OMICs Role in Hereditarian Prostate Cancer †
Abstract
:1. Introduction
2. Materials and Methods
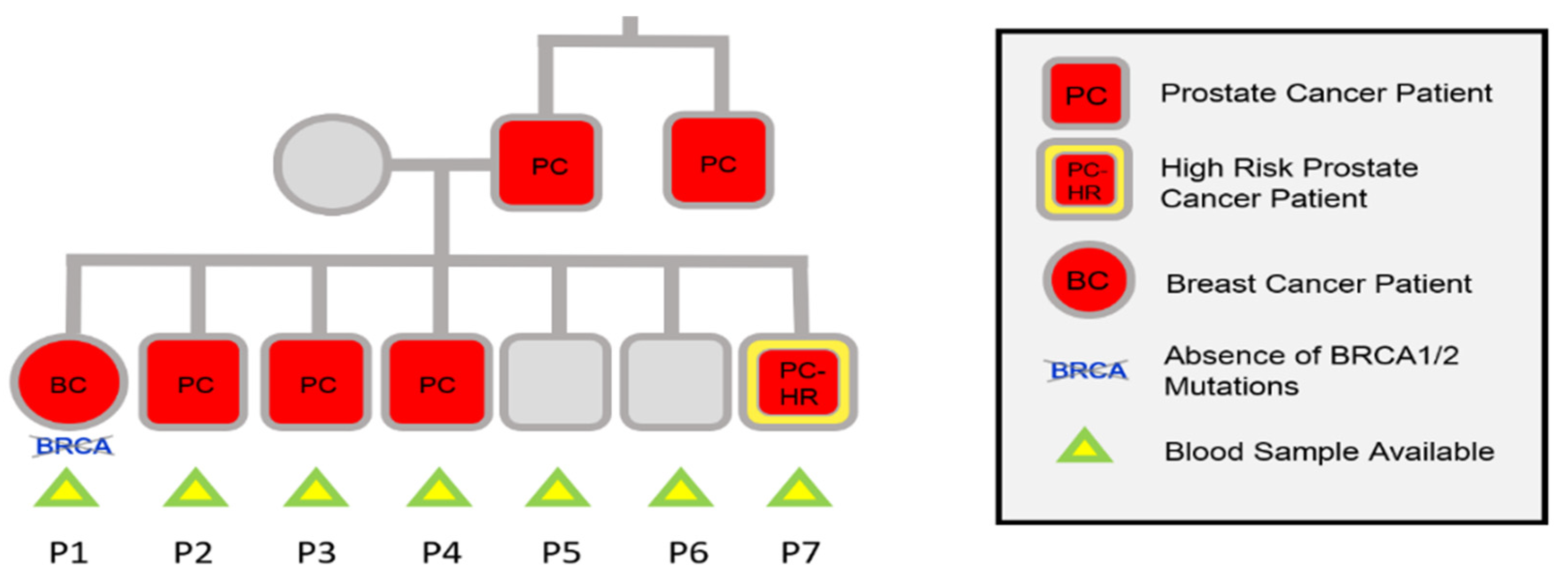
2.1. Samples
2.2. Next-Generation Whole-Exome Sequencing (WES) for Candidate Gene Identification
2.3. Bioinformatic Analysis
3. Results
4. Discussion
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- PDQ Cancer Genetics Editorial Board. Genetics of Prostate Cancer (PDQ®): Health Professional Version; PDQ Cancer Genetics Editorial Board: Bethesda, MD, USA, 2002. [Google Scholar]
- Potter, S.R.; Partin, A.W. Hereditary and familial prostate cancer: Biologic aggressiveness and recurrence. Rev. Urol. 2000, 2, 35–36. [Google Scholar] [PubMed]
- van Dessel, L.F.; van Riet, J.; Smits, M.; Zhu, Y.; Hamberg, P.; van der Heijden, M.S.; Bergman, A.M.; van Oort, I.M.; de Wit, R.; Voest, E.E.; et al. The genomic landscape of metastatic castration-resistant prostate cancers reveals multiple distinct genotypes with potential clinical impact. Nat. Commun. 2019, 10, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Martinez-Gonzalez, L.J.; Antúnez-Rodríguez, A.; Vazquez-Alonso, F.; Hernandez, A.F.; Alvarez-Cubero, M.J. Genetic variants in xenobiotic detoxification enzymes, antioxidant defenses and hormonal pathways as biomarkers of susceptibility to prostate cancer. Sci. Total Environ. 2020, 730, 138314. [Google Scholar] [CrossRef] [PubMed]
- Vaser, R.; Adusumalli, S.; Ngak Leng, S.; Sikic, M.; Ng, P.C. SIFT missense predictions for genomes. Nat. Protoc. 2015, 11, 1. [Google Scholar] [CrossRef] [PubMed]
- Adzhubei, I.A.; Schmidt, S.; Peshkin, L.; Ramensky, V.E.; Gerasimova, A.; Bork, P.; Kondrashov, A.S.; Sunyaev, S.R. A method and server for predicting damaging missense mutations. Nat. Methods 2010, 7, 248–249. [Google Scholar] [CrossRef] [PubMed]
- Graff, J.N.; Puri, S.; Bifulco, C.B.; Fox, B.A.; Beer, T.M. Sustained complete response to CTLA-4 blockade in a patient with metastatic, castration-resistant prostate cancer. Cancer Immunol. Res. 2014, 2, 399–403. [Google Scholar] [CrossRef] [PubMed]
- Russo, J.W.; Gao, C.; Bhasin, S.S.; Voznesensky, O.S.; Calagua, C.; Arai, S.; Nelson, P.S.; Montgomery, B.; Mostaghel, E.A.; Corey, E.; et al. Downregulation of dipeptidyl peptidase 4 accelerates progression to castration-resistant prostate cancer. Cancer Res. 2018, 78, 6354–6362. [Google Scholar] [CrossRef] [PubMed]
- Shan, Y.; Gao, Y.; Jin, W.; Fan, M.; Wang, Y.; Gu, Y.; Shan, C.; Sun, L.; Li, X.; Yu, B.; et al. Targeting HIBCH to reprogram valine metabolism for the treatment of colorectal cancer. Cell Death Dis. 2019, 10, 1–15. [Google Scholar] [CrossRef] [PubMed]
- Stelzer, G.; Rosen, N.; Plaschkes, I.; Zimmerman, S.; Twik, M.; Fishilevich, S.; Iny Stein, T.; Nudel, R.; Lieder, I.; Mazor, Y.; et al. The GeneCards suite: From gene data mining to disease genome sequence analyses. Curr. Protoc. Bioinform. 2016, 54, 1.30.1–1.30.33. [Google Scholar] [CrossRef] [PubMed]
| Variations in All PC Patients | |
|---|---|
| Gene | SNP |
| DPP4 | rs116302758 |
| HIBCH | rs291466 |
| MOK | rs56377169 |
| PPP4R3A | |
| STK31 | |
| Variations in High Risk PC Patient | |
| Gene | SNP |
| ACE | |
| ALDH3B2 | rs7947754 |
| CD63 | |
| EFCAB13 | rs2271803 |
| FAM86HP | rs16834628 |
| FANK1 | |
| MYO15B | rs73998360 |
| SLC25A5 | |
| TUBA3FP | rs2075276 |
| ZNF142 | |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cuenca-Lopez, S.; Porras-Quesada, P.M.; Vazquez-Alonso, F.; Sanchez-Conde, V.; Gomez-Matas, M.d.P.; Aneas-Alaminos, A.; Arenas-Rodriguez, V.; Cano-Gutierrez, B.; Martinez-Gonzalez, L.J.; Alvarez-Cubero, M.J. OMICs Role in Hereditarian Prostate Cancer. Proceedings 2021, 76, 5. https://doi.org/10.3390/IECGE-07147
Cuenca-Lopez S, Porras-Quesada PM, Vazquez-Alonso F, Sanchez-Conde V, Gomez-Matas MdP, Aneas-Alaminos A, Arenas-Rodriguez V, Cano-Gutierrez B, Martinez-Gonzalez LJ, Alvarez-Cubero MJ. OMICs Role in Hereditarian Prostate Cancer. Proceedings. 2021; 76(1):5. https://doi.org/10.3390/IECGE-07147
Chicago/Turabian StyleCuenca-Lopez, Sergio, Patricia Maria Porras-Quesada, Fernando Vazquez-Alonso, Victor Sanchez-Conde, Maria del Pilar Gomez-Matas, Adoracion Aneas-Alaminos, Veronica Arenas-Rodriguez, Blanca Cano-Gutierrez, Luis Javier Martinez-Gonzalez, and Maria Jesus Alvarez-Cubero. 2021. "OMICs Role in Hereditarian Prostate Cancer" Proceedings 76, no. 1: 5. https://doi.org/10.3390/IECGE-07147
APA StyleCuenca-Lopez, S., Porras-Quesada, P. M., Vazquez-Alonso, F., Sanchez-Conde, V., Gomez-Matas, M. d. P., Aneas-Alaminos, A., Arenas-Rodriguez, V., Cano-Gutierrez, B., Martinez-Gonzalez, L. J., & Alvarez-Cubero, M. J. (2021). OMICs Role in Hereditarian Prostate Cancer. Proceedings, 76(1), 5. https://doi.org/10.3390/IECGE-07147








