AIBH: Accurate Identification of Brain Hemorrhage Using Genetic Algorithm Based Feature Selection and Stacking
Abstract
1. Introduction
- (i)
- reducing the human-errors (it is well-known that the performance of human experts can drop below acceptable levels if they are distracted, stressed, overworked, and emotionally unbalanced, etc.),
- (ii)
- reducing the time/effort associated with training and hiring physicians,
- (iii)
- useful in teaching and research purpose as it can be used to train the senior medical student as well as resident doctors, and
- (iv)
- useful in building a context-based medical image retrieval system [3].
2. Proposed Method
2.1. Dataset
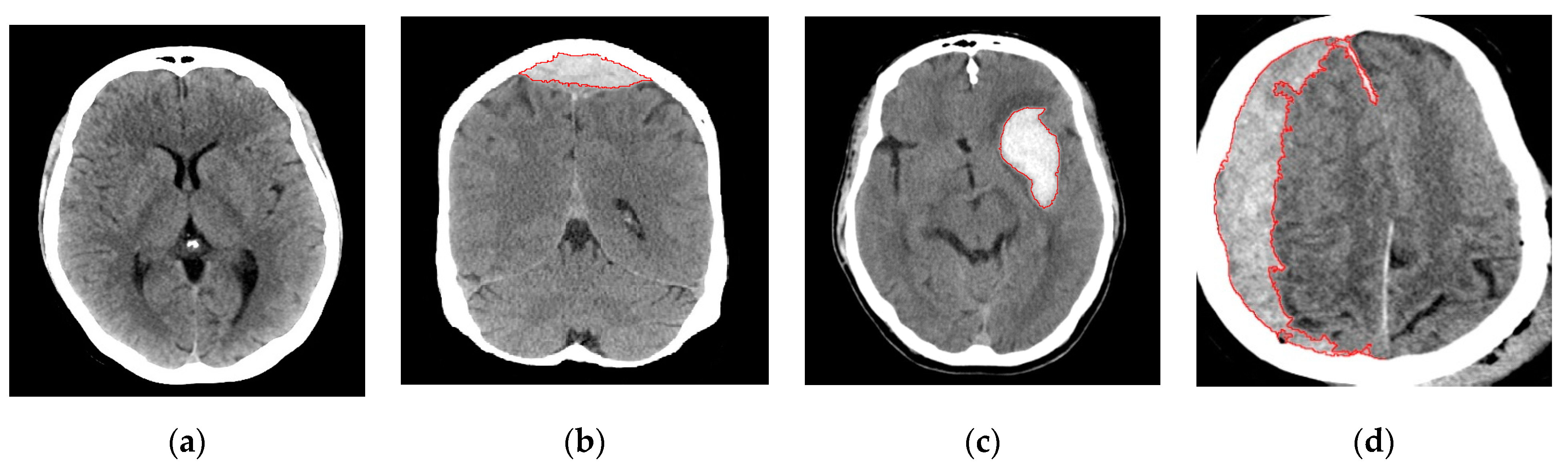
2.2. Image Preprocessing and Segmentation
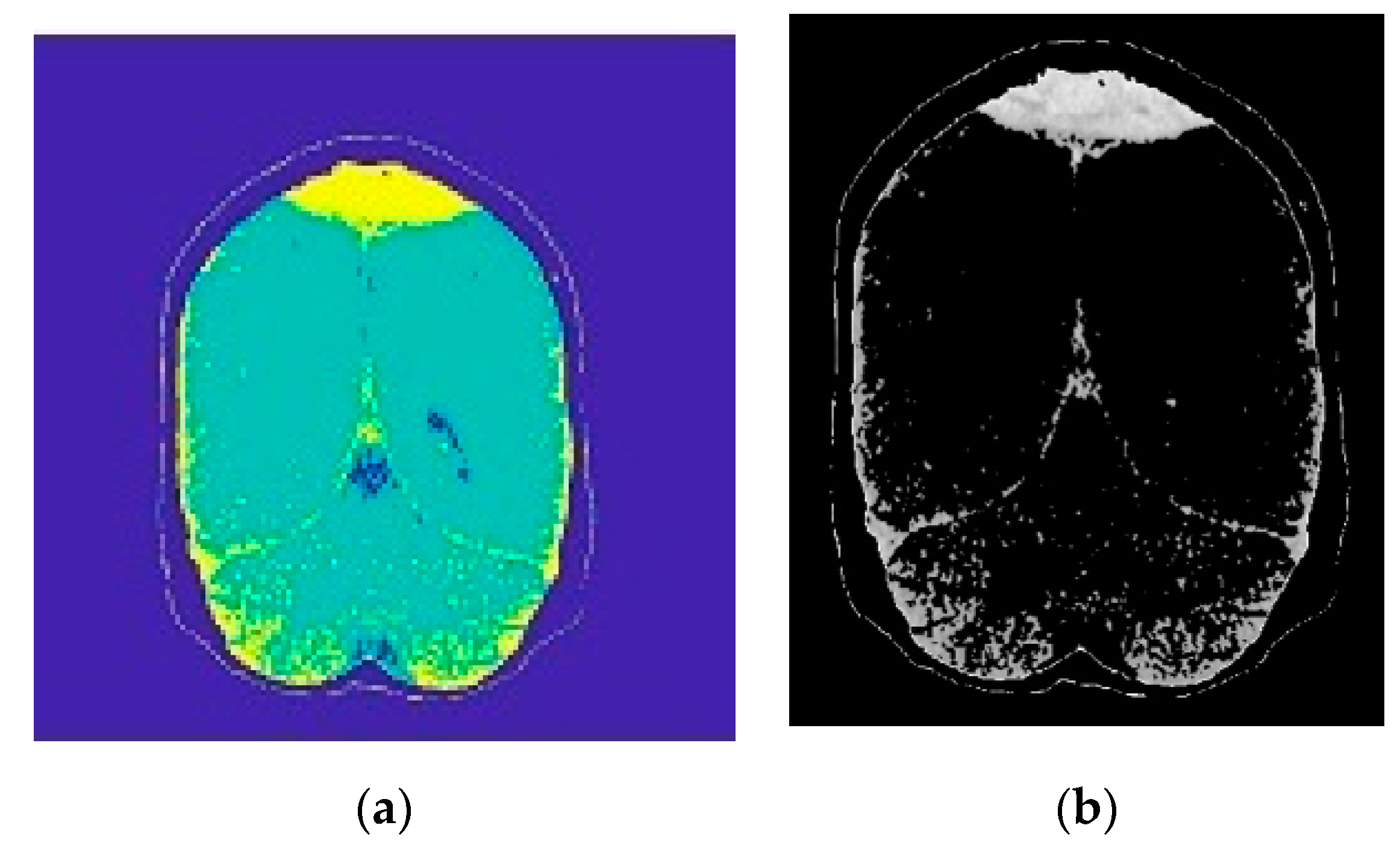
2.2.1. Image Segmentation
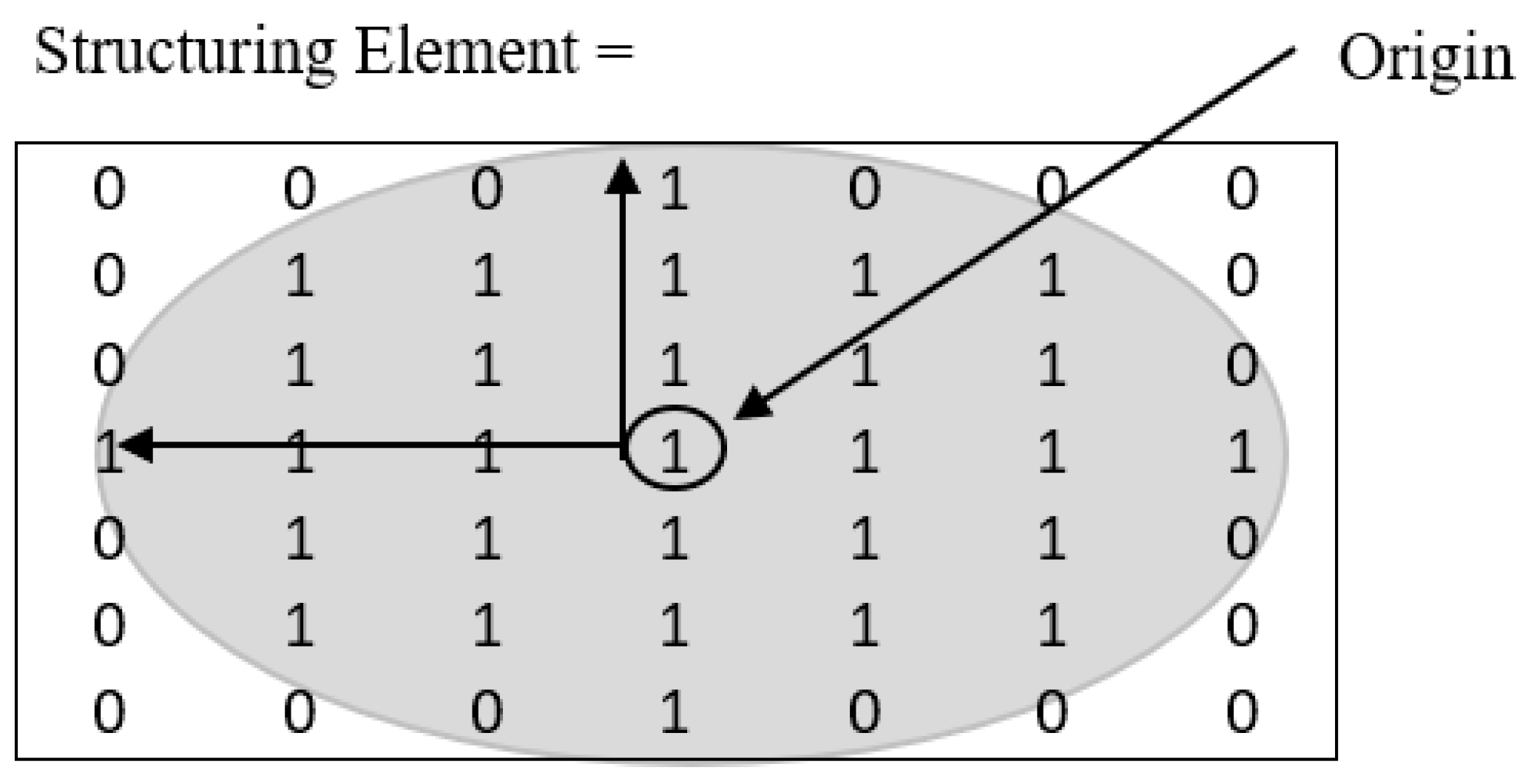
2.2.2. Morphological Operations
2.2.3. Region Growing
2.3. Feature Extraction
- Area: the actual number of pixels in the ROI. The area of the ROI provides a single scalar feature.
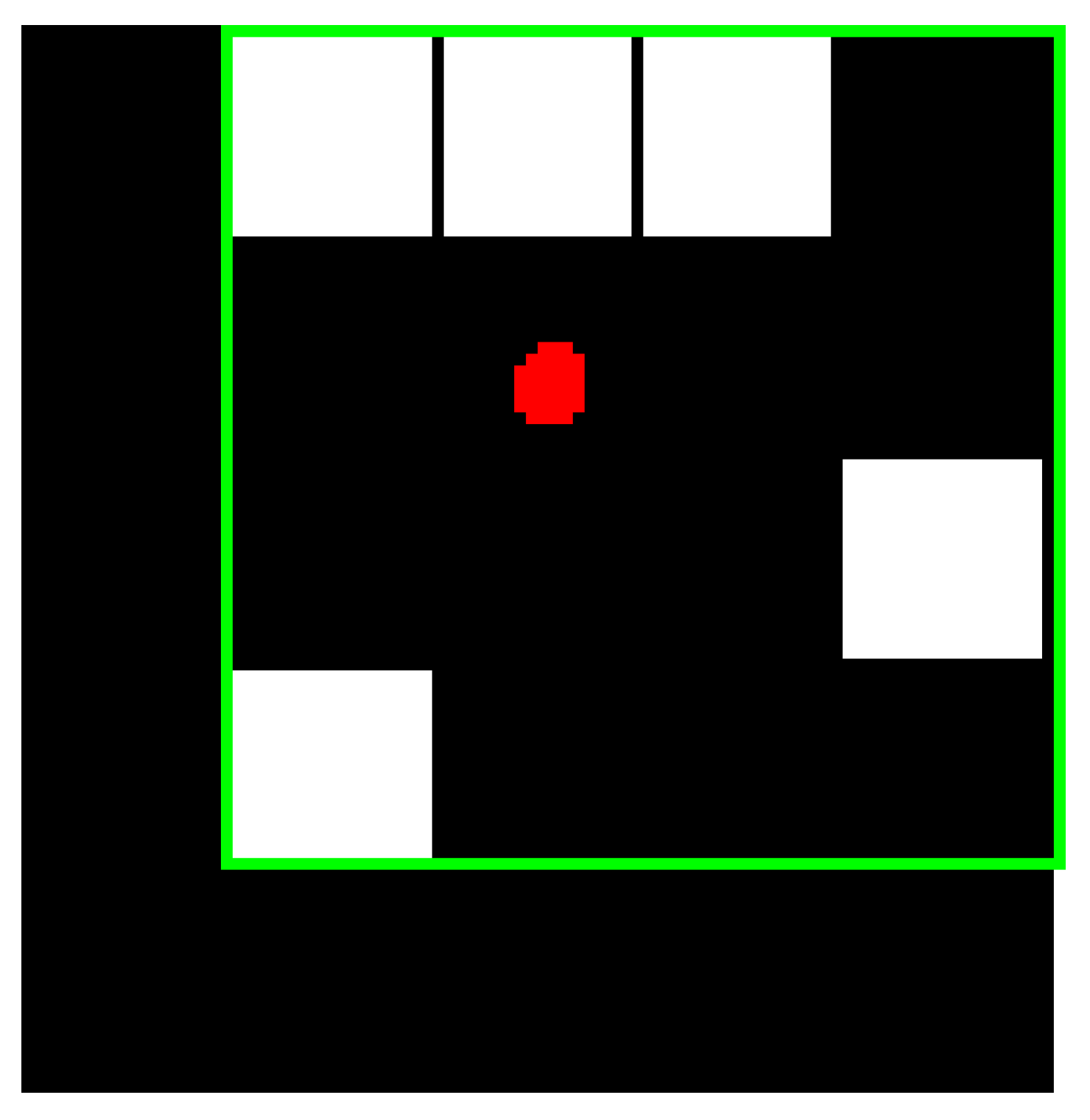
- Bounding Box: the smallest rectangle containing the ROI, which is represented as a 1-by-Q*2 vector, where Q is the number of image dimensions. Suppose that the ROI is represented by the white pixels in Figure 7, then the green box represents the bounding box of the discontinuous ROI. The bounding box of ROI provides four features, which are upper-left corner x-coordinate (ULX), upper-left corner y-coordinate (ULY), width (W), and length (L).
- Centroid: the center of mass of the ROI shown by a red dot in Figure 7. It is represented as a 1-by-Q vector, where Q is the number of image dimensions. The first element of the centroid is the horizontal coordinate or x-coordinate of the center of mass. Likewise, the second element of the centroid is the vertical coordinate or y-coordinate of the center of mass. The centroid of the ROI provides two features (centroid x, centroid y). Figure 7 illustrates the centroid for a discontinuous ROI.
- EquivDiameter: measures the diameter of a circle containing the ROI. The EquivDiameter of ROI provides a single feature and is computed as (11):
- Eccentricity: the eccentricity of an ellipse provides a measure of how nearly circular the ellipse is. It is computed as the ratio of the distance between the foci of the ellipse and its major axis length. In our implementation, eccentricity is used to capture the ROI. It provides a single feature whose value is between 0 and 1. An ellipse whose eccentricity is 0 is actually a circle, while an ellipse whose eccentricity is 1 is a line segment.
- Extent: the ratio of the pixels in the ROI to the pixels in the total bounding box. The extent provides a single feature and is computed as the area divided by the area of the bounding box given, represented as (12):
- Convex Area: the number of pixels in the convex hull of the ROI, where convex hull is the smallest convex polygon that can contain the ROI. The convex area provides a single feature.
- Filled Area: the number of on pixels in the bounding box. The on pixels correspond to the region, with all holes filled in. The filled area provides a single feature.
- Major Axis Length: measures the length (in pixels) of the major axis of the ellipse that includes the ROI. The major axis length provides a single feature.
- Minor Axis Length: measures the length (in pixels) of the minor axis of the ellipse that includes the ROI. The minor axis length provides a single feature.
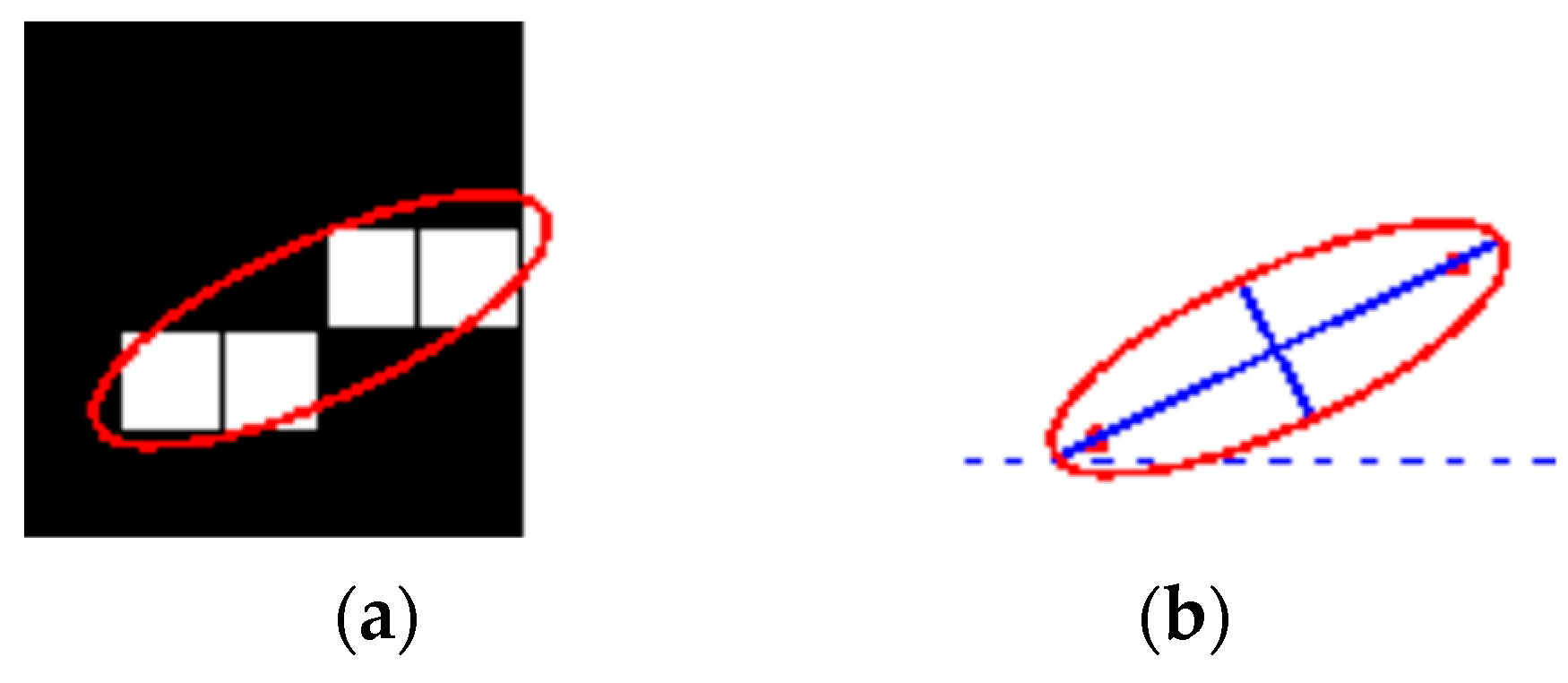
- Orientation: represents the angle (ranging from −90 to 90 degrees) between the x-axis and the major axis of the ellipse that includes the ROI, as shown in Figure 8. Figure 8 illustrates the axes and orientation of the ellipse. The left side of the figure (Figure 8a) shows an image region and its corresponding ellipse. Likewise, the right side of the figure (Figure 8b) shows the same ellipse with the solid blue lines representing the axes. Furthermore, in Figure 8b, the red dots are the foci, and the orientation is the angle between the horizontal dotted line and the major axis. The orientation also provides a single feature
- Perimeter: represents the distance around the boundary of the ROI. It is computed as the distance between each adjoining pair of pixels around the border of the ROI. The perimeter provides a single feature.
- Solidity: Solidity is given as the proportion of the pixels in the convex hull that is also in the ROI. It is computed as the ratio of area and convex area, represented as (13). The solidity also provides a single feature.
2.4. Feature Selection Using Genetic Algorithm (GA)
2.5. Performance Evaluation Metrics
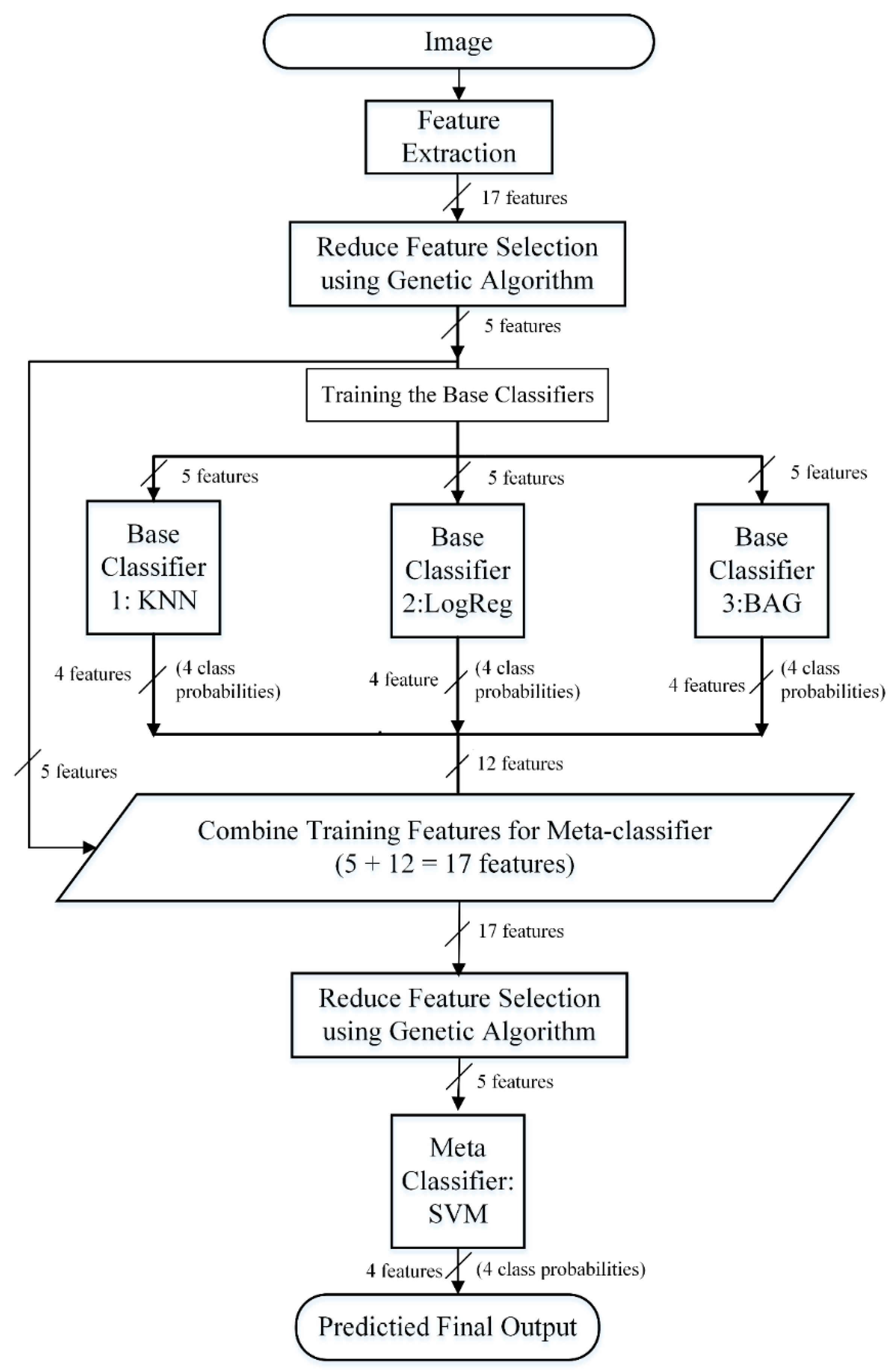
2.6. Prediction Framework
- SVM: considered as an effective algorithm for the binary prediction that minimizes both the empirical classification error in the training phase and generalized error in the test phase. In this research, we explored an RBF-kernel SVM [32,36] as one of the possible candidates to be used in the stacking framework. SVM performs classification task by maximizing the separating hyperplane between two classes and penalizes the instances on the wrong side of the decision boundary using a cost parameter, C. The RBF kernel parameter, γ and the cost parameter, C were optimized to achieve the best accuracy using a grid search [23] approach. The best values of the parameters found are C = 13.4543 and gamma = 1.6817 and used as representative parameter values for the full dataset.We used a grid search to find the best values for c, gamma. This is a search technique that has been widely used in many machine learning applications when it comes to hyperparameter optimization. There are many reasons which let us use grid search: first, the computational time required to find good parameters by grid search is not much more than that by advanced methods since there are only two parameters. Furthermore, grid search can be easily parallelized because each of (C, γ) is independent. Since doing a complete grid search may still be time-consuming, we used a coarse grid first. After identifying a better region on the grid, a finer grid search on that region was conducted [37].
- RDF: The RDF [33] works by creating a large number of decision trees, each of which is trained on a random sub-samples of the training data. The sub-samples that are used to create a decision tree is designed from a given set of observation of training data by taking ‘x’ observations at random and with replacement (also known as bootstrap sampling). Then, the final prediction results are achieved by aggregating the prediction from the individual decision trees. In our configuration, we used bootstrap samples to construct 1000 trees in the forest, and the rest of the parameters were set as default.
- ET: We explored an extremely randomized tree or extra tree [34] as one of the other possible candidates to be used in the stacking framework. ET works by building randomized decision trees on various sub-samples from the original learning sample and uses averaging to improve the prediction accuracy. We constructed the ET model with 1000 trees, and the quality of a split was measured by the Gini impurity index. The rest of the parameters were set as default.
- KNN: KNN [35] is a non-parametric and lazy learning algorithm. It is called non-parametric as it does not make any assumption for underlying data distribution, rather it creates the model directly from the dataset. Additionally, it is called lazy learning as it does not need any training data points for model generation, rather, it directly uses the training data while testing. KNN works by learning from the k closest training samples in the feature space around a target point. The classification decision is produced based on the majority votes coming from the K nearest neighbors. In this research, the value of k was set to 9, and the rest of the parameters were set as default.
- BAG: BAG [38] is an ensemble method, which operates by forming a class of algorithms that creates several instances of a base classifier on random sub-samples of the training samples and subsequently combines their individual predictions to yield a final prediction. Here, the bagging classifier was fit on multiple subsets of data constructed with repetitions using 1000 decision trees, and the rest of the parameters were set as default.
- LogReg: LogReg [21,35], also referred to as logit or MaxEnt, is a machine learning predictor that measures the relationship between the dependent categorical variable and one or more independent variables by creating an estimation probability using logistic regression. In our implementation, all the parameters of the LogReg predictor were set as default.
- SM1: KNN, LogReg, BAG in base-layer and SVM in meta-layer,
- SM2: KNN, LogReg, BAG, SVM in base-layer and SVM in meta-layer,
- SM3: KNN, LogReg, ET, SVM in base-layer and SVM in meta-layer and
- SM4: KNN, LogReg, RDF, SVM in base-layer and SVM in meta-layer
3. Results
3.1. Feature Selection
3.2. Selection of Classifiers for Stacking
3.3. Performance Comparison with Existing Approach
4. Conclusions
Author Contributions
Funding
Conflicts of Interest
Appendix A
| Before Feature Selection | ||||||
| Evaluation Metrics | ||||||
| Classifier | Hemorrhage Type | ACC | PR | Recall | F1-Score | MCC |
| ET | Epidural | 0.88 | 0.88 | 0.88 | 0.88 | 0.84 |
| Subdural | 0.96 | 0.92 | 0.96 | 0.94 | 0.92 | |
| Intraparenchymal | 0.92 | 0.96 | 0.92 | 0.94 | 0.92 | |
| Normal | 1 | 1 | 1 | 1 | 1 | |
| Average | 0.94 | 0.94 | 0.94 | 0.94 | 0.92 | |
| RDF | Epidural | 0.8 | 0.87 | 0.80 | 0.83 | 0.78 |
| Subdural | 0.96 | 0.92 | 0.96 | 0.94 | 0.92 | |
| Intraparenchymal | 0.92 | 0.88 | 0.92 | 0.90 | 0.87 | |
| Normal | 1 | 1 | 1 | 1 | 1 | |
| Average | 0.92 | 0.92 | 0.92 | 0.92 | 0.89 | |
| SVM | Epidural | 0.92 | 0.88 | 0.92 | 0.90 | 0.87 |
| Subdural | 0.96 | 0.96 | 0.96 | 0.96 | 0.95 | |
| Intraparenchymal | 0.92 | 0.96 | 0.92 | 0.94 | 0.92 | |
| Normal | 1 | 1 | 1 | 1 | 1 | |
| Average | 0.95 | 0.95 | 0.95 | 0.95 | 0.93 | |
| LogReg | Epidural | 0.76 | 0.76 | 0.76 | 0.76 | 0.68 |
| Subdural | 0.96 | 0.83 | 0.96 | 0.89 | 0.85 | |
| Intraparenchymal | 0.72 | 0.86 | 0.72 | 0.78 | 0.72 | |
| Normal | 1 | 1 | 1 | 1 | 1 | |
| Average | 0.86 | 0.86 | 0.86 | 0.86 | 0.82 | |
| KNN | Epidural | 0.56 | 0.50 | 0.56 | 0.53 | 0.36 |
| Subdural | 0.88 | 0.79 | 0.88 | 0.83 | 0.77 | |
| Intraparenchymal | 0.32 | 0.44 | 0.32 | 0.37 | 0.21 | |
| Normal | 1 | 0.96 | 1 | 0.98 | 0.97 | |
| Average | 0.69 | 0.67 | 0.69 | 0.68 | 0.59 | |
| BAG | Epidural | 0.8 | 0.91 | 0.80 | 0.85 | 0.81 |
| Subdural | 0.96 | 0.92 | 0.96 | 0.94 | 0.92 | |
| Intraparenchymal | 0.96 | 0.89 | 0.96 | 0.92 | 0.90 | |
| Normal | 1 | 1 | 1 | 1 | 1 | |
| Average | 0.93 | 0.93 | 0.93 | 0.93 | 0.91 | |
| After Feature Selection | ||||||
| ET | Epidural | 0.88 | 1 | 0.88 | 0.94 | 0.92 |
| Subdural | 1 | 0.96 | 1 | 0.98 | 0.97 | |
| Intraparenchymal | 1 | 0.93 | 1 | 0.96 | 0.95 | |
| Normal | 1 | 1 | 1 | 1 | 1 | |
| Average | 0.97 | 0.97 | 0.97 | 0.97 | 0.96 | |
| RDF | Epidural | 0.8 | 0.91 | 0.80 | 0.85 | 0.81 |
| Subdural | 0.96 | 0.92 | 0.96 | 0.94 | 0.92 | |
| Intraparenchymal | 0.96 | 0.89 | 0.96 | 0.92 | 0.90 | |
| Normal | 1 | 1 | 1 | 1 | 1 | |
| Average | 0.93 | 0.93 | 0.93 | 0.93 | 0.91 | |
| SVM | Epidural | 0.92 | 1 | 0.92 | 0.96 | 0.95 |
| Subdural | 1 | 0.93 | 1 | 0.96 | 0.95 | |
| Intraparenchymal | 1 | 1 | 1 | 1 | 1 | |
| Normal | 1 | 1 | 1 | 1 | 1 | |
| Average | 0.98 | 0.98 | 0.98 | 0.98 | 0.97 | |
| LogReg | Epidural | 0.48 | 0.52 | 0.48 | 0.50 | 0.34 |
| Subdural | 1 | 0.76 | 1 | 0.86 | 0.82 | |
| Intraparenchymal | 0.52 | 0.68 | 0.52 | 0.59 | 0.49 | |
| Normal | 1 | 1 | 1 | 1 | 1 | |
| Average | 0.75 | 0.74 | 0.75 | 0.74 | 0.67 | |
| KNN | Epidural | 0.4 | 0.45 | 0.40 | 0.43 | 0.25 |
| Subdural | 0.88 | 0.81 | 0.88 | 0.85 | 0.79 | |
| Intraparenchymal | 0.56 | 0.54 | 0.56 | 0.55 | 0.39 | |
| Normal | 1 | 1 | 1 | 1 | 1 | |
| Average | 0.71 | 0.70 | 0.71 | 0.71 | 0.61 | |
| BAG | Epidural | 0.76 | 0.90 | 0.76 | 0.83 | 0.78 |
| Subdural | 0.96 | 0.89 | 0.96 | 0.92 | 0.90 | |
| Intraparenchymal | 0.96 | 0.89 | 0.96 | 0.92 | 0.90 | |
| Normal | 1 | 1 | 1 | 1 | 1 | |
| Average | 0.92 | 0.92 | 0.92 | 0.92 | 0.89 | |
| Exp1 | ||||||
| Evaluation Metrics | ||||||
| Stacking Models | Hemorrhage Type | ACC | PR | Recall | F1-Score | MCC |
| SM1 | Epidural | 0.930 | 0.846 | 0.880 | 0.851 | 0.863 |
| Subdural | 0.980 | 0.960 | 0.960 | 0.941 | 0.960 | |
| Intraparenchymal | 0.950 | 0.916 | 0.880 | 0.923 | 0.897 | |
| Normal | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | |
| Average | 0.965 | 0.930 | 0.930 | 0.928 | 0.930 | |
| SM2 | Epidural | 0.970 | 0.958 | 0.92 | 0.938 | 0.919 |
| Subdural | 0.980 | 0.960 | 0.96 | 0.96 | 0.946 | |
| Intraparenchymal | 0.990 | 0.961 | 1.00 | 0.980 | 0.974 | |
| Normal | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | |
| Average | 0.985 | 0.969 | 0.970 | 0.969 | 0.959 | |
| SM3 | Epidural | 0.970 | 0.958 | 0.92 | 0.938 | 0.919 |
| Subdural | 0.980 | 0.960 | 0.96 | 0.960 | 0.946 | |
| Intraparenchymal | 0.990 | 0.961 | 1.00 | 0.980 | 0.974 | |
| Normal | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | |
| Average | 0.985 | 0.969 | 0.970 | 0.969 | 0.959 | |
| SM4 | Epidural | 0.970 | 0.958 | 0.92 | 0.938 | 0.919 |
| Subdural | 0.980 | 0.960 | 0.96 | 0.960 | 0.946 | |
| Intraparenchymal | 0.990 | 0.961 | 1.00 | 0.980 | 0.974 | |
| Normal | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | |
| Average | 0.985 | 0.969 | 0.970 | 0.969 | 0.959 | |
| Exp2 | ||||||
| SM1 | Epidural | 0.950 | 0.884 | 0.920 | 0.901 | 0.868 |
| Subdural | 0.980 | 0.960 | 0.960 | 0.960 | 0.946 | |
| Intraparenchymal | 0.970 | 0.958 | 0.920 | 0.938 | 0.919 | |
| Normal | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | |
| Average | 0.975 | 0.950 | 0.950 | 0.950 | 0.933 | |
| SM2 | Epidural | 0.990 | 1.00 | 0.960 | 0.979 | 0.973 |
| Subdural | 0.990 | 0.961 | 1.00 | 0.980 | 0.974 | |
| Intraparenchymal | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | |
| Normal | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | |
| Average | 0.995 | 0.990 | 0.99 | 0.989 | 0.986 | |
| SM3 | Epidural | 0.990 | 1.00 | 0.960 | 0.979 | 0.973 |
| Subdural | 0.990 | 0.961 | 1.00 | 0.980 | 0.974 | |
| Intraparenchymal | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | |
| Normal | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | |
| Average | 0.995 | 0.990 | 0.99 | 0.989 | 0.986 | |
| SM4 | Epidural | 0.990 | 1.00 | 0.960 | 0.979 | 0.973 |
| Subdural | 0.990 | 0.961 | 1.00 | 0.980 | 0.974 | |
| Intraparenchymal | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | |
| Normal | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | |
| Average | 0.995 | 0.990 | 0.99 | 0.989 | 0.986 | |
| Exp3 | ||||||
| SM1 | Epidural | 0.989 | 1.00 | 0.958 | 0.978 | 0.972 |
| Subdural | 0.989 | 0.961 | 1.00 | 0.980 | 0.974 | |
| Intraparenchymal | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | |
| Normal | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | |
| Average | 0.995 | 0.990 | 0.989 | 0.989 | 0.986 | |
| SM2 | Epidural | 0.989 | 1.00 | 0.958 | 0.978 | 0.972 |
| Subdural | 0.989 | 0.961 | 1.00 | 0.980 | 0.974 | |
| Intraparenchymal | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | |
| Normal | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | |
| Average | 0.995 | 0.990 | 0.989 | 0.989 | 0.986 | |
| SM3 | Epidural | 0.989 | 1.00 | 0.958 | 0.978 | 0.972 |
| Subdural | 0.989 | 0.961 | 1.00 | 0.980 | 0.974 | |
| Intraparenchymal | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | |
| Normal | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | |
| Average | 0.995 | 0.990 | 0.989 | 0.989 | 0.986 | |
| SM4 | Epidural | 0.989 | 1.00 | 0.958 | 0.978 | 0.972 |
| Subdural | 0.989 | 0.961 | 1.00 | 0.980 | 0.974 | |
| Intraparenchymal | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | |
| Normal | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | |
| Average | 0.995 | 0.990 | 0.989 | 0.989 | 0.986 | |
| Stacking Models | Meta Layer | Num. of Features in Base-Layer | Num. of Features in Meta-Layer | Evaluation Metrics | |||||
|---|---|---|---|---|---|---|---|---|---|
| ACC | PR | Recall | F1-Score | MCC | |||||
| Exp1 | SM1 | SVM | 17 | 29 | 0.995 | 0.990 | 0.989 | 0.989 | 0.986 |
| SM2 | SVM | 17 | 33 | 0.995 | 0.990 | 0.989 | 0.989 | 0.986 | |
| SM3 | SVM | 17 | 33 | 0.995 | 0.990 | 0.989 | 0.989 | 0.986 | |
| SM4 | SVM | 17 | 33 | 0.995 | 0.990 | 0.989 | 0.989 | 0.986 | |
| Exp4 | SM1 | ET | 17 | 29 | 0.991 | 0.990 | 0.990 | 0.939 | 0.986 |
| SM2 | ET | 17 | 33 | 0.991 | 0.990 | 0.990 | 0.939 | 0.986 | |
| SM3 | ET | 17 | 33 | 0.962 | 0.960 | 0.960 | 0.960 | 0.946 | |
| SM4 | ET | 17 | 33 | 0.991 | 0.990 | 0.990 | 0.939 | 0.986 | |
| Exp5 | SM1 | BAGGING | 17 | 29 | 0.941 | 0.940 | 0.940 | 0.9465 | 0.939 |
| SM2 | BAGGING | 17 | 33 | 0.941 | 0.940 | 0.940 | 0.9465 | 0.939 | |
| SM3 | BAGGING | 17 | 33 | 0.962 | 0.960 | 0.960 | 0.960 | 0.946 | |
| SM4 | BAGGING | 17 | 33 | 0.941 | 0.940 | 0.940 | 0.9465 | 0.939 | |
| Exp6 | SM1 | KNN | 17 | 29 | 0.790 | 0.790 | 0.780 | 0.780 | 0.722 |
| SM2 | KNN | 17 | 33 | 0.790 | 0.790 | 0.780 | 0.780 | 0.722 | |
| SM3 | KNN | 17 | 33 | 0.739 | 0.720 | 0.730 | 0.730 | 0.727 | |
| SM4 | KNN | 17 | 33 | 0.706 | 0.680 | 0.690 | 0.690 | 0.601 | |
| Exp7 | SM1 | LOG | 17 | 29 | 0.840 | 0.840 | 0.840 | 0.843 | 0.788 |
| SM2 | LOG | 17 | 33 | 0.840 | 0.840 | 0.840 | 0.843 | 0.788 | |
| SM3 | LOG | 17 | 33 | 0.945 | 0940 | 0940 | 0940 | 0.919 | |
| SM4 | LOG | 17 | 33 | 0.810 | 0.800 | 0.810 | 0.80 | 0.749 | |
| Exp8 | SM1 | RAND | 17 | 29 | 0.941 | 0.940 | 0.940 | 0.940 | 0.920 |
| SM2 | RAND | 17 | 33 | 0.941 | 0.940 | 0.940 | 0.940 | 0.920 | |
| SM3 | RAND | 17 | 33 | 0.962 | 0.960 | 0.960 | 0.960 | 0.946 | |
| SM4 | RAND | 17 | 33 | 0.941 | 0.940 | 0.940 | 0.940 | 0.920 | |
| Evaluation Metrics | |||||||
|---|---|---|---|---|---|---|---|
| Stacking Models | Hemorrhage Type | ACC | Stacking Models | Hemorrhage Type | ACC | ||
| Exp2 | SM2 | Epidural | 0.990 | 1.00 | 0.960 | 0.979 | 0.973 |
| Subdural | 0.990 | 0.961 | 1.00 | 0.980 | 0.974 | ||
| Intraparenchymal | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | ||
| Normal | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | ||
| Average | 0.995 | 0.990 | 0.99 | 0.989 | 0.986 | ||
| Exp 9 | SM2 | Epidural | 0.990 | 1.00 | 0.960 | 0.979 | 0.99 |
| Subdural | 0.990 | 0.961 | 1.00 | 0.980 | 0.99 | ||
| Intraparenchymal | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | ||
| Normal | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | ||
| Average | 0.995 | 0.990 | 0.99 | 0.989 | 0.995 | ||
References
- Ali Khairat, M.W. Epidural Hematoma. In StatPearls; StatPearls Publishing: Treasure Island, FL, USA, 2019. [Google Scholar]
- CASP12. Available online: http://predictioncenter.org/casp12/index.cgi (accessed on 20 January 2019).
- Gong, T.; Liu, R.; Tan, C.L.; Farzad, N.; Lee, C.K.; Pang, B.C.; Tian, Q.; Tang, S.; Zhang, Z. Classification of CT Brain Images of Head Trauma. In Proceedings of the 2nd IAPR International Conference on Pattern Recognition in Bioinformatics, Singapore, 1–2 October 2007; pp. 401–408. [Google Scholar]
- Sapra, P.; Singh, R.; Khurana, S. Brain tumor detection using Neural Network. Int. J. Sci. Mod. Eng. 2013, 1, 83–88. [Google Scholar]
- Al-Ayyoub, M.; Alawad, D.; Al-Darabsah, K.; Aljarrah, I. Automatic detection and classification of brain hemorrhages. WSEAS Trans. Comput. 2013, 12, 395–405. [Google Scholar]
- Phong, T.D.; Duong, H.N.; Nguyen, H.T.; Trong, N.T.; Nguyen, V.H.; Hoa, T.V.; Snasel, V. Brain Hemorrhage Diagnosis by Using Deep Learning. In Proceedings of the 2017 International Conference on Machine Learning and Soft Computing, Ho Chi Minh City, Vietnam, 13–16 January 2017. [Google Scholar]
- Sharma, B.; Venugopalan, K. Classification of Hematomas in Brain CT Images Using Neural Network. In Proceedings of the 2014 International Conference on Issues and Challenges in Intelligent Computing Techniques (ICICT), Ghaziabad, India, 7–8 February 2014. [Google Scholar]
- Filho, P.P.R.; Sarmento, R.M.; Holanda, G.B.; de Alencar Lima, D. New approach to detect and classify stroke in skull CT images via analysis of brain tissue densities. Comput. Methods Programs Biomed. 2017, 148, 27–43. [Google Scholar] [CrossRef] [PubMed]
- Roy, S.; Saha, A.; Bandyopadhyay, S.K. Brain tumor segmentation and quantification from MRI of brain. J. Glob. Res. Comput. Sci. 2011, 2, 155–160. [Google Scholar]
- Mahajan, R.; Mahajan, P.M. Survey On Diagnosis Of Brain Hemorrhage By Using Artificial Neural Network. Int. J. Sci. Res. Eng. Technol. 2016, 5, 378–381. [Google Scholar]
- Shahangian, B.; Pourghassem, H. Automatic Brain Hemorrhage Segmentation and Classification in CTscan Images. In Proceedings of the 2013 8th Iranian Conference on Machine Vision and Image Processing, Zanjan, Iran, 10–12 September 2013. [Google Scholar]
- Garg, H. A hybrid GSA-GA algorithm for constrained optimization problems. Inf. Sci. 2019, 478, 499–523. [Google Scholar] [CrossRef]
- Shelke, V.R.; Rajwade, R.A.; Kulkarni, M. Intelligent Acute Brain Hemorrhage Diagnosis System. In Proceedings of the International Conference on Advances in Computer Science, AETACS, NCR, India, 13–14 December 2013. [Google Scholar]
- Garg, H.; Kaur, G. Quantifying gesture information in brain hemorrhage patients using probabilistic dual hesitant fuzzy sets with unknown probability information. Comput. Ind. Eng. 2020, 140, 106211. [Google Scholar] [CrossRef]
- Kerekes, Z.; Tóth, Z.; Szénási, S.; Tóth, Z.; Sergyán, S. Colon Cancer Diagnosis on Digital Tissue Images. In Proceedings of the 2013 IEEE 9th International Conference on Computational Cybernetics (ICCC), Tihany, Hungary, 8–10 July 2013. [Google Scholar]
- Al-Darabsah, K.; Al-Ayyoub, M. Breast Cancer Diagnosis Using Machine Learning Based on Statistical and Texture Features Extraction. In Proceedings of the 4th International Conference on Information and Communication Systems (ICICS 2013), Irbid, Jordan, 23–25 April 2013. [Google Scholar]
- Otsu, N. A threshold selection method from gray-level histograms. IEEE Trans. Syst. Man Cybern. 1979, 9, 62–66. [Google Scholar] [CrossRef]
- Srisha, R.; Khan, A. Morphological Operations for Image Processing: Understanding and Its Applications. In Proceedings of the National Conference on VLSI, Signal processing & Communications, Vignans University, Guntur, India, 11–12 December 2013. [Google Scholar]
- Hoque, M.T.; Iqbal, S. Genetic algorithm-based improved sampling for protein structure prediction. Int. J. Bio-Inspired Comput. 2017, 9, 129–141. [Google Scholar] [CrossRef]
- Hoque, M.T.; Chetty, M.; Sattar, A. Protein Folding Prediction in 3D FCC HP Lattice Model Using Genetic Algorithm. In Proceedings of the IEEE Congress on Evolutionary Computation (CEC), Singapore, 25–28 September 2007; pp. 4138–4145. [Google Scholar]
- Hoque, M.T.; Chetty, M.; Lewis, A.; Sattar, A.; Avery, V.M. DFS Generated Pathways in GA Crossover for Protein Structure Prediction. Neurocomputing 2010, 73, 2308–2316. [Google Scholar] [CrossRef]
- Frey, D.J.; Mishra, A.; Hoque, M.T.; Abdelguerfi, M.; Soniat, T. A machine learning approach to determine oyster vessel behavior. Mach. Learn. Knowl. Extr. 2018, 1, 4. [Google Scholar] [CrossRef]
- Bergstra, J.; Bengio, Y. Random search for hyper-parameter optimization. J. Mach. Learn. Res. 2012, 13, 281–305. [Google Scholar]
- Hastie, T.; Tibshirani, R.; Friedman, J. The Elements of Statistical Learning, 2nd ed.; Springer: New York, NY, USA, 2009. [Google Scholar]
- Bishop, C. Pattern Recognition and Machine Learning; Springer: New York, NY, USA, 2009. [Google Scholar]
- Iqbal, S.; Mishra, A.; Hoque, T. Improved Prediction of Accessible Surface Area Results in Efficient Energy Function Application. J. Theor. Biol. 2015, 380, 380–391. [Google Scholar] [CrossRef]
- Wolpert, D.H. Stacked generalization. Neural Netw. 1992, 5, 241–259. [Google Scholar] [CrossRef]
- Mishra, A.; Pokhrel, P.; Hoque, M.T. StackDPPred: A stacking based prediction of DNA-binding protein from sequence. Bioinformatics 2019, 35, 433–441. [Google Scholar] [CrossRef] [PubMed]
- Iqbal, S.; Hoque, M.T. PBRpredict-Suite: A suite of models to predict peptide-recognition domain residues from protein sequence. Bioinformatics 2018, 34, 3289–3299. [Google Scholar] [CrossRef] [PubMed]
- Nagi, S.; Bhattacharyya, D.K. Classification of microarray cancer data using ensemble approach. Netw. Model. Anal. Health Inform. Bioinform. 2013, 2, 159–173. [Google Scholar] [CrossRef]
- Gattani, S.; Mishra, A.; Hoque, M.T. StackCBPred: A stacking based prediction of protein-carbohydrate binding sites from sequence. Carbohydr. Res. 2019, 486, 107857. [Google Scholar] [CrossRef]
- Cortes, C.; Vapnik, V. Support-vector networks. Mach. Learn. 1995, 20, 273–297. [Google Scholar] [CrossRef]
- Ho, T.K. Random Decision Forests. In Proceedings of the Third International Conference on Document Analysis and Recognition, Montreal, QC, Canada, 14–16 August 1995; pp. 278–282. [Google Scholar]
- Geurts, P.; Ernst, D.; Wehenkel, L. Extremely randomized trees. Mach. Learn. 2006, 63, 3–42. [Google Scholar] [CrossRef]
- Altman, N.S. An Introduction to Kernel and Nearest-Neighbor Nonparametric Regression. Am. Stat. 1992, 46, 175–185. [Google Scholar]
- Vapnik, V.N. An overview of statistical learning theory. IEEE Trans. Neural Netw. 1999, 10, 988–999. [Google Scholar] [CrossRef] [PubMed]
- Hsu, C.W.; Chang, C.C.; Lin, C.-J. A Practical Guide to Support Vector Classication; Department of Computer Science, National Taiwan University: Taipei, Taiwan, 2010; pp. 1–12. [Google Scholar]
- Breiman, L. Bagging predictors. Mach. Learn. 1996, 24, 123–140. [Google Scholar] [CrossRef]
- Ma, Z.; Wang, P.; Gao, Z.; Wang, R.; Khalighi, K. Ensemble of machine learning algorithms using the stacked generalization approach to estimate the warfarin dose. PLoS ONE 2018, 13, e0205872. [Google Scholar] [CrossRef]
- Pedregosa, F.; Varoquaux, G.; Gramfort, A.; Michel, V.; Thirion, B.; Grisel, O.; Blondel, M.; Prettenhofer, P.; Weiss, R.; Dubourg, V.; et al. Scikit-learn: Machine Learning in Python. J. Mach. Learn. Res. 2011, 12, 2825–2830. [Google Scholar]
- Xu, R.; Zhou, J.; Wang, H.; He, Y.; Wang, X.; Liu, B. Identifying DNA-binding proteins by combining support vector machine and PSSM distance transformation. BMC Syst. Biol. 2015, 9, S10. [Google Scholar] [CrossRef]
- Taherzadeh, G.; Yang, Y.; Zhang, T.; Liew, A.W.C.; Zhou, Y. Sequence-based prediction of protein–peptide binding sites using support vector machine. J. Comput. Chem. 2016, 37, 1223–1229. [Google Scholar] [CrossRef]
- Liua, H.-L.; Chen, S.-C. Prediction of disulfide connectivity in proteins with support vector machine. J. Chin. Inst. Chem. Eng. 2007, 38, 63–70. [Google Scholar] [CrossRef]
- Kumar, R.; Srivastava, A.; Kumari, B.; Kumar, M. Prediction of β-lactamase and its class by Chou’s pseudo-amino acid composition and support vector machine. J. Theor. Biol. 2015, 365, 96–103. [Google Scholar] [CrossRef]
- Bzdok, D. Classical Statistics and Statistical Learning in Imaging Neuroscience. Front. Neurosci. 2017, 11, 543. [Google Scholar] [CrossRef] [PubMed]
- Bzdok, D.; Altman, N.; Krzywinski, M. Points of significance: Statistics versus machine learning. Nat. Methods 2018, 15, 233–234. [Google Scholar] [CrossRef] [PubMed]



| Name of Metric | Definition |
|---|---|
| True positive (TP) | Correctly predicted type of Brain hemorrhage |
| True negative (TN) | Correctly predicted other type of Brain hemorrhage |
| False positive (FP) | Incorrectly predicted Brain hemorrhage type |
| False negative (FN) | Incorrectly predicted other type of Brain hemorrhage |
| Recall/Sensitivity/True Positive Rate (TPR) | |
| Accuracy (ACC) | |
| Precision | |
| F1 score (Harmonic mean of precision and recall) | |
| Mathews Correlation Coefficient (MCC) |
| Algorithm | Num. of Features | Evaluation Metrics | |||||
|---|---|---|---|---|---|---|---|
| ACC | PR | Recall | F1-Score | MCC | |||
| Before Feature Selection | ET | 17 | 0.94 | 0.94 | 0.94 | 0.94 | 0.92 |
| RDF | 17 | 0.92 | 0.92 | 0.92 | 0.92 | 0.89 | |
| SVM | 17 | 0.95 | 0.95 | 0.95 | 0.95 | 0.93 | |
| LogReg | 17 | 0.86 | 0.86 | 0.86 | 0.86 | 0.82 | |
| KNN | 17 | 0.69 | 0.67 | 0.69 | 0.68 | 0.59 | |
| BAG | 17 | 0.93 | 0.93 | 0.93 | 0.93 | 0.91 | |
| After GA-based Feature Selection | ET | 5 | 0.97 | 0.97 | 0.97 | 0.97 | 0.96 |
| RDF | 5 | 0.93 | 0.93 | 0.93 | 0.93 | 0.91 | |
| SVM | 5 | 0.98 | 0.98 | 0.98 | 0.98 | 0.97 | |
| LogReg | 5 | 0.75 | 0.74 | 0.75 | 0.74 | 0.67 | |
| KNN | 5 | 0.71 | 0.70 | 0.71 | 0.71 | 0.61 | |
| BAG | 5 | 0.92 | 0.92 | 0.92 | 0.92 | 0.89 | |
| Metric/Methods | ET | RDF | SVM | LogReg | KNN | BAG |
|---|---|---|---|---|---|---|
| ACC | 0.97 | 0.93 | 0.98 | 0.75 | 0.71 | 0.92 |
| PR | 0.97 | 0.93 | 0.98 | 0.74 | 0.70 | 0.92 |
| Recall | 0.97 | 0.93 | 0.98 | 0.75 | 0.71 | 0.92 |
| F1-score | 0.97 | 0.93 | 0.98 | 0.74 | 0.71 | 0.92 |
| MCC | 0.96 | 0.91 | 0.97 | 0.67 | 0.61 | 0.89 |
| Stacking Models | Num. of Features in Base-Layer | Num. of Features in Meta-Layer | Evaluation Metrics | |||||
|---|---|---|---|---|---|---|---|---|
| ACC | PR | Recall | F1-Score | MCC | ||||
| Exp1 | SM1 | 17 | 29 | 0.965 | 0.930 | 0.930 | 0.928 | 0.930 |
| SM2 | 17 | 33 | 0.985 | 0.969 | 0.970 | 0.969 | 0.959 | |
| SM3 | 17 | 33 | 0.985 | 0.969 | 0.970 | 0.969 | 0.959 | |
| SM4 | 17 | 33 | 0.985 | 0.969 | 0.970 | 0.969 | 0.959 | |
| Exp2 | SM1 | 5 | 17 | 0.975 | 0.950 | 0.950 | 0.950 | 0.933 |
| SM2 | 5 | 21 | 0.995 | 0.990 | 0.990 | 0.989 | 0.986 | |
| SM3 | 5 | 21 | 0.995 | 0.990 | 0.990 | 0.989 | 0.986 | |
| SM4 | 5 | 21 | 0.995 | 0.990 | 0.990 | 0.989 | 0.986 | |
| Exp3 | SM1 | 5 | 5 | 0.995 | 0.990 | 0.989 | 0.989 | 0.986 |
| SM2 | 5 | 6 | 0.995 | 0.990 | 0.989 | 0.989 | 0.986 | |
| SM3 | 5 | 8 | 0.995 | 0.990 | 0.989 | 0.989 | 0.986 | |
| SM4 | 5 | 7 | 0.995 | 0.990 | 0.989 | 0.989 | 0.986 | |
| Metric/Methods | Al-Ayyoub et al. | AIBH (% imp.) |
|---|---|---|
| ACC | - | 0.995 (-) |
| PR | 0.925 | 0.990 (7.03%) |
| Recall | 0.922 | 0.989 (7.27%) |
| F1-score | 0.921 | 0.989 (7.38%) |
| MCC | - | 0.986 (-) |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Alawad, D.M.; Mishra, A.; Hoque, M.T. AIBH: Accurate Identification of Brain Hemorrhage Using Genetic Algorithm Based Feature Selection and Stacking. Mach. Learn. Knowl. Extr. 2020, 2, 56-77. https://doi.org/10.3390/make2020005
Alawad DM, Mishra A, Hoque MT. AIBH: Accurate Identification of Brain Hemorrhage Using Genetic Algorithm Based Feature Selection and Stacking. Machine Learning and Knowledge Extraction. 2020; 2(2):56-77. https://doi.org/10.3390/make2020005
Chicago/Turabian StyleAlawad, Duaa Mohammad, Avdesh Mishra, and Md Tamjidul Hoque. 2020. "AIBH: Accurate Identification of Brain Hemorrhage Using Genetic Algorithm Based Feature Selection and Stacking" Machine Learning and Knowledge Extraction 2, no. 2: 56-77. https://doi.org/10.3390/make2020005
APA StyleAlawad, D. M., Mishra, A., & Hoque, M. T. (2020). AIBH: Accurate Identification of Brain Hemorrhage Using Genetic Algorithm Based Feature Selection and Stacking. Machine Learning and Knowledge Extraction, 2(2), 56-77. https://doi.org/10.3390/make2020005







