Study on the Differences between the Extraction Results of the Structural Parameters of Individual Trees for Different Tree Species Based on UAV LiDAR and High-Resolution RGB Images
Abstract
1. Introduction
2. Materials and Methods
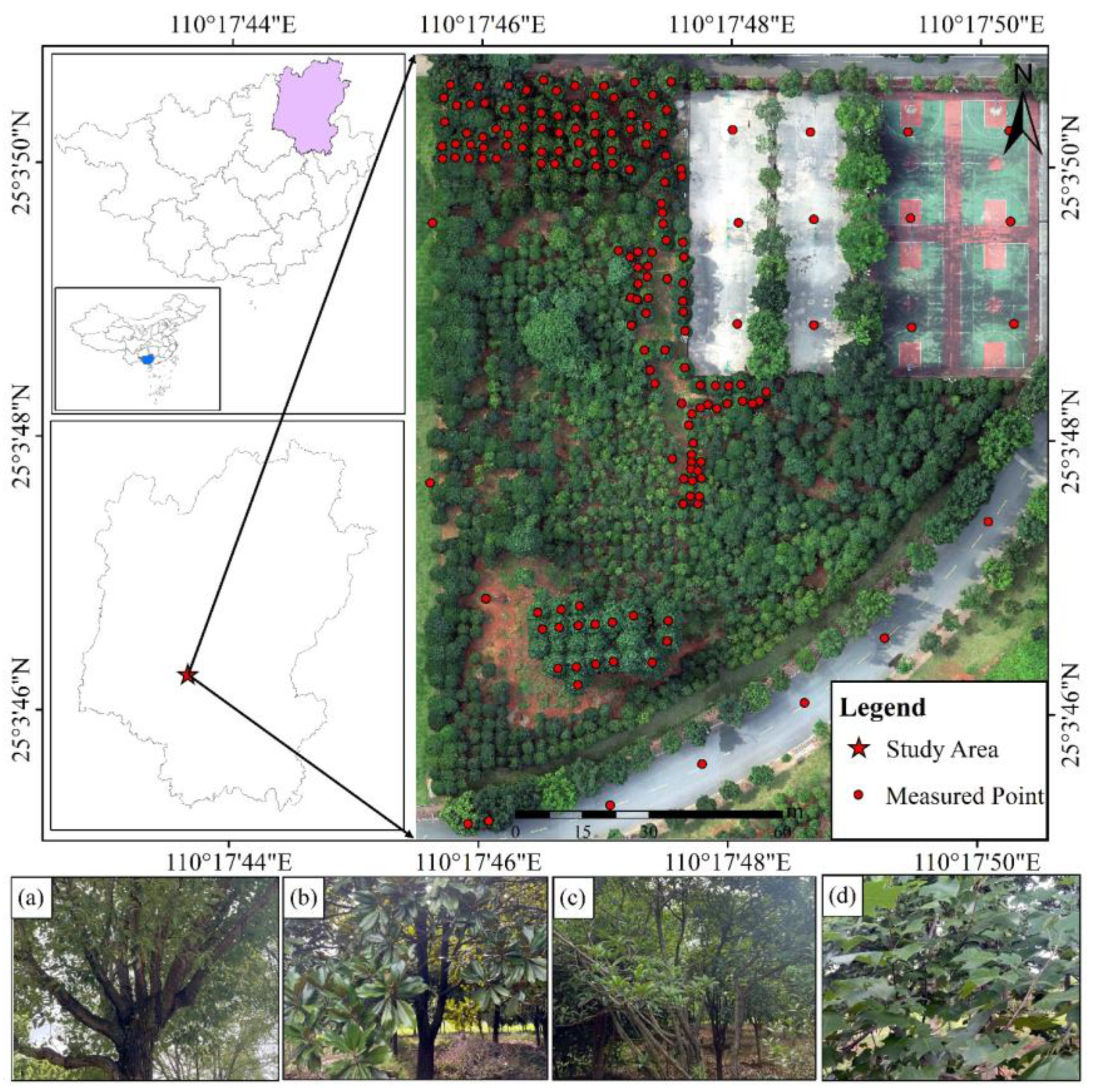
2.1. Study Area
2.2. Data Collection and Processing
- 1.
- Field Data
- 2.
- UAV LiDAR Point Cloud Data
- 3.
- High-resolution RGB Stereo Image-derived Point Cloud Data
2.3. Individual Tree Crown Segmentation
2.4. Individual Tree Structure Parameter Extraction
3. Results
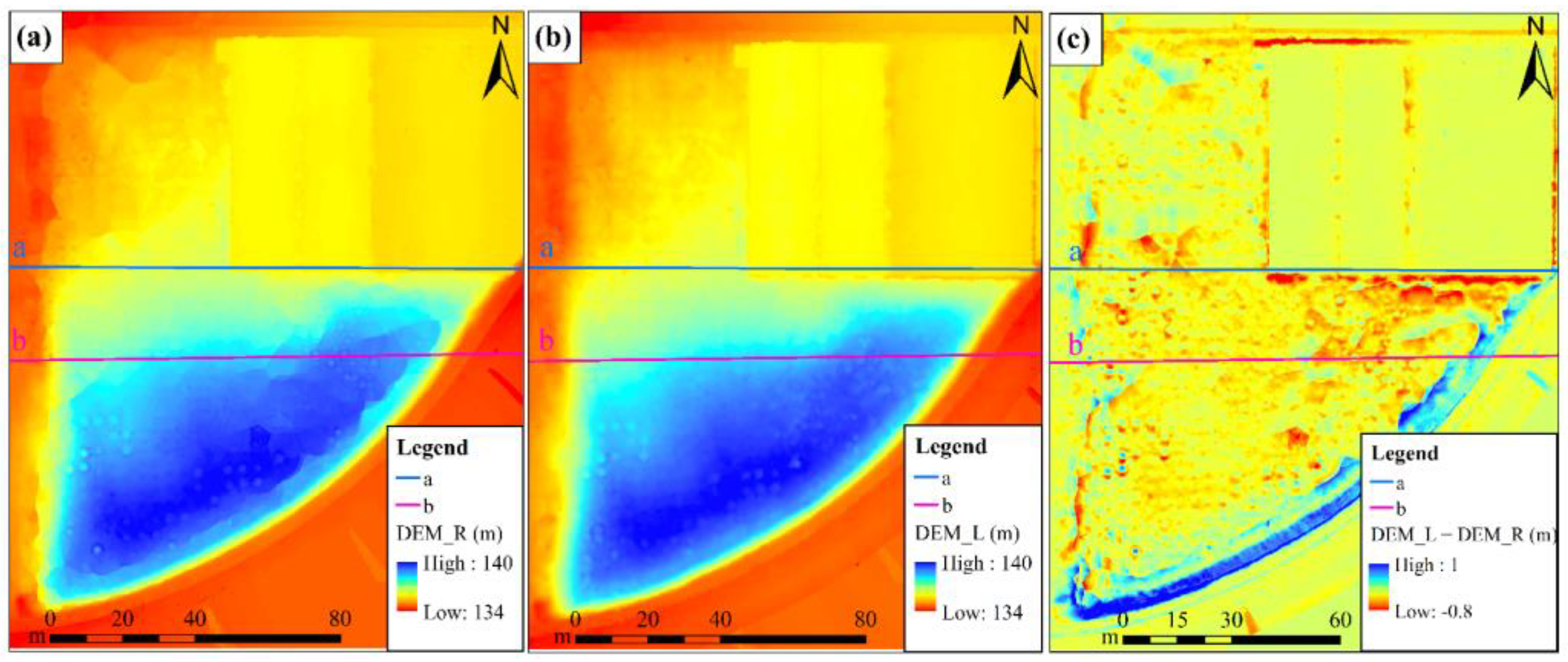
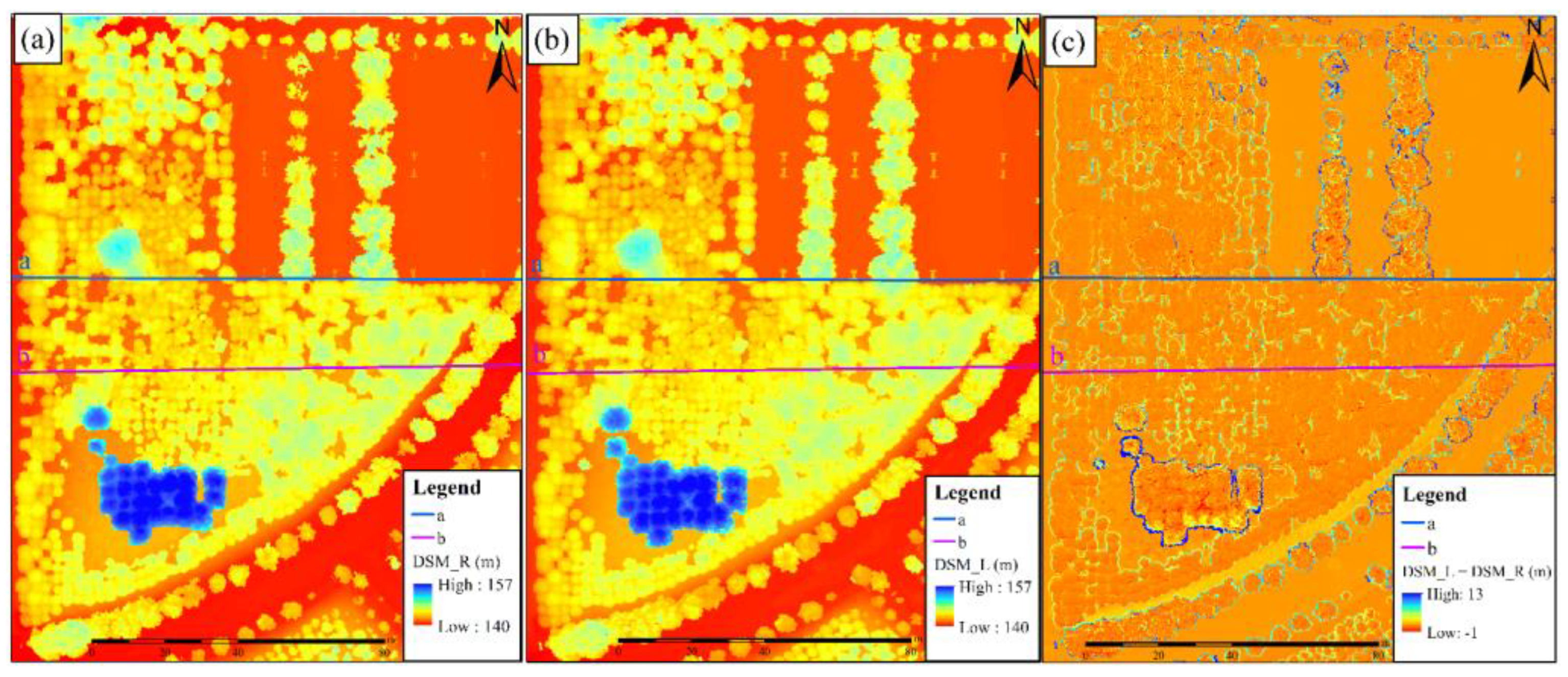
3.1. Analysis of the Generation Results of the DEMs and DSMs
- Results of the DEMs
- 2.
- Results of the DSMs
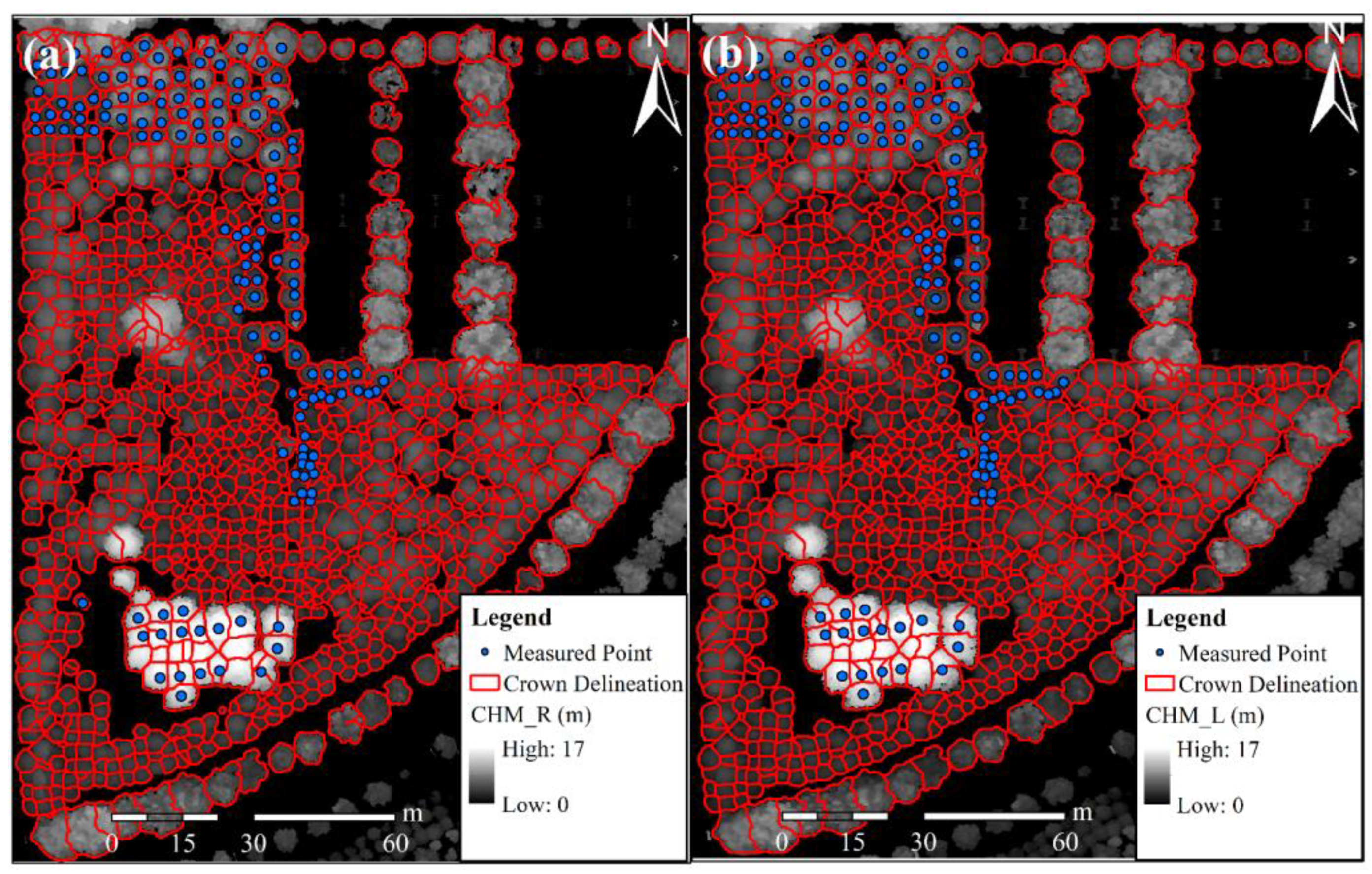
3.2. Analysis of Individual Tree Segmentation Results and Extraction Results for the Structural Parameters of Individual Trees
- 1.
- Segmentation Results for Individual Trees
- 2.
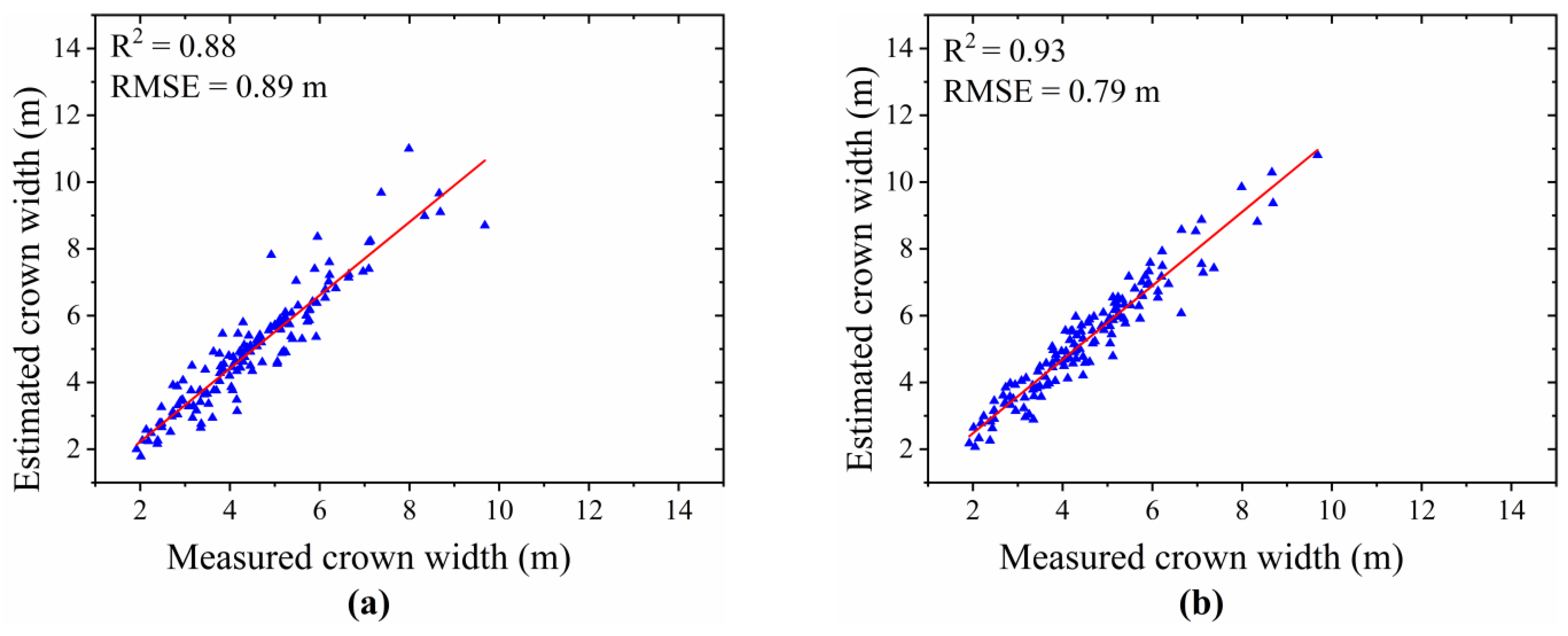
- Result of Tree Crown Width Extraction
- 3.
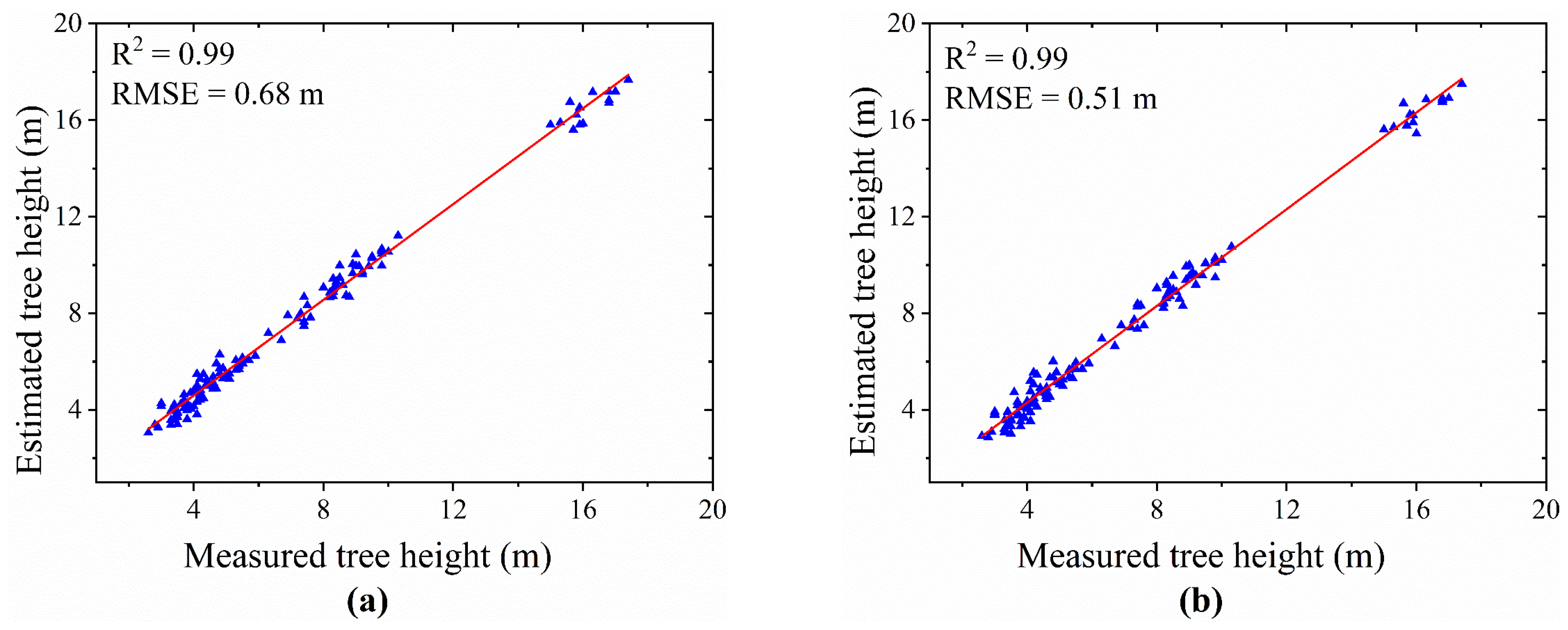
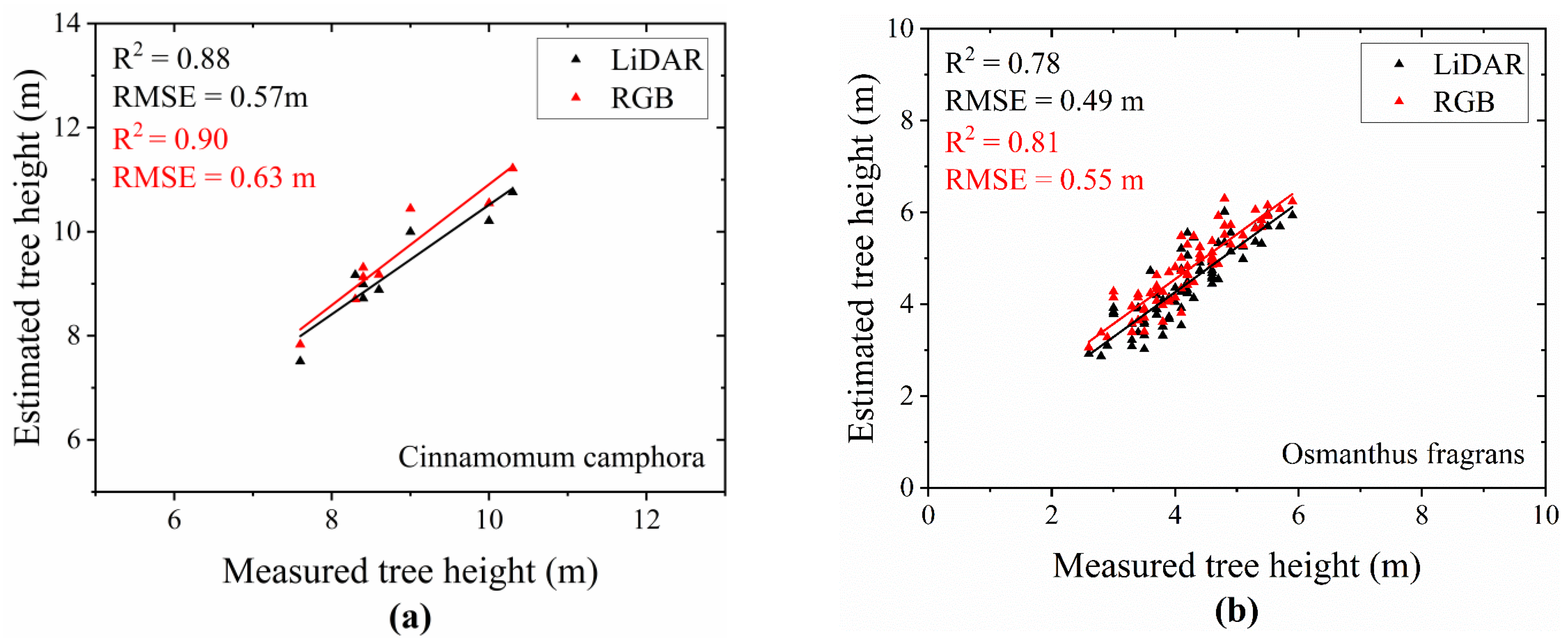
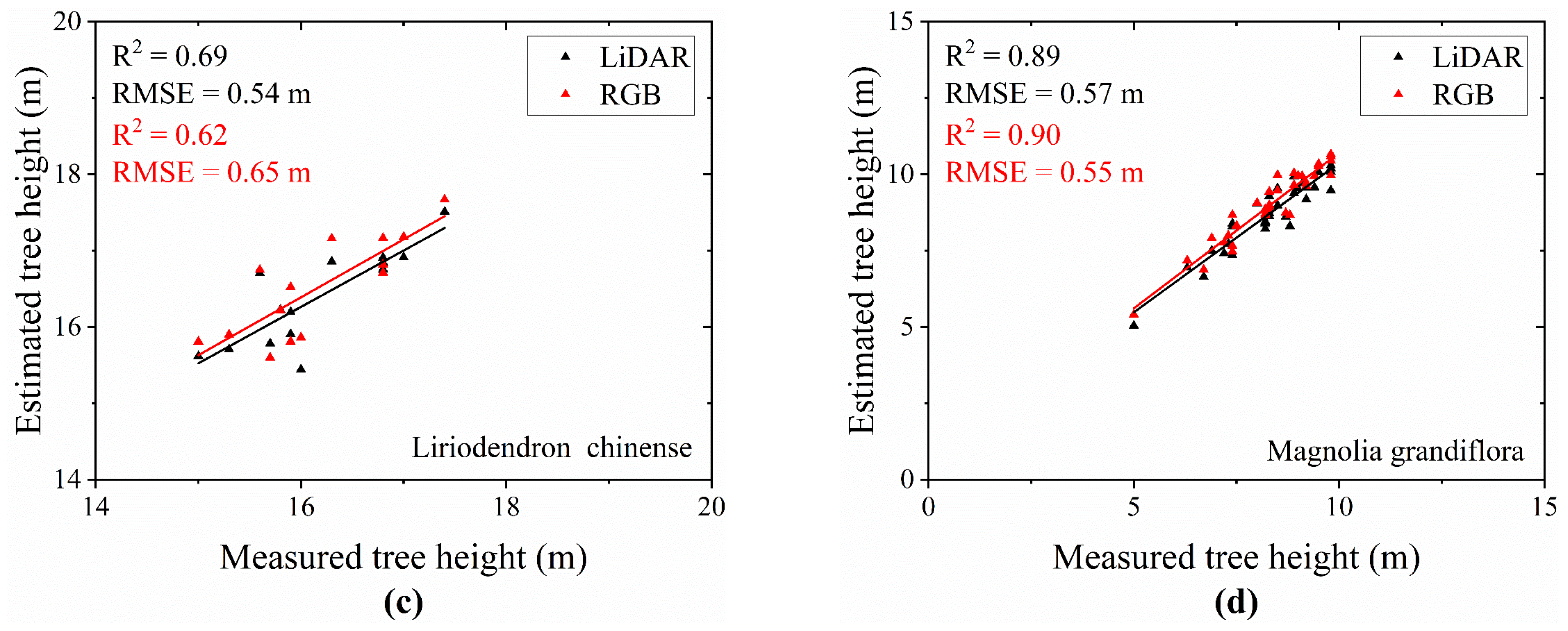
- Result of Tree Height Extraction
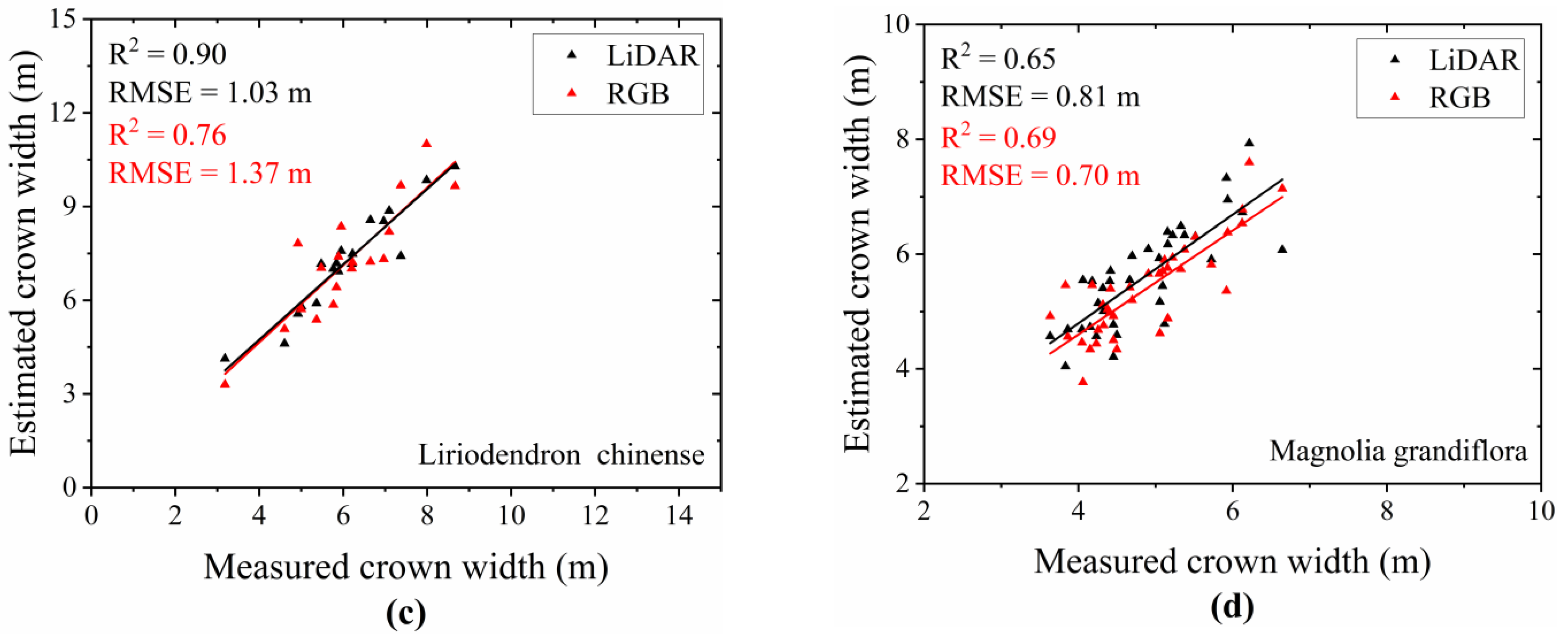
3.3. Comparison of Crown Width and Height Extraction Results for Different Tree Species
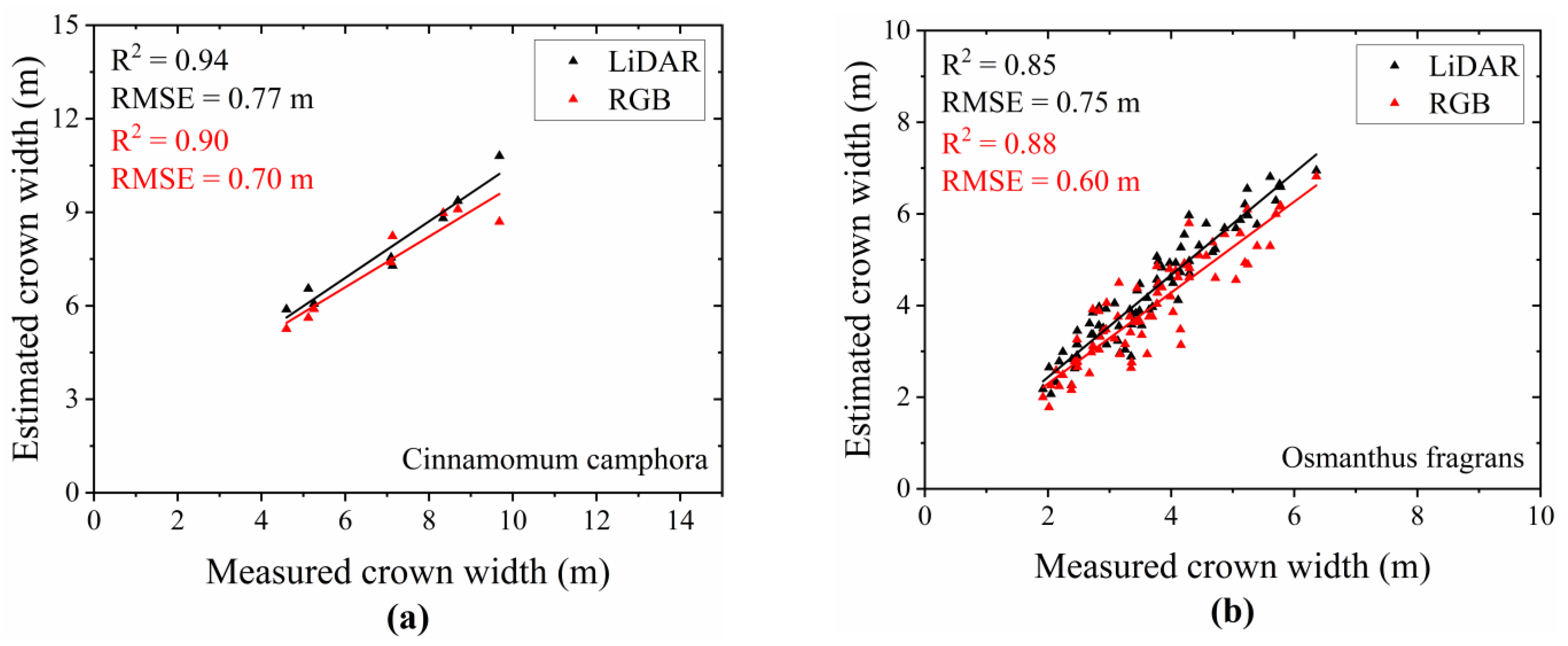
- Extraction Results of Crown Width for Different Tree Species
- 2.
- Extraction Results of Tree Height for Different Tree Species
4. Discussion
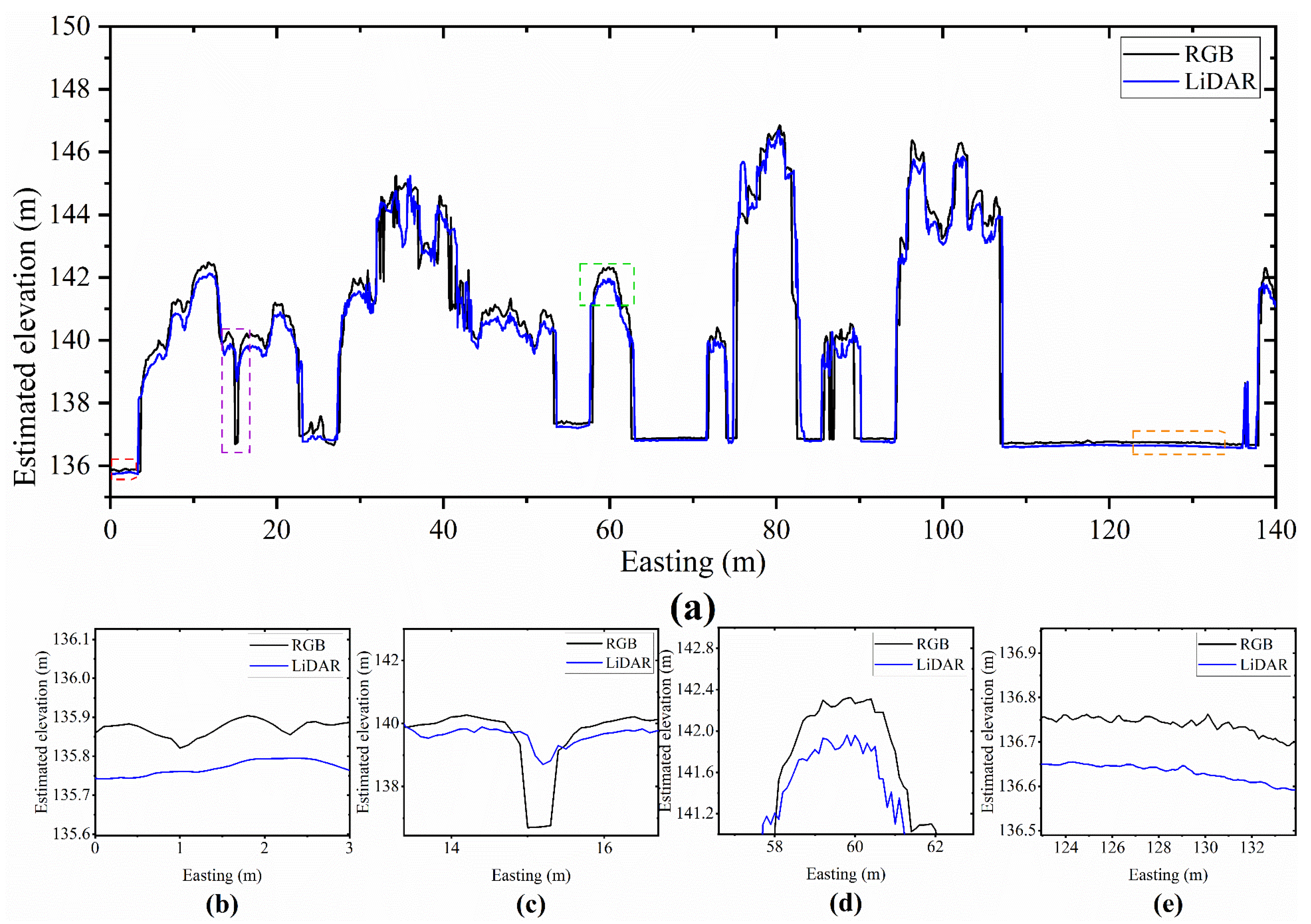
4.1. Differences in Analysis of DEM and DSM Results
4.2. Analyzing the Differences between the Extraction Results of the Structural Parameters of Individual Trees
4.3. Analyzing the Differences between the Results of Structural Parameter Extraction for Different Tree Species
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Dandois, J.P.; Olano, M.; Ellis, E.C. Optimal altitude, overlap, and weather conditions for computer vision UAV estimates of forest structure. Remote Sens. 2015, 7, 13895–13920. [Google Scholar] [CrossRef]
- Clark, M.L.; Clark, D.B.; Roberts, D.A. Small-footprint lidar estimation of sub-canopy elevation and tree height in a tropical rain forest landscape. Remote Sens. Environ. 2004, 91, 68–89. [Google Scholar] [CrossRef]
- Maltamo, M.; Peuhkurinen, J.; Malinen, J.; Vauhkonen, J.; Packalén, P.; Tokola, T. Predicting tree attributes and quality characteristics of Scots pine using airborne laser scanning data. Silva Fenn. 2009, 43, 507–521. [Google Scholar] [CrossRef]
- Latifi, H.; Fassnacht, F.; Koch, B. Forest structure modeling with combined airborne hyperspectral and LiDAR data. Remote Sens. Environ. 2012, 121, 10–25. [Google Scholar] [CrossRef]
- Hudak, A.T.; Crookston, N.L.; Evans, J.S.; Falkowski, M.J.; Smith, A.M.; Gessler, P.E.; Morgan, P. Regression modeling and mapping of coniferous forest basal area and tree density from discrete-return lidar and multispectral satellite data. Can. J. Remote Sens. 2006, 32, 126–138. [Google Scholar] [CrossRef]
- Jaakkola, A.; Hyyppä, J.; Kukko, A.; Yu, X.; Kaartinen, H.; Lehtomäki, M.; Lin, Y. A low-cost multi-sensoral mobile mapping system and its feasibility for tree measurements. ISPRS J. Photogramm. Remote Sens. 2010, 65, 514–522. [Google Scholar] [CrossRef]
- da Cunha Neto, E.M.; Rex, F.E.; Veras, H.F.P.; Moura, M.M.; Sanquetta, C.R.; Käfer, P.S.; Sanquetta, M.N.I.; Zambrano, A.M.A.; Broadbent, E.N.; Dalla Corte, A.P. Using high-density UAV-Lidar for deriving tree height of Araucaria Angustifolia in an Urban Atlantic Rain Forest. Urban For. Urban Green. 2021, 63, 127197. [Google Scholar] [CrossRef]
- Iglhaut, J.; Cabo, C.; Puliti, S.; Piermattei, L.; O’Connor, J.; Rosette, J. Structure from motion photogrammetry in forestry: A review. Curr. For. Rep. 2019, 5, 155–168. [Google Scholar] [CrossRef]
- Chen, J.; Chen, Z.; Huang, R.; You, H.; Han, X.; Yue, T.; Zhou, G. The Effects of Spatial Resolution and Resampling on the Classification Accuracy of Wetland Vegetation Species and Ground Objects: A Study Based on High Spatial Resolution UAV Images. Drones 2023, 7, 61. [Google Scholar] [CrossRef]
- Thiel, C.; Schmullius, C. Comparison of UAV photograph-based and airborne lidar-based point clouds over forest from a forestry application perspective. Int. J. Remote Sens. 2017, 38, 2411–2426. [Google Scholar] [CrossRef]
- Torresan, C.; Berton, A.; Carotenuto, F.; Di Gennaro, S.F.; Gioli, B.; Matese, A.; Miglietta, F.; Vagnoli, C.; Zaldei, A.; Wallace, L. Forestry applications of UAVs in Europe: A review. Int. J. Remote Sens. 2017, 38, 2427–2447. [Google Scholar] [CrossRef]
- Karpina, M.; Jarząbek-Rychard, M.; Tymków, P.; Borkowski, A. UAV-based automatic tree growth measurement for biomass estimation. Int. Arch. Photogramm. Remote Sens. Spat. Inf. Sci 2016, 8, 685–688. [Google Scholar] [CrossRef]
- Guerra-Hernández, J.; González-Ferreiro, E.; Monleón, V.J.; Faias, S.P.; Tomé, M.; Díaz-Varela, R.A. Use of multi-temporal UAV-derived imagery for estimating individual tree growth in Pinus pinea stands. Forests 2017, 8, 300. [Google Scholar] [CrossRef]
- Tang, X.; You, H.; Liu, Y.; You, Q.; Chen, J. Monitoring of Monthly Height Growth of Individual Trees in a Subtropical Mixed Plantation Using UAV Data. Remote Sens. 2023, 15, 326. [Google Scholar] [CrossRef]
- Dempewolf, J.; Nagol, J.; Hein, S.; Thiel, C.; Zimmermann, R. Measurement of within-season tree height growth in a mixed forest stand using UAV imagery. Forests 2017, 8, 231. [Google Scholar] [CrossRef]
- Guerra-Hernández, J.; Cosenza, D.N.; Rodriguez, L.C.E.; Silva, M.; Tomé, M.; Díaz-Varela, R.A.; González-Ferreiro, E. Comparison of ALS-and UAV (SfM)-derived high-density point clouds for individual tree detection in Eucalyptus plantations. Int. J. Remote Sens. 2018, 39, 5211–5235. [Google Scholar] [CrossRef]
- Wallace, L.; Lucieer, A.; Malenovský, Z.; Turner, D.; Vopěnka, P. Assessment of forest structure using two UAV techniques: A comparison of airborne laser scanning and structure from motion (SfM) point clouds. Forests 2016, 7, 62. [Google Scholar] [CrossRef]
- Guerra-Hernández, J.; Cosenza, D.N.; Cardil, A.; Silva, C.A.; Botequim, B.; Soares, P.; Silva, M.; González-Ferreiro, E.; Díaz-Varela, R.A. Predicting growing stock volume of eucalyptus plantations using 3-D point clouds derived from UAV imagery and ALS data. Forests 2019, 10, 905. [Google Scholar] [CrossRef]
- Khosravipour, A.; Skidmore, A.K.; Isenburg, M.; Wang, T.; Hussin, Y.A. Generating Pit-free Canopy Height Models from Airborne Lidar. Photogramm. Eng. Remote Sens. 2014, 80, 863–872. [Google Scholar] [CrossRef]
- Zhao, X.; Guo, Q.; Su, Y.; Xue, B. Improved progressive TIN densification filtering algorithm for airborne LiDAR data in forested areas. ISPRS J. Photogramm. Remote Sens. 2016, 117, 79–91. [Google Scholar] [CrossRef]
- Tanhuanpää, T.; Saarinen, N.; Kankare, V.; Nurminen, K.; Vastaranta, M.; Honkavaara, E.; Karjalainen, M.; Yu, X.; Holopainen, M.; Hyyppä, J. Evaluating the Performance of High-Altitude Aerial Image-Based Digital Surface Models in Detecting Individual Tree Crowns in Mature Boreal Forests. Forests 2016, 7, 143. [Google Scholar] [CrossRef]
- Chen, Q.; Baldocchi, D.; Gong, P.; Kelly, M. Isolating individual trees in a savanna woodland using small footprint lidar data. Photogramm. Eng. Remote Sens. 2006, 72, 923–932. [Google Scholar] [CrossRef]
- Miraki, M.; Sohrabi, H.; Fatehi, P.; Kneubuehler, M. Individual tree crown delineation from high-resolution UAV images in broadleaf forest. Ecol. Inform. 2021, 61, 101207. [Google Scholar] [CrossRef]
- Nuijten, R.J.; Coops, N.C.; Goodbody, T.R.; Pelletier, G. Examining the multi-seasonal consistency of individual tree segmentation on deciduous stands using digital aerial photogrammetry (DAP) and unmanned aerial systems (UAS). Remote Sens. 2019, 11, 739. [Google Scholar] [CrossRef]
- Panagiotidis, D.; Abdollahnejad, A.; Surový, P.; Chiteculo, V. Determining tree height and crown diameter from high-resolution UAV imagery. Int. J. Remote Sens. 2017, 38, 2392–2410. [Google Scholar] [CrossRef]
- Goldbergs, G.; Maier, S.W.; Levick, S.R.; Edwards, A. Efficiency of individual tree detection approaches based on light-weight and low-cost UAS imagery in Australian Savannas. Remote Sens. 2018, 10, 161. [Google Scholar] [CrossRef]
- Yin, D.; Wang, L. How to assess the accuracy of the individual tree-based forest inventory derived from remotely sensed data: A review. Int. J. Remote Sens. 2016, 37, 4521–4553. [Google Scholar] [CrossRef]
- Tilly, N.; Kelterbaum, D.; Zeese, R. Geomorphological Mapping With Terrestrial Laser Scanning And Uav-Based Imaging. Int. Arch. Photogramm. Remote Sens. Spat. Inf. Sci. 2016, 41, 591–597. [Google Scholar] [CrossRef]
- Wilkinson, M.; Jones, R.; Woods, C.; Gilment, S.; McCaffrey, K.; Kokkalas, S.; Long, J. A comparison of terrestrial laser scanning and structure-from-motion photogrammetry as methods for digital outcrop acquisition. Geosphere 2016, 12, 1865–1880. [Google Scholar] [CrossRef]
- Goodbody, T.R.H.; Coops, N.C.; Hermosilla, T.; Tompalski, P.; Pelletier, G. Vegetation Phenology Driving Error Variation in Digital Aerial Photogrammetrically Derived Terrain Models. Remote Sens. 2018, 10, 1554. [Google Scholar] [CrossRef]
- Nikolakopoulos, K.G.; Antonakakis, A.; Kyriou, A.; Koukouvelas, I.; Stefanopoulos, P. Comparison of terrestrial laser scanning and structure-from-motion photogrammetry for steep slope mapping. In Proceedings of the Earth Resources and Environmental Remote Sensing/GIS Applications IX, Berlin, Germany, 11–13 September 2018; pp. 95–105. [Google Scholar]
- Rogers, S.R.; Manning, I.; Livingstone, W. Comparing the Spatial Accuracy of Digital Surface Models from Four Unoccupied Aerial Systems: Photogrammetry Versus LiDAR. Remote Sens. 2020, 12, 2806. [Google Scholar] [CrossRef]
- Zarco-Tejada, P.J.; Diaz-Varela, R.; Angileri, V.; Loudjani, P. Tree height quantification using very high resolution imagery acquired from an unmanned aerial vehicle (UAV) and automatic 3D photo-reconstruction methods. Eur. J. Agron. 2014, 55, 89–99. [Google Scholar] [CrossRef]
- Yu, X.; Hyyppä, J.; Kukko, A.; Maltamo, M.; Kaartinen, H. Change detection techniques for canopy height growth measurements using airborne laser scanner data. Photogramm. Eng. Remote Sens. 2006, 72, 1339–1348. [Google Scholar] [CrossRef]



| Trees Species | Number (Tree) | Mean Tree Height (m) | Mean Crown Width (m) | SD of Tree Height (m) | SD of Crown Width (m) |
|---|---|---|---|---|---|
| Cinnamomum camphora | 8 | 8.83 | 6.99 | 0.75 | 1.03 |
| Osmanthus fragrans | 87 | 4.19 | 3.63 | 0.76 | 0.87 |
| Liriodendron chinense | 14 | 16.16 | 6.06 | 0.68 | 1.25 |
| Magnolia grandiflora | 34 | 8.32 | 4.88 | 0.86 | 0.75 |
| Data Type | Quantity (Trees) | TP | FN | FP | r (%) | p (%) | F (%) |
|---|---|---|---|---|---|---|---|
| UAV-RGB | 143 | 118 | 10 | 15 | 92.19 | 88.72 | 90.42 |
| UAV-LiDAR | 143 | 112 | 13 | 18 | 89.60 | 86.15 | 87.84 |
| - | Data Sources | DF | Square Sum | Mean Square | F | p |
|---|---|---|---|---|---|---|
| Model | LiDAR points | 3 | 193.43 | 64.48 | 41.03 | 0 |
| Error | 140 | 207.41 | 1.57 | - | - | |
| Total | 143 | 400.84 | - | - | - | |
| Model | Image points | 3 | 219.28 | 73.09 | 48.75 | 0 |
| Error | 140 | 197.71 | 1.50 | - | - | |
| Total | 143 | 417.18 | - | - | - |
| Data Sources | Species | Mean Square | SEM | q | p | Alpha | Sig |
|---|---|---|---|---|---|---|---|
| LiDAR points | a and b | 3.49 | 0.47 | 10.58 | 0 | 0.05 | 1 |
| a and c | 1.56 | 0.53 | 1.50 | 0 | 0.05 | 1 | |
| a and d | 2.17 | 0.49 | 6.27 | 0 | 0.05 | 1 | |
| b and c | 2.93 | 0.33 | 12.56 | 0 | 0.05 | 1 | |
| b and d | 1.33 | 0.25 | 7.42 | 0 | 0.05 | 1 | |
| c and d | −1.60 | 0.36 | 6.29 | 0 | 0.05 | 1 | |
| Image points | a and b | 3.45 | 0.46 | 10.69 | 0 | 0.05 | 1 |
| a and c | 1.59 | 0.52 | 0.53 | 0 | 0.05 | 1 | |
| a and d | 2.00 | 0.48 | 5.94 | 0 | 0.05 | 1 | |
| b and c | 3.25 | 0.32 | 14.27 | 0 | 0.05 | 1 | |
| b and d | 1.44 | 0.25 | 8.25 | 0 | 0.05 | 1 | |
| c and d | −1.81 | 0.35 | 7.28 | 0 | 0.05 | 1 |
| - | Data Sources | DF | Square Sum | Mean Square | F | p |
|---|---|---|---|---|---|---|
| Model | LiDAR points | 3 | 1807.52 | 602.51 | 674.85 | 0 |
| Error | 140 | 107.14 | 0.89 | - | - | |
| Total | 143 | 1914.66 | - | - | - | |
| Model | Image points | 3 | 1766.07 | 588.69 | 649.71 | 0 |
| Error | 140 | 108.73 | 0.91 | - | - | |
| Total | 143 | 1874.80 | - | - | - |
| Data Sources | Species | Mean Square | SEM | q | p | Alpha | Sig |
|---|---|---|---|---|---|---|---|
| LiDAR points | a and b | 4.83 | 0.35 | 19.32 | 0 | 0.05 | 1 |
| a and c | −7.10 | 0.42 | 23.99 | 0 | 0.05 | 1 | |
| a and d | 3.54 | 0.37 | 12.07 | 0 | 0.05 | 1 | |
| b and c | 11.93 | 0.28 | 60.84 | 0 | 0.05 | 1 | |
| b and d | 44.28 | 0.20 | 30.51 | 0 | 0.05 | 1 | |
| c and d | −7.65 | 0.30 | 36.05 | 0 | 0.05 | 1 | |
| Image points | a and b | 4.80 | 0.36 | 19.11 | 0 | 0.05 | 1 |
| a and c | −6.97 | 0.42 | 23.36 | 0 | 0.05 | 1 | |
| a and d | 3.71 | 0.37 | 12.13 | 0 | 0.05 | 1 | |
| b and c | 11.78 | 0.28 | 59.61 | 0 | 0.05 | 1 | |
| b and d | 4.26 | 0.20 | 30.16 | 0 | 0.05 | 1 | |
| c and d | −7.51 | 0.30 | 35.15 | 0 | 0.05 | 1 |
| Tree Species/ Structural Parameters | Crown Width | Tree Height |
|---|---|---|
| Cinnamomum camphora | UAV-LiDAR | UAV-RGB |
| Osmanthus fragrans | UAV-RGB | UAV-RGB |
| Liriodendron chinense | UAV-LiDAR | UAV-LiDAR |
| Magnolia grandiflora | UAV-RGB | UAV-RGB |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
You, H.; Tang, X.; You, Q.; Liu, Y.; Chen, J.; Wang, F. Study on the Differences between the Extraction Results of the Structural Parameters of Individual Trees for Different Tree Species Based on UAV LiDAR and High-Resolution RGB Images. Drones 2023, 7, 317. https://doi.org/10.3390/drones7050317
You H, Tang X, You Q, Liu Y, Chen J, Wang F. Study on the Differences between the Extraction Results of the Structural Parameters of Individual Trees for Different Tree Species Based on UAV LiDAR and High-Resolution RGB Images. Drones. 2023; 7(5):317. https://doi.org/10.3390/drones7050317
Chicago/Turabian StyleYou, Haotian, Xu Tang, Qixu You, Yao Liu, Jianjun Chen, and Feng Wang. 2023. "Study on the Differences between the Extraction Results of the Structural Parameters of Individual Trees for Different Tree Species Based on UAV LiDAR and High-Resolution RGB Images" Drones 7, no. 5: 317. https://doi.org/10.3390/drones7050317
APA StyleYou, H., Tang, X., You, Q., Liu, Y., Chen, J., & Wang, F. (2023). Study on the Differences between the Extraction Results of the Structural Parameters of Individual Trees for Different Tree Species Based on UAV LiDAR and High-Resolution RGB Images. Drones, 7(5), 317. https://doi.org/10.3390/drones7050317






