Structural Component Phenotypic Traits from Individual Maize Skeletonization by UAS-Based Structure-from-Motion Photogrammetry
Abstract
:1. Introduction
2. Materials and Methods
2.1. Experimental Setup and Data Acquisition
2.2. Imagery-Based Point Cloud
2.3. Individual Maize Extraction
2.4. Curve-Skeleton Extraction
2.5. Phenotypic Traits of Structural Components
2.6. Accuracy Assessment
3. Results
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Roitsch, T.; Cabrera-Bosquet, L.; Fournier, A.; Ghamkhar, K.; Jiménez-Berni, J.; Pinto, F.; Ober, E.S. New sensors and data-driven approaches—A path to next generation phenomics. Plant Sci. 2019, 282, 2–10. [Google Scholar] [CrossRef] [PubMed]
- Herrero-Huerta, M.; Bucksch, A.; Puttonen, E.; Rainey, K.M. Canopy roughness: A new phenotypic trait to estimate above-ground biomass from unmanned aerial system. Plant Phenomics 2020, 2020, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Herrero-Huerta, M.; Rodriguez-Gonzalvez, P.; Rainey, K.M. Yield prediction by machine learning from UAS-based multi-sensor data fusion in soybean. Plant Methods 2020, 16, 78. [Google Scholar] [CrossRef] [PubMed]
- Symonova, O.; Topp, C.N.; Edelsbrunner, H. DynamicRoots: A software platform for the reconstruction and analysis of growing plant roots. PLoS ONE 2015, 10, e0127657. [Google Scholar] [CrossRef] [PubMed]
- Moreira, F.F.; Hearst, A.A.; Cherkauer, K.A.; Rainey, K.M. Improving the efficiency of soybean breeding with high-throughput canopy phenotyping. Plant Methods 2019, 15, 139–148. [Google Scholar] [CrossRef]
- Herrero-Huerta, M.; Meline, V.; Iyer-Pascuzzi, A.S.; Souza, A.M.; Tuinstra, M.R.; Yang, Y. 4D Structural root architecture modeling from digital twins by X-Ray Computed Tomography. Plant Methods 2021, 17, 1–12. [Google Scholar] [CrossRef]
- Gerth, S.; Claußen, J.; Eggert, A.; Wörlein, N.; Waininger, M.; Wittenberg, T.; Uhlmann, N. Semiautomated 3D root segmentation and evaluation based on X-Ray CT imagery. Plant Phenomics 2021, 2021, 8747930. [Google Scholar] [CrossRef]
- Jin, S.; Su, Y.; Gao, S.; Wu, F.; Ma, Q.; Xu, K.; Guo, Q. Separating the structural components of maize for field phenotyping using terrestrial lidar data and deep convolutional neural networks. IEEE Trans. Geosci. Remote Sens. 2019, 58, 2644–2658. [Google Scholar] [CrossRef]
- York, L.M. Functional phenomics: An emerging field integrating high-throughput phenotyping, physiology, and bioinformatics. J. Exp. Bot. 2019, 70, 379–386. [Google Scholar] [CrossRef]
- Bucksch, A.; Fleck, S. Automated detection of branch dimensions in woody skeletons of fruit tree canopies. Photogramm. Eng. Remote Sens. 2011, 77, 229–240. [Google Scholar] [CrossRef]
- Bucksch, A. A practical introduction to skeletons for the plant sciences. Appl. Plant Sci. 2014, 2, 1400005. [Google Scholar] [CrossRef] [PubMed]
- Cornea, N.D.; Silver, D.; Min, P. Curve-skeleton properties, applications, and algorithms. IEEE Trans. Vis. Comput. Graph. 2007, 13, 530. [Google Scholar] [CrossRef] [PubMed]
- Au, O.K.C.; Tai, C.L.; Chu, H.K.; Cohen-Or, D.; Lee, T.Y. Skeleton extraction by mesh contraction. ACM Trans. Graph. 2008, 27, 1–10. [Google Scholar] [CrossRef]
- Brandt, J.W.; Algazi, V.R. Continuous skeleton computation by Voronoi diagram. CVGIP Image Underst. 1992, 55, 329–338. [Google Scholar] [CrossRef]
- Marie, R.; Labbani-Igbida, O.; Mouaddib, E.M. The delta medial axis: A fast and robust algorithm for filtered skeleton extraction. Pattern Recognit. 2016, 56, 26–39. [Google Scholar] [CrossRef]
- Mohamed, W.; Hamza, A.B. Reeb graph path dissimilarity for 3D object matching and retrieval. Vis. Comput. 2012, 28, 305–318. [Google Scholar] [CrossRef]
- Herrero-Huerta, M.; Lindenbergh, R.; Gard, W. Leaf Movements of indoor plants monitored by Terrestrial LiDAR. Front. Plant Sci. 2018, 9, 189. [Google Scholar] [CrossRef]
- Moeslund, J.E.; Clausen, K.K.; Dalby, L.; Fløjgaard, C.; Pärtel, M.; Pfeifer, N.; Brunbjerg, A.K. Using airborne lidar to characterize North European terrestrial high-dark-diversity habitats. Remote Sens. Ecol. Conserv. 2022. [Google Scholar] [CrossRef]
- Li, N.; Kähler, O.; Pfeifer, N. A comparison of deep learning methods for airborne lidar point clouds classification. IEEE J. Sel. Top. Appl. Earth Obs. Remote Sens. 2021, 14, 6467–6486. [Google Scholar] [CrossRef]
- Campos, M.B.; Litkey, P.; Wang, Y.; Chen, Y.; Hyyti, H.; Hyyppä, J.; Puttonen, E. A long-term terrestrial laser scanning measurement station to continuously monitor structural and phenological dynamics of boreal forest canopy. Front. Plant Sci. 2021, 11, 606752. [Google Scholar] [CrossRef]
- Lei, L.; Li, Z.; Wu, J.; Zhang, C.; Zhu, Y.; Chen, R.; Yang, G. Extraction of maize leaf base and inclination angles using Terrestrial Laser Scanning (TLS) data. IEEE Trans. Geosci. Remote Sens. 2022, 60, 1–17. [Google Scholar] [CrossRef]
- Lin, C.; Hu, F.; Peng, J.; Wang, J.; Zhai, R. Segmentation and stratification methods of field maize terrestrial LiDAR point cloud. Agriculture 2022, 12, 1450. [Google Scholar] [CrossRef]
- Wu, S.; Wen, W.; Xiao, B.; Guo, X.; Du, J.; Wang, C.; Wang, Y. An accurate skeleton extraction approach from 3D point clouds of maize plants. Front. Plant Sci. 2019, 10, 248. [Google Scholar] [CrossRef] [PubMed]
- Miao, T.; Zhu, C.; Xu, T.; Yang, T.; Li, N.; Zhou, Y.; Deng, H. Automatic stem-leaf segmentation of maize shoots using three-dimensional point cloud. Comput. Electron. Agric. 2021, 187, 106310. [Google Scholar] [CrossRef]
- Xin, X.; Iuricich, F.; Calders, K.; Armston, J.; De Floriani, L. Topology-based individual tree segmentation for automated processing of terrestrial laser scanning point clouds. Int. J. Appl. Earth Obs. Geoinf. 2023, 116, 103145. [Google Scholar]
- Li, M.; Shamshiri, R.R.; Schirrmann, M.; Weltzien, C.; Shafian, S.; Laursen, M.S. UAV oblique imagery with an adaptive micro-terrain model for estimation of leaf area index and height of maize canopy from 3D point clouds. Remote Sens. 2022, 14, 585. [Google Scholar] [CrossRef]
- Tirado, S.B.; Hirsch, C.N.; Springer, N.M. UAV-based imaging platform for monitoring maize growth throughout development. Plant Direct 2020, 4, e00230. [Google Scholar] [CrossRef]
- Du, L.; Yang, H.; Song, X.; Wei, N.; Yu, C.; Wang, W.; Zhao, Y. Estimating leaf area index of maize using UAV-based digital imagery and machine learning methods. Sci. Rep. 2022, 12, 15937. [Google Scholar] [CrossRef]
- Qi, C.R.; Su, H.; Mo, K.; Guibas, L.J. Pointnet: Deep learning on point sets for 3d classification and segmentation. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Honolulu, HI, USA, 21–26 July 2017; pp. 652–660. [Google Scholar]
- Krizhevsky, A.; Sutskever, I.; Hinton, G.E. Imagenet classification with deep convolutional neural networks. Adv. Neural Inf. Process. Syst. 2012, 25, 1097–1105. [Google Scholar] [CrossRef]
- Su, Y.-T.; Bethel, J.; Hu, s. Octree-based segmentation for terrestrial LiDAR point cloud data in industrial applications. ISPRS J. Photogramm. Remote Sens. 2016, 113, 59–74. [Google Scholar] [CrossRef]
- Cao, J.; Tagliasacchi, A.; Olson, M.; Zhang, H.; Su, Z. Point Cloud Skeletons via Laplacian Based Contraction. In Proceedings of 2010 Shape Modeling International Conference, Aix-en-Provence, France, 21–23 June 2010; pp. 187–197. [Google Scholar]
- Herrero-Huerta, M.; Meline, V.; Iyer-Pascuzzi, A.S.; Souza, A.M.; Tuinstra, M.R.; Yang, Y. Root phenotyping from X-ray Computed Tomography: Skeleton extraction. Int. Arch. Photogramm. Remote Sens. Spatial Inf. Sc 2021, XLIII-B4–2021, 417–422. [Google Scholar] [CrossRef]
- Jin, S.; Su, Y.; Zhang, Y.; Song, S.; Li, Q.; Liu, Z.; Ma, Q.; Ge, Y.; Liu, L.; Ding, Y.; et al. Exploring Seasonal and Circadian Rhythms in Structural Traits of Field Maize from LiDAR Time Series. Plant Phenomics 2021, 2021, 9895241. [Google Scholar] [CrossRef] [PubMed]
- Russ, T.; Boehnen, C.; Peters, T. 3D Face Recognition Using 3D Alignment for PCA. In Proceedings of IEEE Computer Society Conference on Computer Vision and Pattern Recognition (CVPR′06), New York, NY, USA, 17–22 June 2006; pp. 1391–1398. [CrossRef]
- Jin, S.; Su, Y.; Wu, F.; Pang, S.; Gao, S.; Hu, T.; Guo, Q. Stem–leaf segmentation and phenotypic trait extraction of individual maize using terrestrial LiDAR data. IEEE Trans. Geosci. Remote Sens. 2018, 57, 1336–1346. [Google Scholar] [CrossRef]
- Zhou, L.; Gu, X.; Cheng, S.; Yang, G.; Shu, M.; Sun, Q. Analysis of plant height changes of lodged maize using UAV-LiDAR data. Agriculture 2020, 10, 146. [Google Scholar] [CrossRef]
- Elarab, M.; Ticlavilca, A.M.; Torres-Rua, A.F.; Maslova, I.; McKee, M. Estimating chlorophyll with thermal and roadband multispectral high-resolution imagery from an unmanned aerial system using relevance vector machines for precision agriculture. Int. J. Appl Earth Obs 2015, 43, 32–42. [Google Scholar]
- Herrero-Huerta, M.; Rodriguez-Gonzalvez, P.; Lindenbergh, R. Automatic Tree Parameter Extraction by Mobile Lidar System in an Urban Context. PLoS ONE 2018, 13, e0196004. [Google Scholar] [CrossRef]




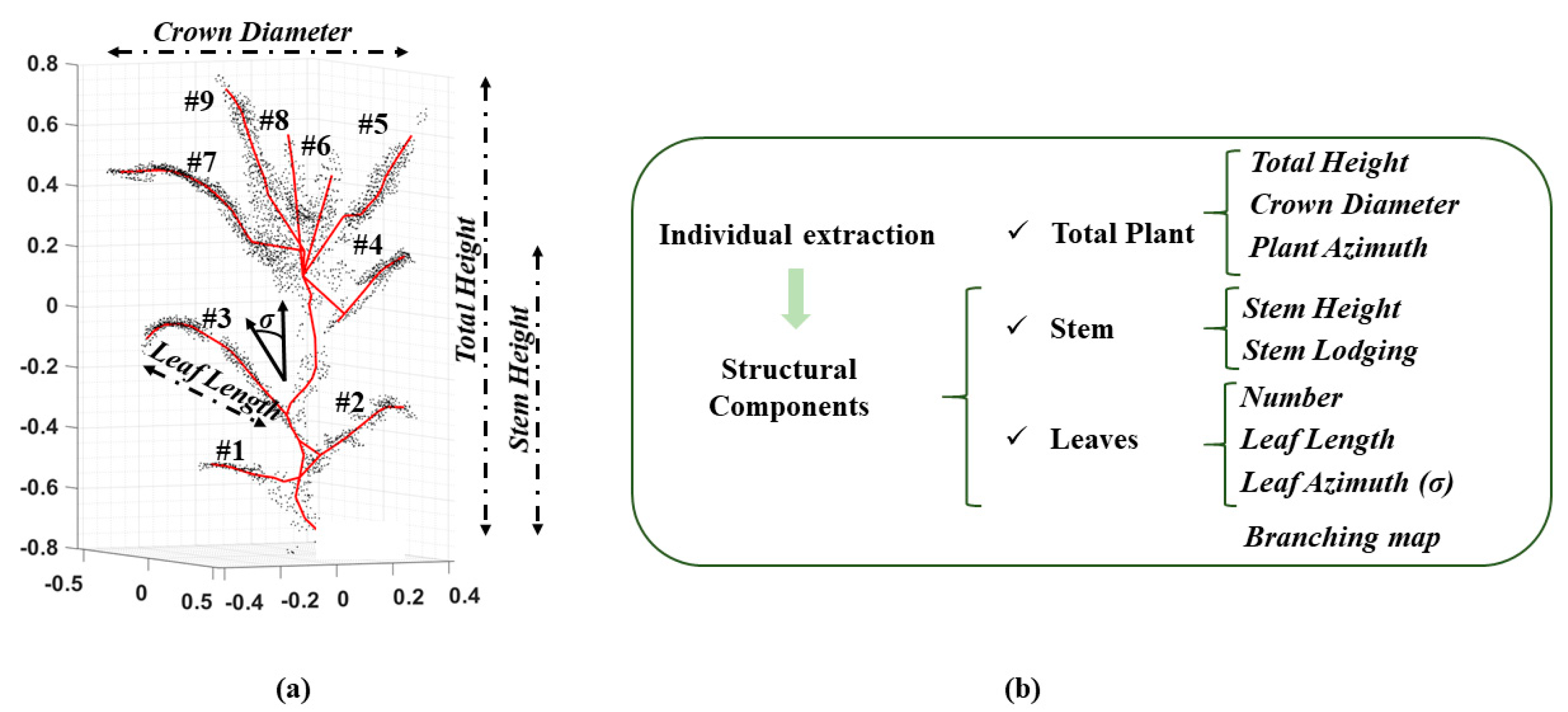
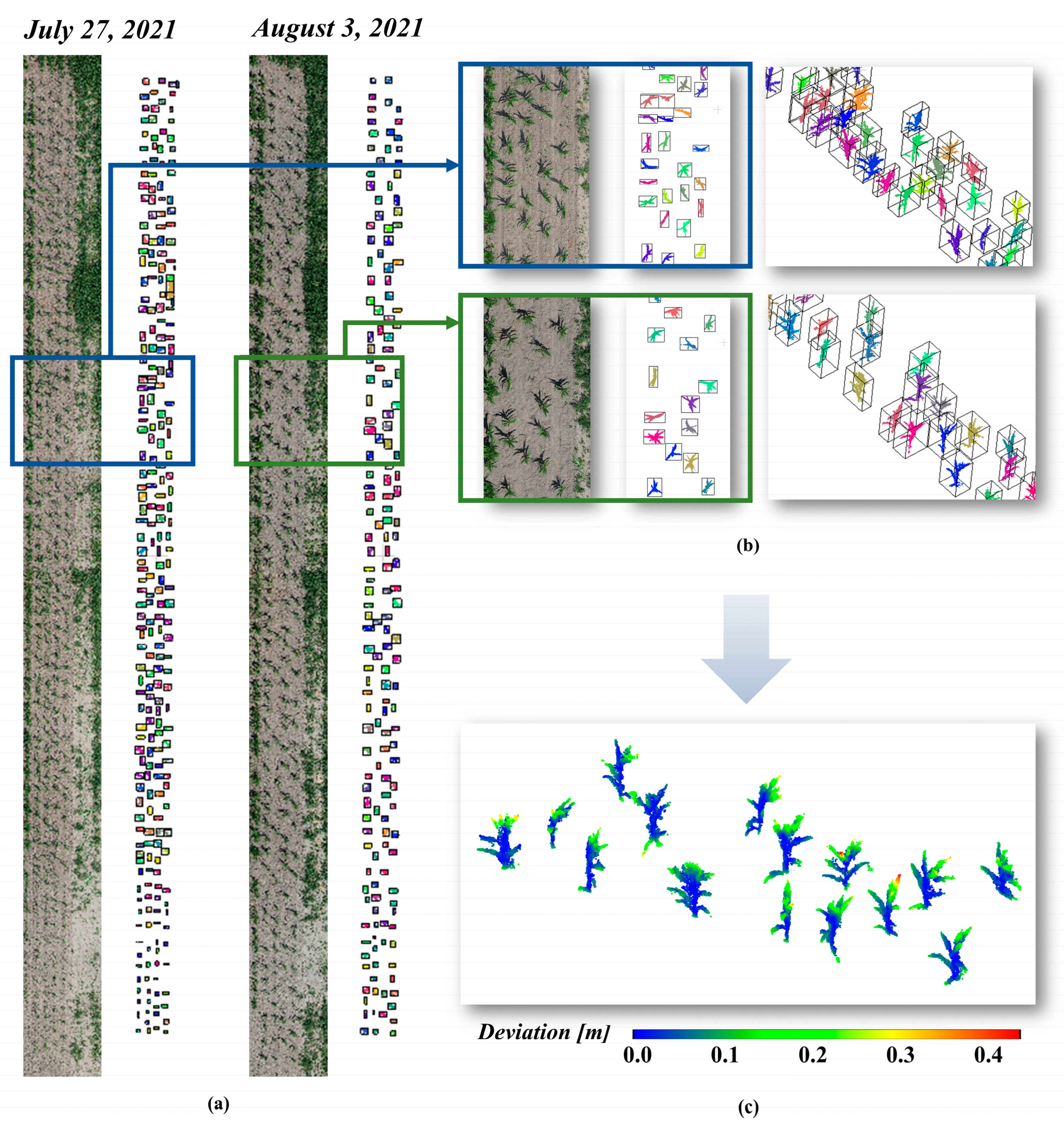
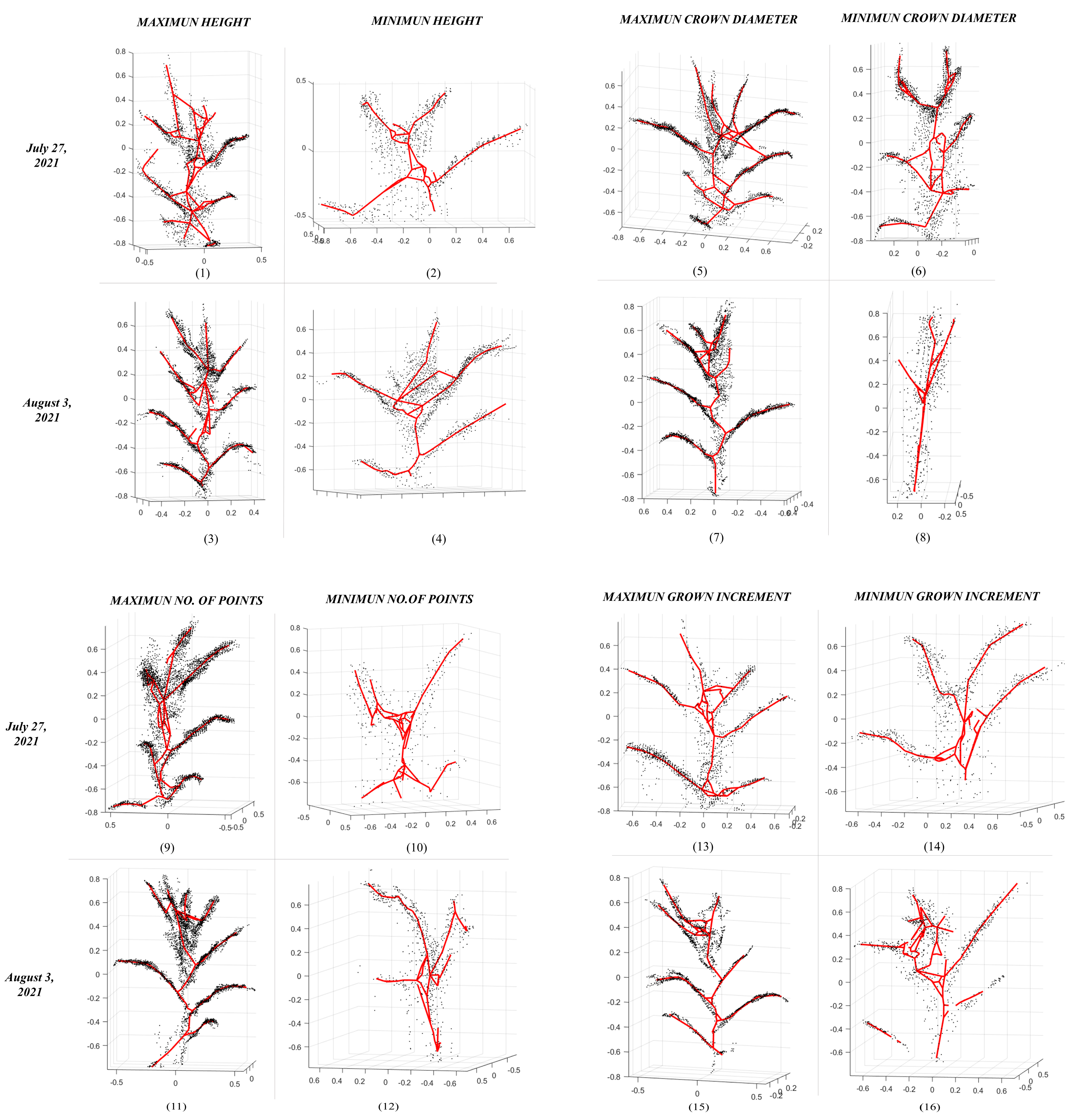
| Plant ID | Point Cloud | Skeleton | |||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Nº. Points | UTM Coord. Center | Bounding Box (m) | Trait Extraction | ||||||||||||
| AX+500,000 | AY+4,480,000 | AZ+0 | X | Y | Z | #Leaves | Height (m) | Crow Diam. (m) | Plant Azimuth (º) | Lodging (º) | Stem Height (m) | Mean LA (º) | Mean LL (m) | ||
| 1 | 2429 | 224.21 | 202.40 | 181.93 | 0.66 | 0.30 | 1.04 | 9 | 0.97 | 0.69 | 339.4 | 351.0 | 0.58 | 344.4 | 0.21 |
| 2 | 956 | 225.36 | 156.35 | 180.90 | 0.38 | 0.26 | 0.25 | 4 | 0.22 | 0.37 | 338.2 | 341.1 | 0.16 | 340.7 | 0.13 |
| 3 | 3481 | 223.47 | 205.84 | 182.20 | 0.43 | 0.82 | 1.32 | 8 | 1.29 | 0.78 | 1.1 | 3.2 | 0.87 | 1.4 | 0.34 |
| 4 | 1729 | 223.11 | 156.49 | 181.19 | 0.58 | 0.34 | 0.32 | 5 | 0.29 | 0.52 | 27.8 | 19.4 | 0.24 | 24.3 | 0.20 |
| 5 | 3929 | 223.33 | 190.44 | 181.87 | 0.90 | 0.31 | 0.83 | 8 | 0.81 | 0.88 | 6.7 | 8.4 | 0.56 | 8.3 | 0.24 |
| 6 | 2259 | 225.88 | 221.57 | 181.49 | 0.16 | 0.23 | 0.81 | 7 | 0.79 | 0.24 | 358.1 | 5.5 | 0.61 | 1.7 | 0.12 |
| 7 | 3891 | 223.3 | 172.99 | 181.98 | 0.89 | 0.64 | 1.15 | 7 | 1.12 | 0.82 | 3.9 | 2.5 | 0.89 | 2.7 | 0.30 |
| 8 | 740 | 223.79 | 149.17 | 180.86 | 0.139 | 0.31 | 0.39 | 3 | 0.36 | 0.29 | 19.8 | 23.3 | 0.16 | 22.0 | 0.07 |
| 9 | 4641 | 224.96 | 199.35 | 181.90 | 0.59 | 0.59 | 0.87 | 7 | 0.85 | 0.57 | 18.8 | 8.1 | 0.60 | 15.6 | 0.21 |
| 10 | 369 | 223.85 | 149.24 | 180.68 | 0.44 | 0.40 | 0.26 | 5 | 0.23 | 0.47 | 9.4 | 12.4 | 0.12 | 10.8 | 0.06 |
| 11 | 5045 | 223.44 | 197.97 | 182.17 | 0.88 | 0.58 | 1.22 | 9 | 1.20 | 0.81 | 357.6 | 1.2 | 0.89 | 259.6 | 0.32 |
| 12 | 547 | 223.22 | 158.67 | 181.20 | 0.36 | 0.42 | 0.40 | 4 | 0.35 | 0.40 | 345.6 | 354.8 | 0.13 | 347.2 | 0.10 |
| 13 | 2115 | 225.82 | 212.08 | 181.68 | 0.86 | 0.45 | 1.00 | 6 | 0.97 | 0.79 | 17 | 4.0 | 0.49 | 4.9 | 0.23 |
| 14 | 947 | 223.87 | 153.23 | 180.80 | 0.40 | 0.43 | 0.45 | 4 | 0.39 | 0.41 | 2.9 | 6.2 | 0.16 | 5.8 | 0.15 |
| 15 | 1636 | 225.72 | 212.19 | 181.68 | 0.74 | 0.23 | 0.82 | 8 | 0.79 | 0.68 | 349.2 | 356.1 | 0.50 | 355.6 | 0.38 |
| 16 | 847 | 223.85 | 153.22 | 180.84 | 0.30 | 0.34 | 0.56 | 8 | 0.51 | 0.38 | 6.1 | 4.2 | 0.14 | 6.7 | 0.11 |
| #Leaf | Height (cm) | Crown Diam. (cm) | Azimuth (º) | Lodging (º) | Hstem (cm) | Mean LA (º) | Mean LL (cm) | |
|---|---|---|---|---|---|---|---|---|
| Mean | 5.98 | 70.16 | 54.66 | 1.18 | 4.56 | 42.76 | −4.94 | 19.19 |
| Std | 1.40 | 34.84 | 21.43 | 15.61 | 8.48 | 28.12 | 26.82 | 9.61 |
| Median | 7 | 79.81 | 54.91 | 1.71 | 4.66 | 51.57 | 3.54 | 20.55 |
| NMAD | 2.43 | 44.84 | 28.45 | 14.45 | 13.2 | 35.80 | 23.13 | 11.44 |
| BwMv | 0.25 | 82.66 | 31.43 | 12.22 | 4.82 | 53.80 | 24.65 | 6.22 |
| R2 (%) | 90.9 | 99.8 | 99.7 | 99.8 | 99.9 | 99.4 | 99.7 | 68.8 |
| RMSE | 0.661 | 1.769 | 1.137 | 8.456 | 4.650 | 2.341 | 11.054 | 8.231 |
| nRMSE (%) | 10.5 | 2.5 | 2.0 | 6.1 | 4.9 | 5.2 | 6.1 | 32.7 |
| MBE | 0.063 | −0.431 | −0.150 | −3.375 | −2.125 | −0.544 | −3.563 | −5.000 |
| AMBE | 0.438 | 1.244 | 0.888 | 6.000 | 3.875 | 1.906 | 10.063 | 5.613 |
| RE | 0.781 | −0.026 | −0.052 | −0.429 | −0.122 | −0.333 | −0.294 | −2.780 |
| AE | 0.781 | 0.026 | 0.104 | 0.429 | 0.122 | 0.333 | 0.294 | 14.053 |
| Ƞ | 0.879 | 0.997 | 0.997 | 0.997 | 0.999 | 0.993 | 0.996 | 0.267 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Herrero-Huerta, M.; Gonzalez-Aguilera, D.; Yang, Y. Structural Component Phenotypic Traits from Individual Maize Skeletonization by UAS-Based Structure-from-Motion Photogrammetry. Drones 2023, 7, 108. https://doi.org/10.3390/drones7020108
Herrero-Huerta M, Gonzalez-Aguilera D, Yang Y. Structural Component Phenotypic Traits from Individual Maize Skeletonization by UAS-Based Structure-from-Motion Photogrammetry. Drones. 2023; 7(2):108. https://doi.org/10.3390/drones7020108
Chicago/Turabian StyleHerrero-Huerta, Monica, Diego Gonzalez-Aguilera, and Yang Yang. 2023. "Structural Component Phenotypic Traits from Individual Maize Skeletonization by UAS-Based Structure-from-Motion Photogrammetry" Drones 7, no. 2: 108. https://doi.org/10.3390/drones7020108
APA StyleHerrero-Huerta, M., Gonzalez-Aguilera, D., & Yang, Y. (2023). Structural Component Phenotypic Traits from Individual Maize Skeletonization by UAS-Based Structure-from-Motion Photogrammetry. Drones, 7(2), 108. https://doi.org/10.3390/drones7020108








