diverse datasets or on previously unseen disease classes.
Catal Reis et al. [
22] used machine learning algorithms such as support vector machine (SVM), logistic regression (LR), Random Forest (RF), and adaptive boosting (AdaBoost) algorithms to propose the multi-head attention mechanism depthwise separable convolution inception reduction network (MDSCIRNet), which overcomes common issues encountered during the training process of deep neural networks, such as gradient vanishing, underfitting, and overfitting. This method achieved an accuracy of up to 99.33% in identifying potato leaf diseases on the Potato Leaf Disease dataset. Nevertheless, the deployment of conventional machine learning algorithms, such as support vector machines (SVM) and Random Forest, while effective for small to medium-sized datasets, may limit scalability and performance when applied to larger and more complex datasets. Furthermore, the study did not specify the dimensions and computational efficiency of the model, which is crucial for practical agricultural applications.
Shabrina et al. [
23] proposed a novel dataset for potato leaf diseases collected in uncontrolled environments, with images captured in real agricultural fields under various weather conditions. The dataset comprises 3076 images categorized into seven distinct classes. The authors conducted experiments using five different deep learning models, with the EfficientNetV2B3 model attaining the highest accuracy of 73.63% among the models compared. The creation of datasets in uncontrolled environments is a valuable contribution, as it more accurately reflects real-world scenarios where lobar diseases can be more challenging to detect. However, the relatively low accuracy rates suggest that the existing deep learning models may struggle to generalize effectively under such challenging conditions. Further research into more sophisticated data augmentation techniques or advanced model architectures may be necessary to enhance performance on realistic datasets.
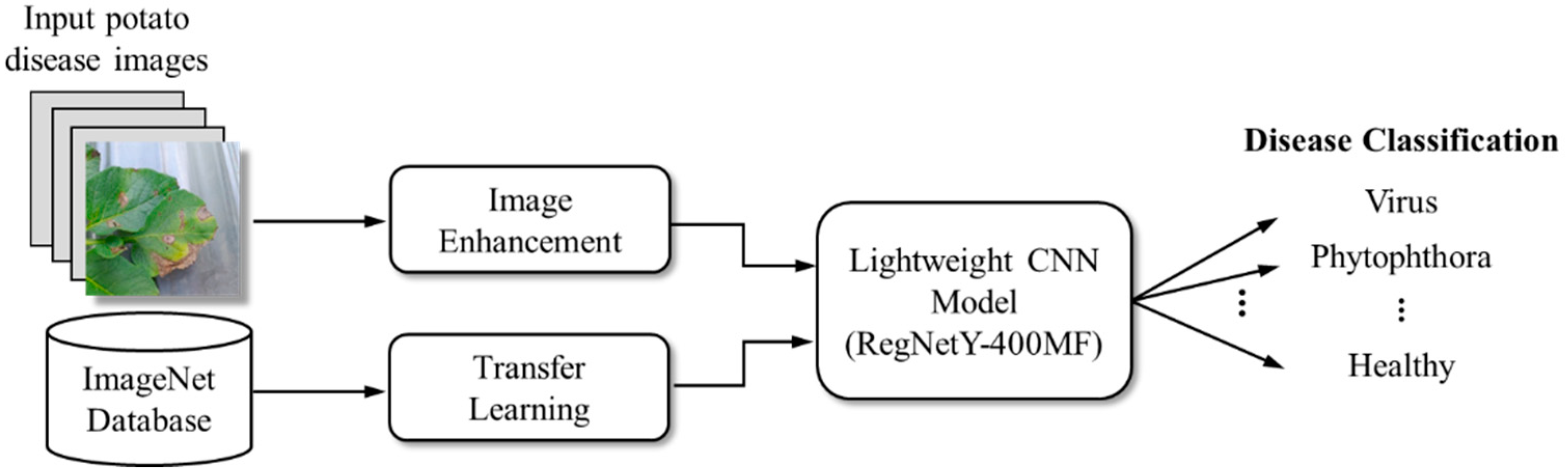
The implementation of deep learning models has the potential to significantly improve the accuracy of potato leaf disease diagnosis while simultaneously reducing the time required for diagnosis. This advancement will enable growers to take prompt action and effectively mitigate the losses caused by these diseases. As of early 2024, most research using deep learning for potato leaf disease identification has primarily focused on datasets with three or five disease categories and has demonstrated commendable performance. However, studies using datasets with seven categories still show significant potential for improving classification accuracy. Furthermore, it has been observed that the deep learning models discussed in the literature typically require significant computational resources and storage capacity. This poses significant challenges for deployment in resource-constrained environments, such as portable mobile devices (e.g., smartphones and tablets), where the computational power required for these models may not be feasible. To address these practical challenges, this study proposes the use of the RegNetY-400MF model [
24] for potato leaf disease detection. RegNetY-400MF is a lightweight and efficient deep learning architecture that achieves high accuracy while significantly reducing computational and storage requirements. This makes it well suited for use on portable mobile devices, making disease detection more accessible and practical in agricultural settings.
3. Experimental Results
3.1. Experimental Platform and Model Parameters
The experimental environment consisted of an 8-core computer with an Intel Core i7-10700 CPU model, a NVIDIA GeForce RTX 2070 Super GPU model, and a memory size of 16 GB. In terms of software, the operating system was Windows 11 Professional, the development environment was Anaconda 24.3.0 coupled with Python 3.11.8, using the PyTorch 2.2.2 deep learning framework and CUDA 12.2 and cuDNN 9.0.0 GPU drivers.
The initial parameters used in the experiment were as follows: The learning rate and batch size were set to 0.0001 and 64, respectively, and the model was trained for 50 epochs. The optimizer used was Adam, a widely used optimizer in deep learning that speeds up model convergence. For the loss function, cross entropy was chosen due to its effectiveness in calculating the loss between predicted probability distributions and true labels in classification tasks. Compared to other loss functions, such as mean squared error (MSE), cross-entropy function is better suited to classification problems, allowing for faster and more accurate model convergence.
3.2. Dataset Construction
In order to accurately assess the identification performance of the proposed method, we used 10-fold cross-validation to evaluate the effectiveness of the system [
29], using the Potato Leaf Disease in Uncontrolled Environment dataset as a test subject. This dataset consists of 3076 images of potato leaf diseases categorized into seven classes. The images have a resolution of 1500 × 1500 pixels and were captured under varying weather conditions, including sunny, cloudy, and partly cloudy skies, between 8 a.m. and 3 p.m. The distances from the subject ranged from approximately 5 to 15 cm, due to differences in photographic positioning. As a result, the backgrounds and occlusions vary, making the dataset a realistic representation of potato leaf diseases. Detailed information about this dataset is provided in
Table 1.
The primary reason for this data imbalance is the nature of data collection. Since agricultural research and datasets often focus on identifying and addressing disease problems that significantly threaten crop yield and quality, more emphasis is placed on collecting images of diseased plants during data collection, resulting in a higher representation of disease categories in the dataset.
3.3. Performance Metrics
In this study, we used several evaluation metrics, including accuracy, precision, recall, and F1 score. Accuracy served as the primary metric for assessing the overall performance of the model across the seven categories. Precision measures the model’s ability to minimize false positives, while recall assesses its effectiveness in identifying all positive cases. The F1 score, calculated as the harmonic mean of precision and recall, provides a balanced assessment of these two metrics.
In the context of these metrics, true positive (
TP) refers to cases where a positive sample is correctly predicted to be positive; a false positive (
FP) occurs when a negative sample is incorrectly predicted to be positive; a false negative (
FN) indicates that a positive sample is incorrectly predicted to be negative; and true negative (
TN) refers to cases where a negative sample is correctly predicted as negative. The equations used to calculate the accuracy, precision, recall, and
F1 score are as follows:
3.4. The Impact of Transfer Learning
In this experiment, we compared the performance of the model with and without the application of transfer learning to assess its impact. The results are shown in
Table 2. The data show that with the same parameters, transfer learning resulted in significant improvements in all performance metrics. Consequently, we decided to include transfer learning in the subsequent experiments.
3.5. Different Combinations of Training Parameters
In this experiment, we evaluated the impact of different batch sizes and learning rates on the model performance by testing different combinations of these parameters. The results are shown in
Table 3. The data show that a batch size of 16 and a learning rate of 0.001 gave the best performance in terms of both training stability and accuracy. In addition, the results suggest that batch sizes that are too large or learning rates that are too small can negatively affect model performance. Therefore, we chose a batch size of 16 and a learning rate of 0.001 as the optimal parameter combination for our experiments.
3.6. Data Enhancement
To further improve the performance of the model, we used a number of image enhancement techniques during the training process, including random cropping, brightness adjustment, horizontal and vertical flipping, and random rotation. These enhancement techniques can increase the robustness of the training, thereby improving the generalization ability of the model in different environments. The experimental results are shown in
Table 4, which shows that, with the same settings for other parameters, the application of data enhancement techniques led to significant improvements in all evaluation metrics of the model during testing.
To assess the impact of image enhancement and data diversity on performance, we analyzed the validation loss function curve. The experimental results are shown in
Figure 5. In
Figure 5a, the blue curve converges close to 0, while the validation accuracy shown in
Figure 5b shows no improvement, suggesting overfitting. Overfitting typically occurs when the training data lack sufficient diversity, causing the model to learn specific features of the training data rather than general patterns, which affects its performance on the validation data [
30]. This observation highlights the effectiveness of data augmentation techniques in improving model performance and increasing training robustness by mitigating overfitting.
3.7. Performance Comparison with Other Methods
The optimal combinations of batch size and learning rate identified in our initial experiments were then integrated with various data augmentation techniques. To ensure robust results, we ran ten tests and calculated average performance metrics. This approach yielded favorable results for several key performance indicators, including test accuracy, precision, recall, and
F1 score. In particular, the highest test accuracy achieved was 90.68%, as shown in
Table 5.
In order to more effectively assess the true performance of the model, this experiment used 10-fold cross-validation to compare with other lightweight models. The 10-fold cross-validation method is a technique used to evaluate the performance of machine learning models by randomly dividing the dataset into 10 equally sized subsets. Each time, one subset is chosen as the validation set, while the remaining 9 subsets are used to train the model. This process is repeated 10 times, resulting in 10 best validation accuracies, from which the mean and standard deviation are calculated as indicators to evaluate the overall performance of the model. In situations with limited datasets, this method allows a more uniform use of all samples in the dataset. For a fair comparison, we selected several mainstream lightweight models, including SqueezeNet 1.1 [
31], MNASNet1_3 [
32], ShuffleNetV2_x2_0 [
33], and EfficientNet_B1 [
34]. The experimental results are shown in
Table 6.
The experimental results show that the RegNetY-400MF model achieved the highest average accuracy of all the models evaluated. It also exhibited a significantly lower standard deviation compared to other lightweight models, indicating greater consistency in its performance.
4. Discussion
This section presents a comparative analysis of the RegNetY-400MF model against other lightweight models, based on various evaluated metrics. The number of floating-point operations (FLOPs) required for a single forward pass was used as a measure of computational complexity. FLOPs, typically expressed in gigaflops (GFLOPs), serve as a key indicator of computational demand—an essential factor for applications requiring real-time processing. Additionally, the model’s memory footprint was measured in megabytes (MB), reflecting the amount of storage space the model occupies. The number of parameters in the model is another crucial performance indicator, as an increase in parameters corresponds to greater model complexity, higher memory usage, and increased computational requirements. A detailed comparison of the experimental results is presented in
Table 7.
Based on the experimental results in
Table 7, we can observe that the RegNetY-400MF model was only worse than the SqueezeNet 1.1 model in the lightweight metrics evaluation. However, when we consider the results in
Table 6, the average accuracy of RegNetY-400MF was not only the highest, but also significantly greater than that of the SqueezeNet 1.1 model. This demonstrates that RegNetY-400MF has a clear advantage in balancing model size and performance, making it the best choice for lightweight models in resource-constrained environments.
However, the results also suggest that there is potential for further performance improvement. To facilitate a deeper analysis of the model’s performance, we generated a confusion matrix of the test results, as shown in
Figure 6. This matrix provides additional insights that could guide future improvements.
The confusion matrix shows that the model we used achieved relatively high accuracy in identifying both the Bacteria disease category and the Healthy category. The model also performed well in identifying the Fungi disease category, with only four images misclassified as belonging to the Pest disease category. On closer inspection of the images where Fungi images were misclassified as Pest, it is apparent that the visual characteristics of these two disease categories are quite similar, as shown in
Figure 7 and
Figure 8. This similarity of features is likely contributing to the misclassification. In the Pest disease category, 55 samples were correctly classified and 4 were misclassified as Fungi. This further suggests that the overlapping features between the Pest and Fungi categories can lead to confusion, causing the model to occasionally misclassify between these two categories.
For the detection of Phytophthora symptoms, 5 out of 35 samples were misclassified as Fungi, as shown in
Figure 9. The possible reason for this is that Fungi images have several characteristics that can easily lead to the misclassification of other disease categories as this type. The characteristics of the Fungi category include “circular patterns or slightly sunken leaf spots along leaf margins with yellow edges and a concentric ring appearance, or powdery spots on yellow leaves”. As these characteristics are not necessarily all present at the same time, this has a negative impact on the training of classification models, leading to identification errors.
Most of the identification errors were concentrated in the Virus disease category. Of the 54 test images, 9 were misidentified, with the most common misclassification being Virus disease images misidentified as Healthy. The characteristics of virus diseases—such as reduced leaf size, wrinkling, slight spotting or variegation, and necrosis—overlap significantly with those of other diseases. This overlap highlights the inherent difficulty in distinguishing virus diseases from other categories, leading to frequent misclassification. In addition, characteristics such as reduced leaf size and light spotting are subtle and may not be readily apparent, increasing the likelihood of these cases being misclassified as Healthy, as shown in
Figure 10.
To further enhance the classification effectiveness of the model, it is proposed that improvements focus on the attention mechanism. Incorporating the sophisticated efficient channel attention (ECA) mechanism allows the model to concentrate more effectively on the critical characteristics inherent in the input data, thereby improving the accuracy of plant disease recognition. ECA calculates the channel weights, enabling the model to automatically emphasize the features that have the greatest impact on classification. Additionally, it can capture the subtle changes caused by the disease. These improvements will make the model more sensitive to similar diseases, thus reducing misclassification and ultimately improving model accuracy.
5. Conclusions
Traditional deep learning models typically require a large amount of computational resources and storage space. Typically, training areas are limited by geography and the difficulty of setting up computational environments, resulting in limited resources. Applying traditional deep learning models can be a significant challenge. This study successfully applied a lightweight deep learning model, RegNetY-400MF, for potato leaf disease detection. Through transfer learning techniques, optimized parameter combinations, and data augmentation techniques, the model demonstrated high accuracy and efficiency. The experimental results showed that the model can achieve a best test accuracy of 90.68% on the Potato Leaf Disease in Uncontrolled Environment dataset, highlighting the strong ability of our method to discriminate between different types of potato leaf diseases. The proposed approach demonstrated superior performance compared to existing lightweight models, achieving the highest average accuracy of 89.87% with the lowest standard deviation of 1.28. This indicates a high degree of consistency in performance. The model’s compact size of 16.8 MB and low computational requirement of 0.40 GFLOPs make it highly suitable for deployment on resource-constrained devices, enabling real-time disease detection in field conditions.
In future work, we aim to explore the potential of incorporating sensors into the model to enhance its utility in realistic agricultural environments. The deployment of cameras, drone monitoring, and sensors for temperature, humidity, and soil moisture will enable real-time data collection on crop-growing conditions, thereby improving our ability to predict and understand disease occurrence. Moreover, we will expand the dataset and conduct tests on various plant diseases to validate the model’s applicability. Future studies will also include Receiver Operating Characteristic Area Under the Curve (ROC-AUC) evaluation metrics to provide a more comprehensive assessment of the model’s performance. Additionally, we will measure inference time once the model is formally ported to an edge device, offering a more accurate performance metric. To further improve the model, we will refine its architecture, with a specific focus on enhancing the attention mechanism. We are considering replacing this with the Efficient Channel Attention (ECA) method, which could lead to more accurate disease predictions and offer timely farm management advice for farmers. Ultimately, these improvements will contribute to improving productivity and crop health.