Classifying Breast Tumors in Digital Tomosynthesis by Combining Image Quality-Aware Features and Tumor Texture Descriptors
Abstract
1. Introduction
- Presenting a novel deep learning-based framework for breast tumor classification in DBT images. The unique merit of this model is the combination of DBT image quality-aware features with tumor texture descriptors, which helps accurately categorize the breast tumor as benign or malignant.
- Proposing the TomoQA deep learning model to extract quality-aware features from DBT images.
- Conducting an extensive experimental analysis on a publicly available DBT dataset and providing comparisons with existing methods to demonstrate the superiority of the proposed method. The implementation code for our proposed method from this study is publicly available on GitHub at https://github.com/loaysh2010/Classifying-Breast-Tumors-in-DBT-Images-based-on-Image-Quality-Aware-Features, accessed on 6 March 2024.
2. Related Work
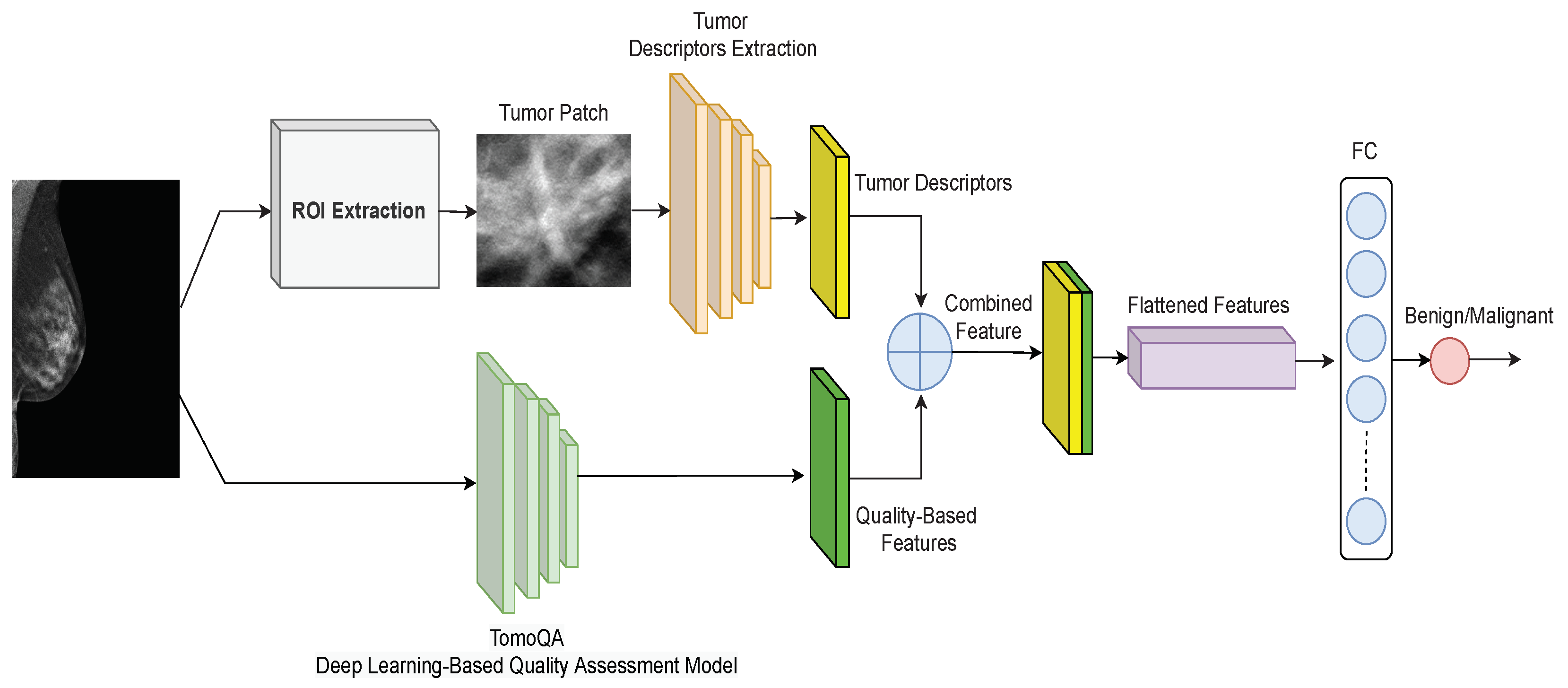
3. Proposed Breast Tumor Classification Method
3.1. Upper Branch: Extracting Tumor Texture Descriptors
3.2. Lower Branch: Extracting Image Quality-Aware Features
4. Implementation Details of the Proposed Framework
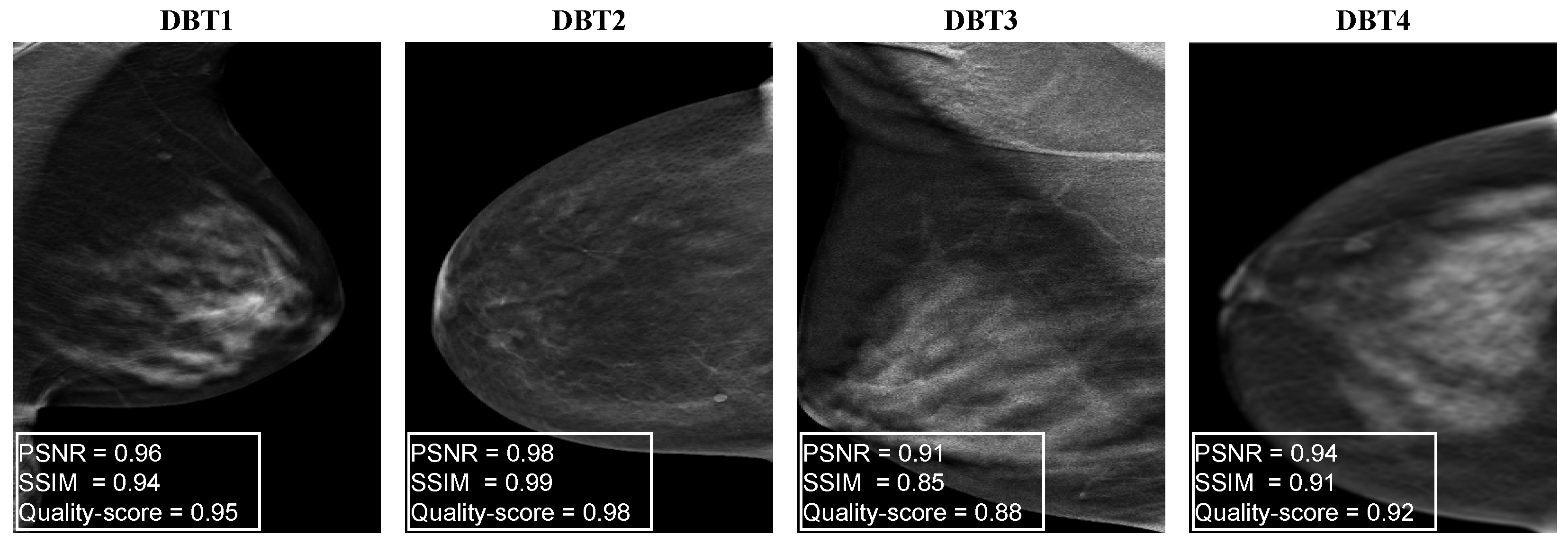
4.1. Generating Synthetic DBT Image Quality Assessment Dataset
- To generate blurred DBT images, we use the mean filter technique that takes the average of all the pixels under the kernel area to replace the central element. Each image is blurred using a mean filter with a random kernel size ranging from to with a step of 2:where m and n represent the kernel size, and represents the pixel value of our DBT image at location inside the kernel window.
- To generate a set of distorted DBT images that simulate different dose levels, we apply gamma correction to adjust the overall brightness of an input DBT image :where is the output distorted DBT image. To simulate different dose levels, 10 random values ranging from to are used for .
- To simulate bright or dark pixel artifacts in DBT images, we generate a set of distorted DBT images by adding speckle noise as follows:where represents the speckle noise of unit mean. and are the input DBT image and the output distorted image. A total of 10 random values for ranging from to are used to generate the distorted DBT images.
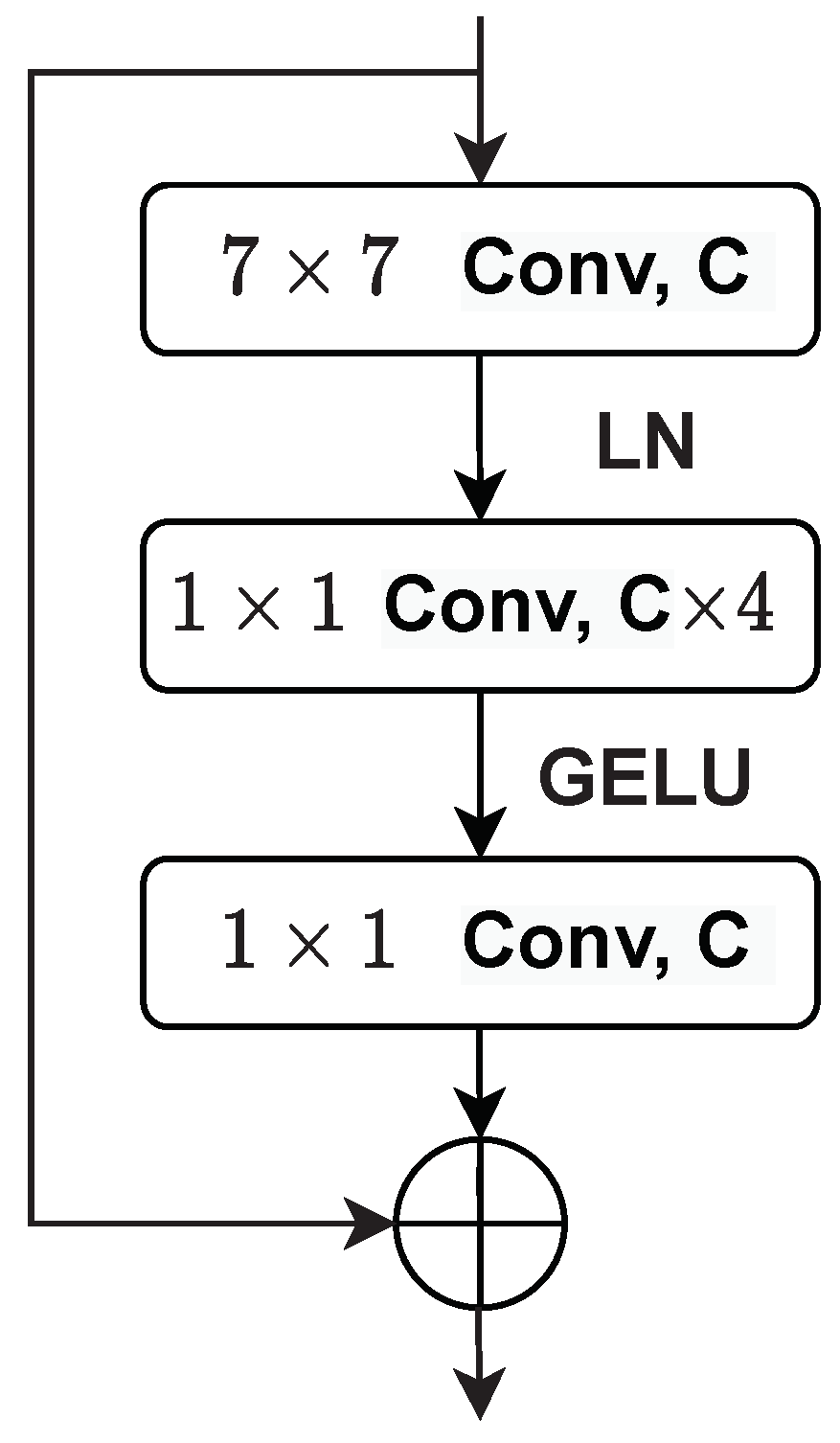
4.2. Implementation Details of the Proposed DBT IQA Model
4.3. Implementation Details of the Tumor Texture Descriptors Extraction Model
5. Experimental Results and Analysis
5.1. Dataset
5.2. Evaluation Metrics
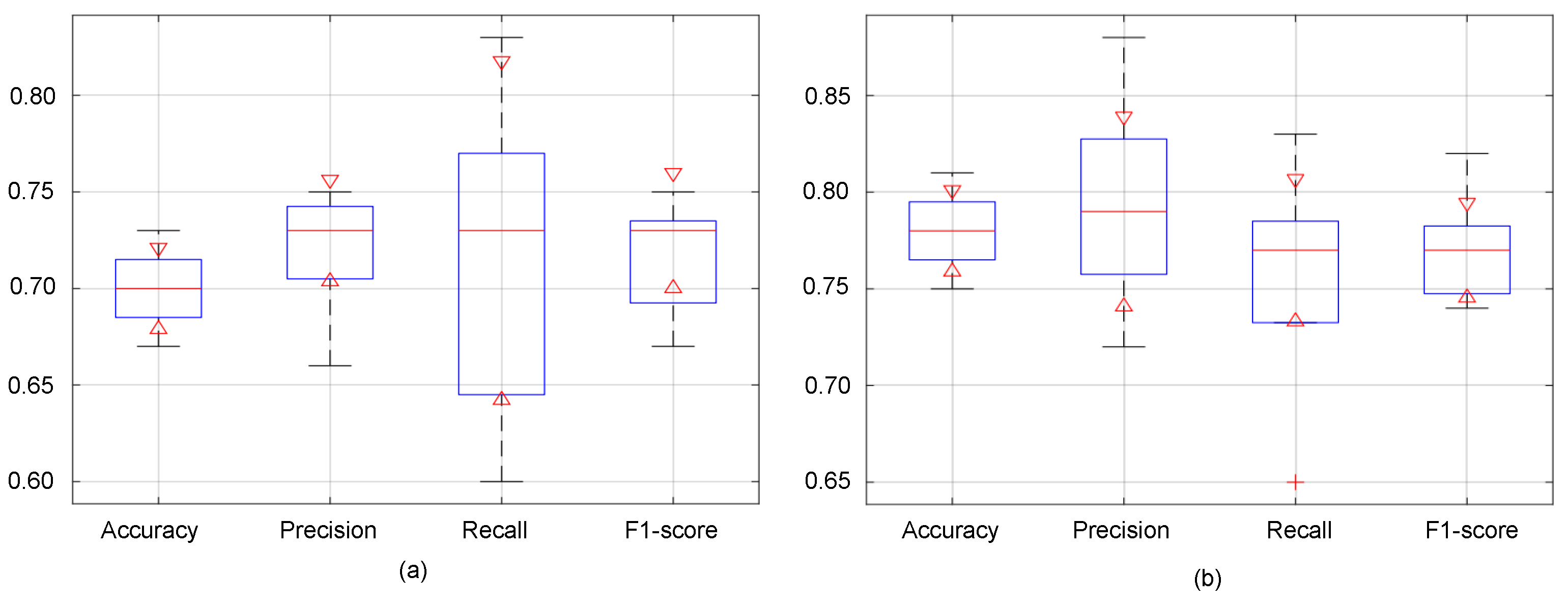
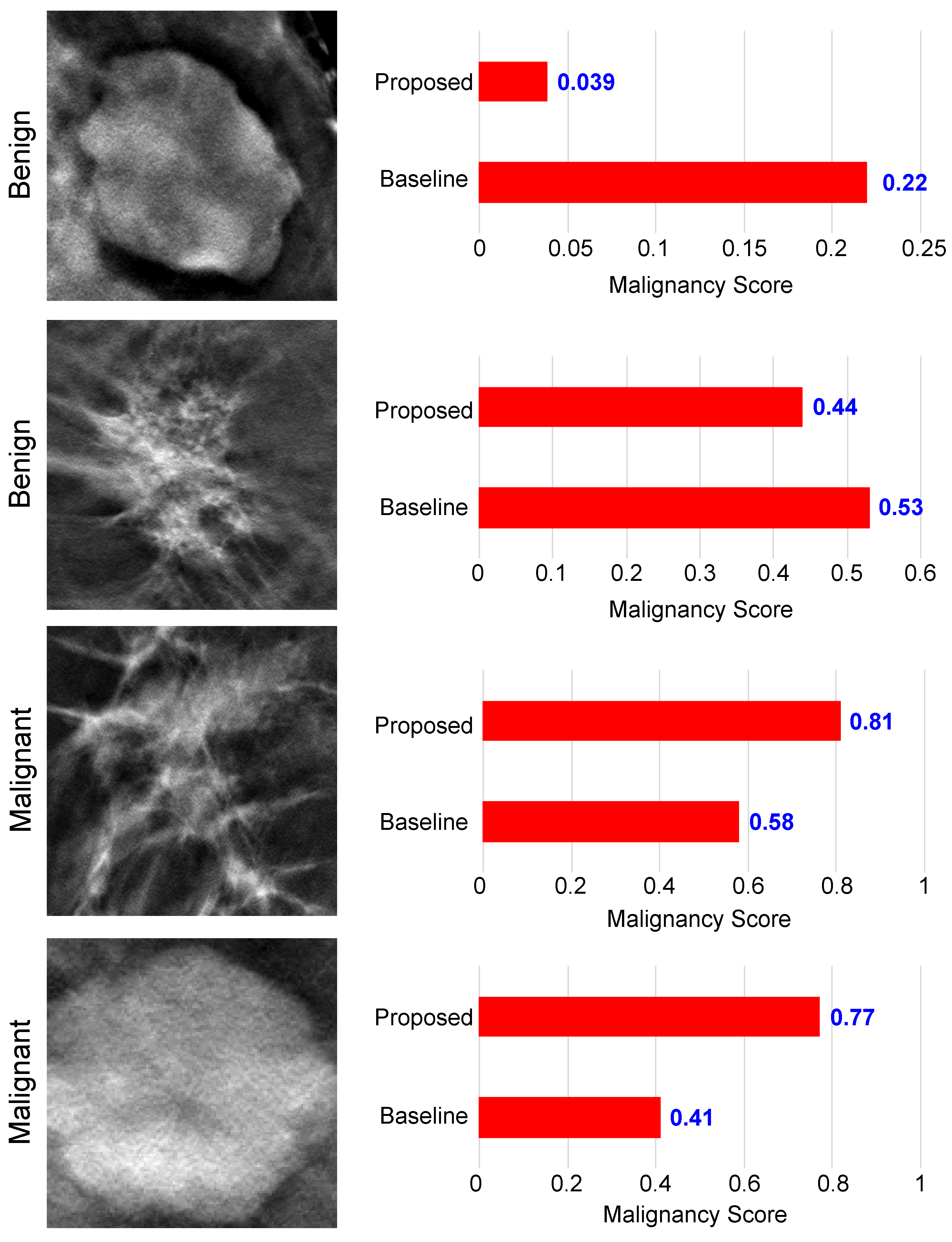
5.3. Performance Evaluation of the Proposed Malignancy Prediction Approach
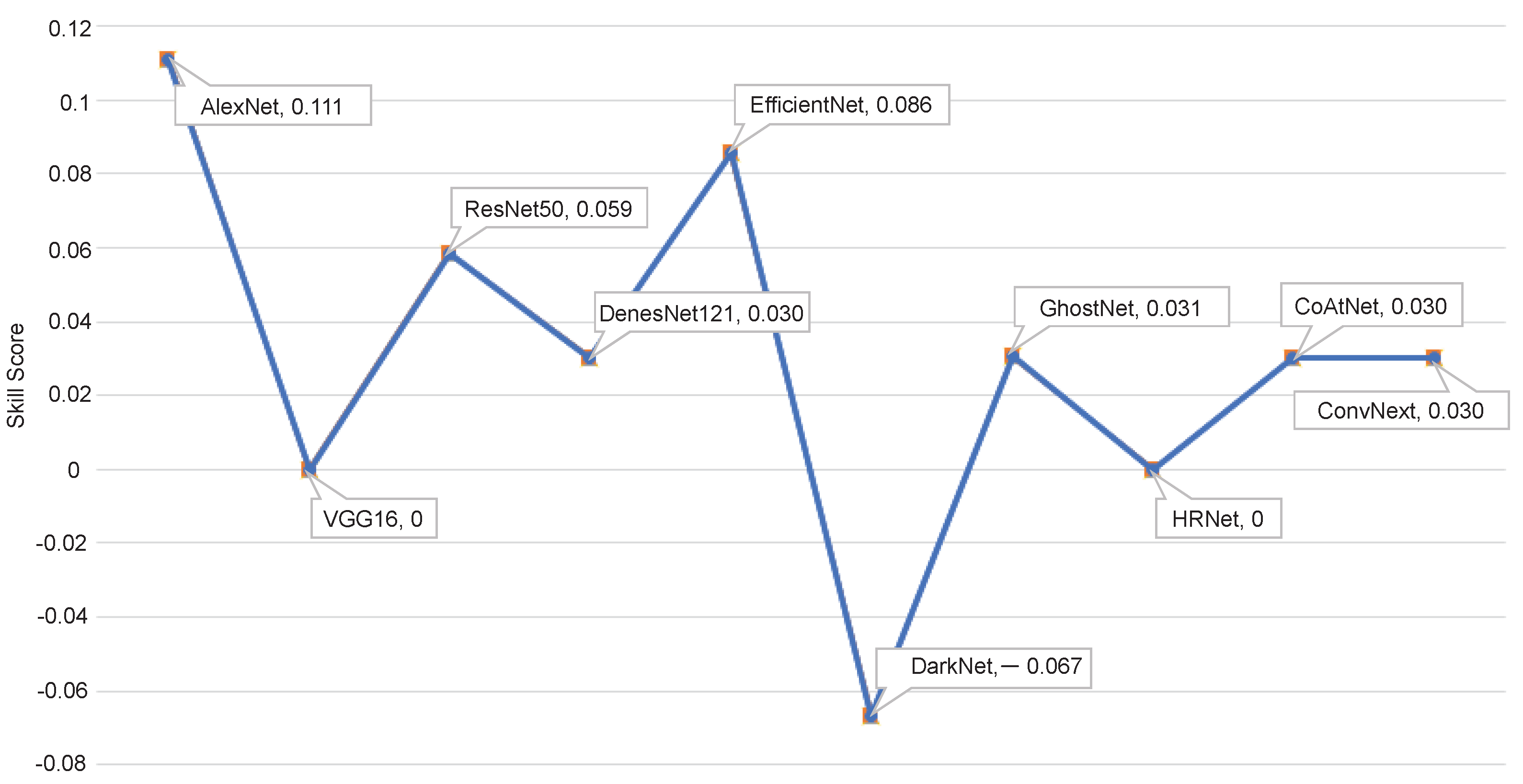
5.4. Ablation Study
5.5. Analyzing the Computational Efficiency of the Proposed Method
5.6. Enhancing Classification Accuracy through Ensemble Models
5.7. Evaluating the Proposed Method on the Breast Mammography Modality
6. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Appendix A
Appendix A.1. Evaluation Metrics for IQA Models
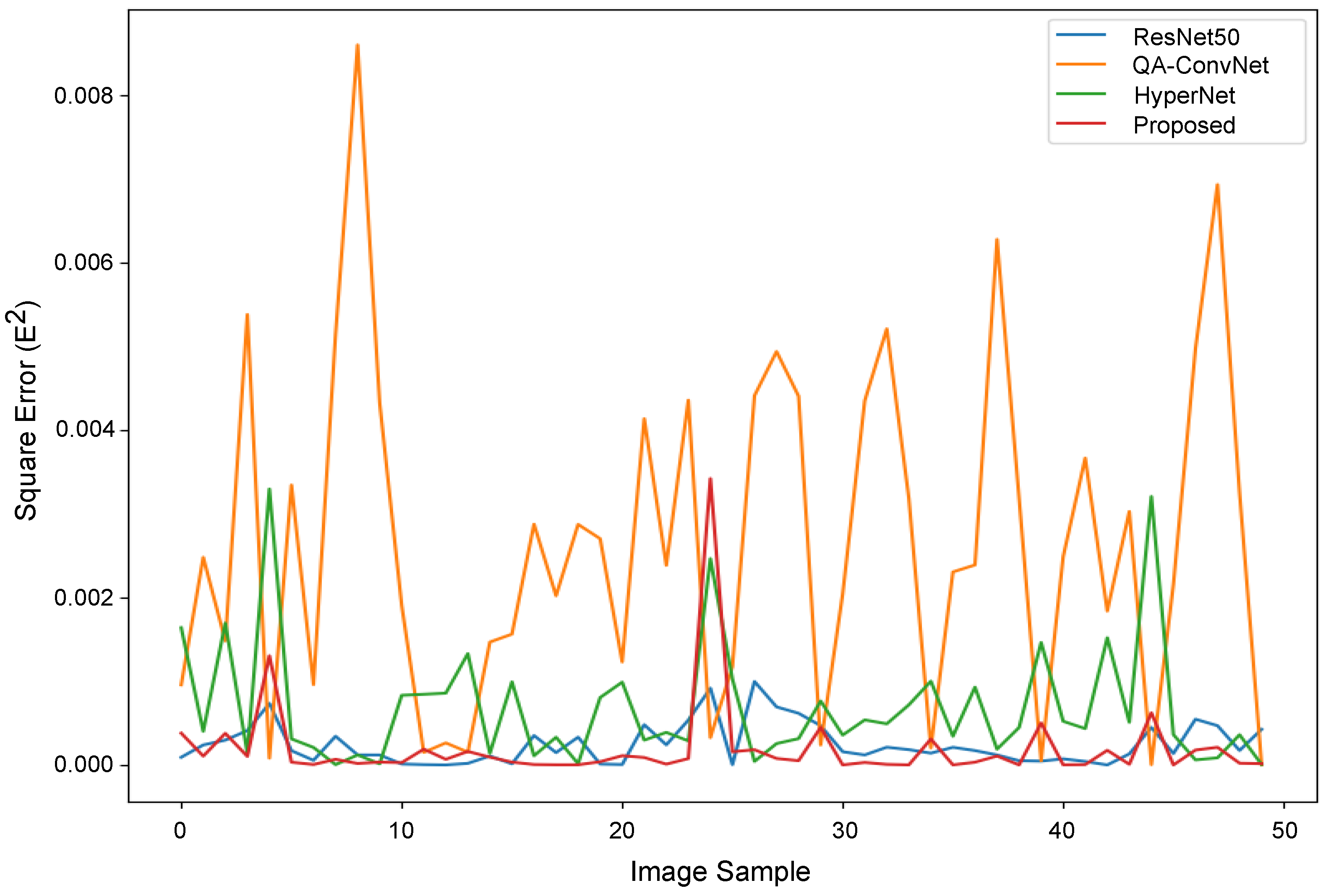
Appendix A.2. Performance Evaluation of the Proposed DBT NR-IQA Method
| PLCC ↑ | SROCC ↑ | RMSE ↓ | |
|---|---|---|---|
| Proposed | 0.9029 | 0.8885 | 0.0141 |
| QA-ConvNet [47] | 0.8608 | 0.8092 | 0.0517 |
| ResNet50 [30] | 0.8742 | 0.8692 | 0.0157 |
| HyperNet [48] | 0.8856 | 0.8673 | 0.0263 |


References
- Jemal, A.; Siegel, R.; Ward, E.; Hao, Y.; Xu, J.; Murray, T.; Thun, M.J. Cancer Statistics, 2008. CA Cancer J. Clin. 2008, 58, 71–96. [Google Scholar] [CrossRef]
- Mridha, M.F.; Hamid, M.A.; Monowar, M.M.; Keya, A.J.; Ohi, A.Q.; Islam, M.R.; Kim, J.M. A Comprehensive Survey on Deep-Learning-Based Breast Cancer Diagnosis. Cancers 2021, 13, 6116. [Google Scholar] [CrossRef] [PubMed]
- Jasti, V.D.P.; Zamani, A.S.; Arumugam, K.; Naved, M.; Pallathadka, H.; Sammy, F.; Raghuvanshi, A.; Kaliyaperumal, K. Computational Technique Based on Machine Learning and Image Processing for Medical Image Analysis of Breast Cancer Diagnosis. Secur. Commun. Netw. 2022, 2022, 1918379. [Google Scholar] [CrossRef]
- Guy, C.; Ffytche, D. An Introduction to the Principles of Medical Imaging; World Scientific Publishing Co.: Singapore, 2005. [Google Scholar] [CrossRef]
- Iranmakani, S.; Mortezazadeh, T.; Sajadian, F.; Ghaziani, M.F.; Ghafari, A.; Khezerloo, D.; Musa, A.E. A review of various modalities in breast imaging: Technical aspects and clinical outcomes. Egypt. J. Radiol. Nucl. Med. 2020, 51, 57. [Google Scholar] [CrossRef]
- Heywang-Köbrunner, S.H.; Hacker, A.; Sedlacek, S. Advantages and Disadvantages of Mammography Screening. Breast Care 2011, 6, 2. [Google Scholar] [CrossRef] [PubMed]
- Dhamija, E.; Gulati, M.; Deo, S.V.S.; Gogia, A.; Hari, S. Digital Breast Tomosynthesis: An Overview. Indian J. Surg. Oncol. 2021, 12, 315–329. [Google Scholar] [CrossRef]
- Helvie, M.A. Digital Mammography Imaging: Breast Tomosynthesis and Advanced Applications. Radiol. Clin. N. Am. 2010, 48, 917–929. [Google Scholar] [CrossRef] [PubMed]
- Haas, B.M.; Kalra, V.; Geisel, J.; Raghu, M.; Durand, M.; Philpotts, L.E. Comparison of Tomosynthesis Plus Digital Mammography and Digital Mammography Alone for Breast Cancer Screening. Radiology 2013, 269, 694–700. [Google Scholar] [CrossRef]
- Jalalian, A.; Mashohor, S.; Mahmud, R.; Karasfi, B.; Saripan, M.I.B.; Ramli, A.R.B. Foundation and methodologies in computer-aided diagnosis systems for breast cancer detection. EXCLI J. 2017, 16, 113–137. [Google Scholar] [CrossRef]
- Amrane, M.; Oukid, S.; Gagaoua, I.; Ensari, T. Breast cancer classification using machine learning. In Proceedings of the 2018 Electric Electronics, Computer Science, Biomedical Engineerings’ Meeting (EBBT), Istanbul, Turkey, 8–19 April 2018; IEEE: Piscataway, NJ, USA, 2018; pp. 1–4. [Google Scholar] [CrossRef]
- Omondiagbe, D.A.; Veeramani, S.; Sidhu, A.S. Machine Learning Classification Techniques for Breast Cancer Diagnosis. IOP Conf. Ser. Mater. Sci. Eng. 2019, 495, 012033. [Google Scholar] [CrossRef]
- Li, H.; Chen, D.; Nailon, W.H.; Davies, M.E.; Laurenson, D.I. Dual Convolutional Neural Networks for Breast Mass Segmentation and Diagnosis in Mammography. IEEE Trans. Med. Imaging 2022, 41, 3–13. [Google Scholar] [CrossRef]
- El-Shazli, A.M.A.; Youssef, S.M.; Soliman, A.H. Intelligent Computer-Aided Model for Efficient Diagnosis of Digital Breast Tomosynthesis 3D Imaging Using Deep Learning. Appl. Sci. 2022, 12, 5736. [Google Scholar] [CrossRef]
- Bai, J.; Posner, R.; Wang, T.; Yang, C.; Nabavi, S. Applying deep learning in digital breast tomosynthesis for automatic breast cancer detection: A review. Med. Image Anal. 2021, 71, 102049. [Google Scholar] [CrossRef] [PubMed]
- Debelee, T.G.; Schwenker, F.; Ibenthal, A.; Yohannes, D. Survey of deep learning in breast cancer image analysis. Evol. Syst. 2019, 11, 143–163. [Google Scholar] [CrossRef]
- Pinto, M.C.; Rodriguez-Ruiz, A.; Pedersen, K.; Hofvind, S.; Wicklein, J.; Kappler, S.; Mann, R.M.; Sechopoulos, I. Impact of Artificial Intelligence Decision Support Using Deep Learning on Breast Cancer Screening Interpretation with Single-View Wide-Angle Digital Breast Tomosynthesis. Radiology 2021, 300, 529–536. [Google Scholar] [CrossRef]
- Geiser, W.R.; Einstein, S.A.; Yang, W.T. Artifacts in Digital Breast Tomosynthesis. Am. J. Roentgenol. 2018, 211, 926–932. [Google Scholar] [CrossRef]
- Yeh, J.Y.; Chan, S. CNN-Based CAD for Breast Cancer Classification in Digital Breast Tomosynthesis. In Proceedings of the 2nd International Conference on Graphics and Signal Processing—ICGSP18, Sydney, Australia, 6–8 October 2018; ACM Press: New York, NY, USA, 2018; pp. 26–30. [Google Scholar] [CrossRef]
- Ricciardi, R.; Mettivier, G.; Staffa, M.; Sarno, A.; Acampora, G.; Minelli, S.; Santoro, A.; Antignani, E.; Orientale, A.; Pilotti, I.; et al. A deep learning classifier for digital breast tomosynthesis. Phys. Med. 2021, 83, 184–193. [Google Scholar] [CrossRef]
- Lee, W.; Lee, H.; Lee, H.; Park, E.K.; Nam, H.; Kooi, T. Transformer-based Deep Neural Network for Breast Cancer Classification on Digital Breast Tomosynthesis Images. Radiol. Artif. Intell. 2023, 5, e220159. [Google Scholar] [CrossRef]
- Moghadam, F.S.; Rashidi, S. Classification of benign and malignant tumors in Digital Breast Tomosynthesis images using Radiomic-based methods. In Proceedings of the 2023 13th International Conference on Computer and Knowledge Engineering (ICCKE), Mashhad, Iran, 1–2 November 2023; IEEE: Piscataway, NJ, USA, 2023; pp. 203–208. [Google Scholar] [CrossRef]
- SPIE-AAPM-NCI DAIR Digital Breast Tomosynthesis Lesion Detection Challenge. Available online: https://www.aapm.org/GrandChallenge/DBTex2/ (accessed on 24 January 2023).
- Zhang, Y.; Wang, X.; Blanton, H.; Liang, G.; Xing, X.; Jacobs, N. 2D Convolutional Neural Networks for 3D Digital Breast Tomosynthesis Classification. In Proceedings of the 2019 IEEE International Conference on Bioinformatics and Biomedicine (BIBM), San Diego, CA, USA, 18–21 November 2019; IEEE: Piscataway, NJ, USA, 2019; pp. 1013–1017. [Google Scholar] [CrossRef]
- Doganay, E.; Li, P.; Luo, Y.; Chai, R.; Guo, Y.; Wu, S. Breast cancer classification from digital breast tomosynthesis using 3D multi-subvolume approach. In Proceedings of the Medical Imaging 2020: Imaging Informatics for Healthcare, Research, and Applications, Houston, TX, USA, 16–17 February 2020; SPIE: Wallisellen, Switzerland, 2020; Volume 11318, p. 113180D. [Google Scholar] [CrossRef]
- Samala, R.K.; Chan, H.P.; Hadjiiski, L.; Helvie, M.A.; Richter, C.D.; Cha, K.H. Breast Cancer Diagnosis in Digital Breast Tomosynthesis: Effects of Training Sample Size on Multi-Stage Transfer Learning Using Deep Neural Nets. IEEE Trans. Med. Imaging 2019, 38, 686–696. [Google Scholar] [CrossRef]
- Domenec, P. Lesion Detection in Breast Tomosynthesis Using Efficient Deep Learning and Data Augmentation Techniques. In Artificial Intelligence Research and Development, Proceedings of the 23rd International Conference of the Catalan Association for Artificial Intelligence, Online, 20–22 October 2021; IOS Press: Amsterdam, The Netherlands, 2021; Volume 339, p. 315. [Google Scholar]
- Krizhevsky, A.; Sutskever, I.; Hinton, G.E. ImageNet classification with deep convolutional neural networks. Commun. ACM 2012, 60, 84–90. [Google Scholar] [CrossRef]
- Liu, S.; Deng, W. Very deep convolutional neural network based image classification using small training sample size. In Proceedings of the 2015 3rd IAPR Asian Conference on Pattern Recognition (ACPR), Kuala Lumpur, Malaysia, 3–6 November 2015; IEEE: Piscataway, NJ, USA, 2015; pp. 730–734. [Google Scholar] [CrossRef]
- He, K.; Zhang, X.; Ren, S.; Sun, J. Deep Residual Learning for Image Recognition. In Proceedings of the 2016 IEEE Conference on Computer Vision and Pattern Recognition (CVPR), Las Vegas, NV, USA, 27–30 June 2016; IEEE: Piscataway, NJ, USA, 2016; pp. 770–778. [Google Scholar] [CrossRef]
- Huang, G.; Liu, Z.; Maaten, L.V.D.; Weinberger, K.Q. Densely Connected Convolutional Networks. In Proceedings of the 2017 IEEE Conference on Computer Vision and Pattern Recognition (CVPR), Honolulu, HI, USA, 21–26 July 2017; IEEE: Piscataway, NJ, USA, 2017; pp. 2261–2269. [Google Scholar] [CrossRef]
- Tan, M.; Le, Q. EfficientNet: Rethinking Model Scaling for Convolutional Neural Networks. In Proceedings of the 36th International Conference on Machine Learning, PMLR, Long Beach, CA, USA, 9–15 June 2019; Volume 97, pp. 6105–6114. [Google Scholar] [CrossRef]
- Redmon, J.; Farhadi, A. YOLOv3: An Incremental Improvement. arXiv 2018, arXiv:1804.02767. [Google Scholar] [CrossRef]
- Han, K.; Wang, Y.; Tian, Q.; Guo, J.; Xu, C.; Xu, C. GhostNet: More Features From Cheap Operations. In Proceedings of the 2020 IEEE/CVF Conference on Computer Vision and Pattern Recognition (CVPR), Seattle, WA, USA, 13–19 June 2020; IEEE: Piscataway, NJ, USA, 2020; pp. 1577–1586. [Google Scholar] [CrossRef]
- Wang, J.; Sun, K.; Cheng, T.; Jiang, B.; Deng, C.; Zhao, Y.; Liu, D.; Mu, Y.; Tan, M.; Wang, X.; et al. Deep High-Resolution Representation Learning for Visual Recognition. IEEE Trans. Pattern Anal. Mach. Intell. 2021, 43, 3349–3364. [Google Scholar] [CrossRef] [PubMed]
- Dai, Z.; Liu, H.; Le, Q.V.; Tan, M. CoAtNet: Marrying Convolution and Attention for All Data Sizes. Adv. Neural Inf. Process. Syst. 2021, 34, 3965–3977. [Google Scholar] [CrossRef]
- Liu, Z.; Mao, H.; Wu, C.Y.; Feichtenhofer, C.; Darrell, T.; Xie, S. A ConvNet for the 2020s. In Proceedings of the 2022 IEEE/CVF Conference on Computer Vision and Pattern Recognition (CVPR), New Orleans, LA, USA, 18–24 June 2022; pp. 11966–11976. [Google Scholar] [CrossRef]
- Hendrycks, D.; Gimpel, K. Gaussian Error Linear Units (GELUs). arXiv 2016, arXiv:1606.08415. [Google Scholar] [CrossRef]
- Tirada, N.; Li, G.; Dreizin, D.; Robinson, L.; Khorjekar, G.; Dromi, S.; Ernst, T. Digital Breast Tomosynthesis: Physics, Artifacts, and Quality Control Considerations. RadioGraphics 2019, 39, 413–426. [Google Scholar] [CrossRef]
- Gao, Q.; Li, S.; Zhu, M.; Li, D.; Bian, Z.; Lyu, Q.; Zeng, D.; Ma, J. Blind CT Image Quality Assessment via Deep Learning Framework. In Proceedings of the 2019 IEEE Nuclear Science Symposium and Medical Imaging Conference (NSS/MIC), Manchester, UK, 26 October–2 November 2019; IEEE: Piscataway, NJ, USA, 2019; pp. 1–4. [Google Scholar] [CrossRef]
- Gao, Q.; Li, S.; Zhu, M.; Li, D.; Bian, Z.; Lv, Q.; Zeng, D.; Ma, J. Combined global and local information for blind CT image quality assessment via deep learning. In Proceedings of the Medical Imaging 2020: Image Perception, Observer Performance, and Technology Assessment, Houston, TX, USA, 15–20 February 2020; SPIE: Wallisellen, Switzerland, 2020; Volume 11316, p. 1131615. [Google Scholar] [CrossRef]
- Baig, M.A.; Moinuddin, A.A.; Khan, E. PSNR of Highest Distortion Region: An Effective Image Quality Assessment Method. In Proceedings of the 2019 International Conference on Electrical, Electronics and Computer Engineering (UPCON), Aligarh, India, 8–10 November 2019; pp. 1–4. [Google Scholar] [CrossRef]
- Wang, Z.; Bovik, A.; Sheikh, H.; Simoncelli, E. Image Quality Assessment: From Error Visibility to Structural Similarity. IEEE Trans. Image Process. 2004, 13, 600–612. [Google Scholar] [CrossRef]
- Song, P.X.-K. Correlated Data Analysis: Modeling, Analytics, and Applications, 1st ed.; Springer: New York, NY, USA, 2007. [Google Scholar] [CrossRef]
- Gautheir, T.D. Detecting Trends Using Spearman’s Rank Correlation Coefficient. Environ. Forensics 2001, 2, 359–362. [Google Scholar] [CrossRef]
- Moreira, I.C.; Amaral, I.; Domingues, I.; Cardoso, A.; Cardoso, M.J.; Cardoso, J.S. INbreast: Toward a full-field digital mammographic database. Acad. Radiol. 2012, 19, 236–248. [Google Scholar] [CrossRef]
- Bosse, S.; Maniry, D.; Wiegand, T.; Samek, W. A deep neural network for image quality assessment. In Proceedings of the 2016 IEEE International Conference on Image Processing (ICIP), Phoenix, AZ, USA, 25–28 September 2016; IEEE: Piscataway, NJ, USA, 2016; pp. 3773–3777. [Google Scholar] [CrossRef]
- Su, S.; Yan, Q.; Zhu, Y.; Zhang, C.; Ge, X.; Sun, J.; Zhang, Y. Blindly Assess Image Quality in the Wild Guided by a Self-Adaptive Hyper Network. In Proceedings of the 2020 IEEE/CVF Conference on Computer Vision and Pattern Recognition (CVPR), Seattle, WA, USA, 13–19 June 2020; IEEE: Piscataway, NJ, USA, 2020; pp. 3664–3673. [Google Scholar] [CrossRef]






| Train | Test | |
|---|---|---|
| No. of Patient | 40 | 10 |
| Blurred | 40 | 10 |
| Gamma Correction | 80 | 20 |
| Speckle Noise | 40 | 10 |
| Total Images | 200 | 50 |
| No. of Patients | No. of Tumor Patches | Augmented Patches | ||||
|---|---|---|---|---|---|---|
| Train | Test | Train | Test | Train | Test | |
| Benign | 50 | 12 | 114 | 23 | 114 | 23 |
| Malignant | 27 | 12 | 63 | 23 | 126 | 23 |
| Total | 77 | 24 | 177 | 46 | 240 | 46 |
| Backbone | Accuracy (%) | Precision (%) | Recall (%) | F1-Score (%) | |
|---|---|---|---|---|---|
| Baseline | AlexNet | 69.57 | 65.52 | 82.61 | 73.08 |
| Proposed | AlexNet + TomoQA | 78.26 | 88.24 | 65.22 | 75.00 |
| Method | Backbone Model | Accuracy (%) | Precision (%) | Recall (%) | F1-Score (%) |
|---|---|---|---|---|---|
| Yeh et al. [19] | LeNet | 59.11 | 87.50 | 66.67 | 75.00 |
| Ricciardi et al. [20] | DBT-DCNN | 64.54 | 73.90 | 65.42 | 69.39 |
| Doganay et al. [25] | Vgg16 | 65.57 | 73.08 | 82.60 | 77.56 |
| Samala et al. [26] | AlexNet | 74.66 | 78.95 | 78.95 | 79.00 |
| Moghadam et al. [22] | ML approach | 80.67 | 75.11 | 77.12 | 76.10 |
| Proposed | AlexNet + TomoQA | 78.26 | 88.24 | 65.22 | 75.00 |
| Proposec vs. Baseline | Best Accuracy Achieved (%) by the Proposed Method | Best Accuracy Achieved (%) by the Baseline | p-Value |
|---|---|---|---|
| AlexNet | 78.26 | 69.57 | <0.003 |
| VGG16 | 69.57 | 65.22 | <0.05 |
| ResNet50 | 73.91 | 67.39 | <0.05 |
| DenseNet121 | 71.74 | 67.39 | <0.02 |
| EfficientNet | 76.08 | 67.39 | <0.007 |
| DarkNet53 | 65.22 | 63.04 | <0.05 |
| GhostNet | 71.74 | 69.57 | <0.009 |
| HRNet18 | 69.57 | 65.22 | <0.05 |
| CoAtNet | 71.74 | 67.39 | <0.02 |
| ConvNext | 71.74 | 63.04 | <0.03 |
| Accuracy (%) | Precision (%) | Recall (%) | F1-Score (%) | |
|---|---|---|---|---|
| Average Aggregation | 84.78 | 90.11 | 82.61 | 82.61 |
| Median Aggregation | 86.96 | 90.48 | 82.61 | 86.36 |
| Backbone | Accuracy (%) | Precision (%) | Recall (%) | F1-Score (%) | |
|---|---|---|---|---|---|
| Baseline | AlexNet | 77.1 | 81.8 | 87.9 | 84.7 |
| Proposed | AlexNet + TomoQA | 85.0 | 84.1 | 90.0 | 86.9 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hassan, L.; Abdel-Nasser, M.; Saleh, A.; Puig, D. Classifying Breast Tumors in Digital Tomosynthesis by Combining Image Quality-Aware Features and Tumor Texture Descriptors. Mach. Learn. Knowl. Extr. 2024, 6, 619-641. https://doi.org/10.3390/make6010029
Hassan L, Abdel-Nasser M, Saleh A, Puig D. Classifying Breast Tumors in Digital Tomosynthesis by Combining Image Quality-Aware Features and Tumor Texture Descriptors. Machine Learning and Knowledge Extraction. 2024; 6(1):619-641. https://doi.org/10.3390/make6010029
Chicago/Turabian StyleHassan, Loay, Mohamed Abdel-Nasser, Adel Saleh, and Domenec Puig. 2024. "Classifying Breast Tumors in Digital Tomosynthesis by Combining Image Quality-Aware Features and Tumor Texture Descriptors" Machine Learning and Knowledge Extraction 6, no. 1: 619-641. https://doi.org/10.3390/make6010029
APA StyleHassan, L., Abdel-Nasser, M., Saleh, A., & Puig, D. (2024). Classifying Breast Tumors in Digital Tomosynthesis by Combining Image Quality-Aware Features and Tumor Texture Descriptors. Machine Learning and Knowledge Extraction, 6(1), 619-641. https://doi.org/10.3390/make6010029







