Abstract
5-enolpyruvylshikimate 3-phosphate synthase (EPSPS) is the central enzyme of the shikimate pathway to synthesize three aromatic amino acids in fungi, plants and prokaryotes. This enzyme is the target of the herbicide glyphosate. In most plants and prokaryotes, the EPSPS protein is constituted by a single domain, whereas in fungi, it contains several EPSPS-associated domains. Here, we perform a comprehensive analysis of 390 EPSPS proteins of fungi to determine the distribution and the evolution of the EPSPS-associated domains. The results of this study will be useful to determine the potential differential impact of glyphosate on alternative domain architectures in fungi.
Published: 2 November 2020
1. Introduction
Glyphosate, the most used herbicide against weeds, and glyphosate-based products (GBPs), target the enzyme 5-enolpyruvylshikimate-3-phosphate synthase (EPSPS) [1,2]. EPSPS (also known as aroA) is the central enzyme in the shikimate pathway for the synthesis of three essential amino acids [3]. The enzyme is present in plants and prokaryotes as a single-domain protein, and in fungi as a multi-domain protein [4,5]. As the enzyme is not found in animals, the use of glyphosate is supposed to be safe for human health. Even if this is the case, the herbicide may still affect the biodiversity of free-living and host-associated microorganisms [6,7,8,9,10]. Thus, the study of the glyphosate target enzyme will provide clues about the potential effect of the herbicide. In recent studies, we analyzed the distribution of the EPSPS protein in organisms and we estimated the differential sensitivity of organisms to the herbicide [7,8].
In this study, we analyze the distribution and evolution of the EPSPS and EPSPS-associated domains in fungi. In plants and prokaryotes, EPSPS has a single domain, thus the potential sensitivity to the herbicide glyphosate depends, in addition to levels of gene expression and other cell factors [4,11,12,13], on the type of amino acids in the EPSPS protein. However, the effect of the herbicide on the multi-domain structure of EPSPS in fungi is yet unclear. This survey of the EPSPS and EPSPS-associated domains will be helpful to determine the potential effect of glyphosate on fungal species.
2. Material and Methods
We obtained protein data of the EPSPS and EPSPS-associated domains from the PFAM database (http://pfam.xfam.org (accessed on 1 September 2019)) [14]. First, we analyzed frequencies of EPSPS-associated domains across 390 fungal species, and then we summarized the main domain function in a Venn diagram. We also analyzed the distribution of the associated domains by taxonomical groups in fungi. A phylogenetic tree of the EPSPS domain was built with the program FastTree2 [15] after aligning the protein sequences with the programs MUSCLE [16] and Gblocks [17]. The program Cytoscape [18] was used to reconstruct a bipartite network of protein domains and fungal species. The resultant network was used to visualize the presence and absence of domains and the distribution of the different architectures of the EPSPS multi-domain. The analysis of the evolution of the EPSPS-associated domains in fungi was performed by Dollon parsimony with the program Count [19].
3. Results
3.1. Functional and Domain Characterization
The EPSPS domain is approximately 450 amino acids long (~1350 nucleotides) and is present (by definition) in all EPSPS proteins [7]. The EPSPS-associated domains can be classified into four partially overlapping groups (Figure 1): shikimate (shikimate pathway proteins), enzymes (proteins with catalytic function), expression (domains whose products are needed in controlling gene expression) and structural function (proteins that do not have a catalytic function). The function of the EPSPS enzyme is to transfer alkyl and aryl (other than methyl) groups. It transfers enolpyruvate from phosphoenolpyruvate to 3-phosphoshikimate [20]. The analysis of the EPSPS-associated domains across fungal species shows that they are mostly involved in the shikimate pathway (e.g., SKI, DHQ_synthase, DHquinase_I) and in the synthesis of aromatic amino acids (e.g., Shikimate_dh_N, PDH). There are also some promiscuous domains (e.g., HTH_3) associated with EPSPS in some fungal species. The least frequent domains, mainly involved in DNA modification and gene expression, are amply distributed across fungal species.
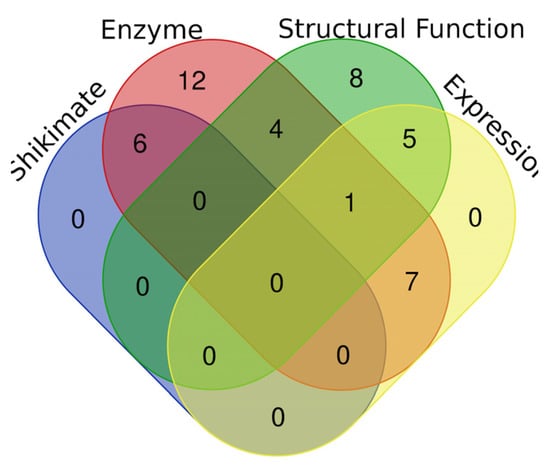
Figure 1.
A Venn diagram of the 5-enolpyruvylshikimate 3-phosphate synthase (EPSPS)-associated domains. Blue is shikimate pathway, red is enzymes, green is structural function and yellow is gene expression. The diagram was made with the following tool: http://bioinformatics.psb.ugent.be/webtools/Venn/ (accessed on 1 October 2019).
3.2. A Bipartite Network of Species and EPSPS-Associated Domains
A total of 22 domains are associated with the EPSPS protein in fungal EPSPS protein architectures, across 390 fungal species. The most frequent EPSPS-associated domains, which are involved in the shikimate pathway, are: Shikimate kinase (SKI) (n = 374); 3-dehydroquinate synthase (DHQ synthase) (n = 374); 3-dehydroquinate dehydratase (DHquinase) (n = 367); shikimate dehydrogenase substrate-binding domain (shikimate dh N) (n = 366); and shikimate/quinate 5-dehydrogenase (shikimate DH) (n = 136). There are also 16 domains that are only present in one or two proteins.
The bipartite network (Figure 2) connects fungal species divided by order (ascomycota on the left and basidiomycota, mucoromycota, chytridiomycota, blastocladiomycota, zoopagomycota and unknown on the right). EPSPS and the most common EPSPS-associated domains are located in the center of the figure in the same order as in domains. Other domains were set on the outside of the network. Ascomycota seems to be the most variable phylum, in terms of EPSPS-associated domains, and contains several infrequent domains. Moreover, this is the phylum that contains the least number of shikimate DH domains, e.g., the domain is infrequent in Eurotiomycetes (APE), Dothideomycetes (APD) and Leotiomycetes (APL). The majority of multi-domain architectures (n = 220) are five domains long, which are involved in the shikimate pathway and approximately 1/3 of the sequences (n = 134) have all six domains of the shikimate pathway. Overall, the bipartite network shows that the EPSPS domain in fungi is mostly associated to other domains of the shikimate pathway. However, there are a few exceptions (e.g., Dothideomycetes).
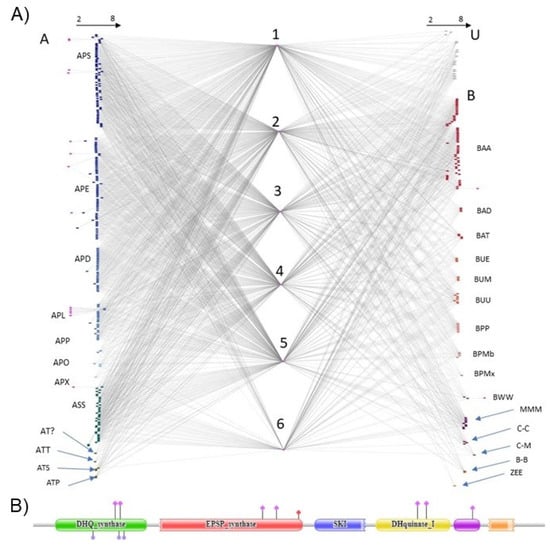
Figure 2.
A bipartite network of fungal species and EPSPS-associated domains. (A) A bipartite network of fungal samples. Letters stands for different phyla and classes of fungi. On the left side there are fungi of the phylum Ascomycota (A) and on the right side there are fungi of the phyla Basidiomycota (B) Mucoromycota (M), Chytridiomycota (C), Blastocladiomycota (Bl) and Zoopagomygota (Z) and unknown taxa (U). Numbers correspond to fungal multi-domain: (1) DH synthase, (2) EPSP synthase, (3) SKI, (4) DH quinase type 1, (5) shikimate dh N and (6) shikimate DH. Arrows on the top of the figure note the number of domains in the multi-domain, which ranges between 2 to 8 domains. Subphyla and classes starting from the upper left are Pezizomycotina: Sordariomycetes (APS), Eurotiomycetes (APE), Dothideomycetes (APD), Leotiomycetes (APL), Pezizomycetes (APP), Orbiliomycetes (APO) and Xylonomycetes (APX); Saccharomycotina: Saccharomycetes (ASS); Taphrinomycotina: incertae sedis (AT?), Taphrinomycetes (ATT), Schizosaccharomycetes (ATS) and Pneumocystidomycetes (ATP); Agariomycotiina: Agaricomycetes (BAA), Dacrymycetes (BAD) and Tremellomycetes (BAT); Ustilaginomycotina: Exobasidiomycetes (BUE), Malasseziomycetes (BUM) and Ustilaginomycetes (BUU); Pucciniomycotina: Pucciniomycetes (BPP), Microbotryomycetes (BPMb) and Mixiomycetes (BPMx); Wallemiomycotina: Wallemiomycetes (BWW); Mucoromycotina: Mucoromycetes (MMM); Chytridiomycota: Chytridiomycetes (C-C) and Monoblepharidomycetes (C-M); Blastocladiomycota: Blastocladiomycetes (B-B); Entomophthoromycotina: Entomophthoromycetes (ZEE). A detailed analysis of the bipartite network can be found in Appendix III. (B) Example of a multi-domain EPSPS protein of 1616 amino acids (protein A0A060S9A7_PYCCI from Pycnoporus cinnabarinus). In this example, the SKI and shikimate DH are fractured.
3.3. Maximum Parsimony Analysis of EPSPS-Associated Domains
Given that the shikimate DH domain is found in most species, it is likely that it was present in the common ancestor of fungi and lost in some lineages. Moreover, rarer domains are late inclusions in the multi-domain structure. We analyzed the evolution of all EPSPS-associated domains with Dollon parsimony (Figure 3).
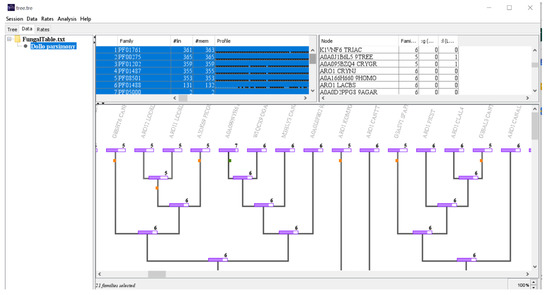
Figure 3.
Screenshot of the output of the program Count [19]. The image shows a part of phylogenetic tree (center); domain family profile (top left) and node information (top right). The multi-domain structure of EPSPS in fungi is ancestral (6 domains) and there are recent domain gains and losses.
The shikimate DH domain has been lost multiple times in different branches of the phylogenetic tree (Figure 3). In addition, many domains have been lost during the evolution of fungi and some new domains have been acquired recently, and independently, in a few species, and others were acquired in internal lineages and subsequently lost in a few branches. Notice that our analysis does not take into account domain duplications. The conclusion from this Dollon parsimony analysis is that a six-domain structure (i.e., a protein sequence with all six most abundant domains) was the original EPSPS sequence in fungi (Table 1). Moreover, we suggest the hypothesis that sequences with the five domains originated by loss of shikimate DH independently in different branches of the evolutionary tree. These losses occurred at early and late stages in the evolution of fungi. In some EPSPS proteins, the shikimate DH (located at the C-terminal of the protein) is truncated. It is possible that this domain was lost in a crossing over event without affecting the shikimate pathway, however, we do not know if the fragment is found in any other part of genome. Additionally, smaller multi-domains originated from the loss of other shikimate domains, usually in higher branches. Moreover, the inclusion of rare domains also occurred in higher branches.

Table 1.
EPSPS and most frequent EPSPS-associated domains.
4. Conclusions
The EPSPS multi-domain structure in fungi ranges between two and eight domains. An evolutionary analysis shows that the ancestral state of the EPSPS protein included six domains (DHQ synthase, EPSPS, SKI, DHquinase I, shikimate DH N and shikimate DH). The current multi-domain structure of EPSPS in fungi is the product of multiple independent domain gains and losses throughout evolution. Further analyses should determine the importance of the EPSPS-associated domains on the sensitivity/resistance of the EPSPS enzyme to glyphosate.
Author Contributions
Conceptualization, P.P.; methodology, P.P.; formal analysis, T.T.; investigation, T.T. and P.P.; resources, P.P and T.T.; writing—original draft preparation, T.T. and P.P.; writing—review and editing, T.T. and P.P.; visualization, T.T.; supervision, P.P. Both authors have read and agreed to the published version of the manuscript.
Funding
This research was supported by funds from the Turku Collegium for Science and Medicine (PP).
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Protein sequence data was obtained from publicly accessible repository http://pfam.xfam.org.
Conflicts of Interest
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript, or in the decision to publish the results.
References
- Bentley, R.; Haslam, E. The shikimate pathway A metabolic tree with many branche. Crit. Rev. Biochem. Mol. Biol. 1990, 25, 307–384. [Google Scholar] [CrossRef] [PubMed]
- Richards, T.A.; Dacks, J.B.; Campbell, S.A.; Blanchard, J.L.; Foster, P.G.; McLeod, R.; Roberts, C.W. Evolutionary origins of the eukaryotic shikimate pathway: Gene fusions, horizontal gene transfer, and endosymbiotic replacements. Eukaryot. Cell. 2006, 5, 1517–1531. [Google Scholar] [CrossRef] [PubMed]
- Maeda, H.; Dudareva, N. The Shikimate Pathway and Aromatic Amino Acid Biosynthesis in Plants. Annu. Rev. Plant Biol. 2012, 63, 73–105. [Google Scholar] [CrossRef] [PubMed]
- Steinrücken, H.C.; Amrhein, N. The herbicide glyphosate is a potent inhibitor of 5-enolpyruvylshikimic acid-3-phosphate synthase. Biochem. Biophys. Res. Commun. 1980, 94, 1207–1212. [Google Scholar] [CrossRef]
- Lumsden, J.; Coggins, J.R. The Subunit Structure of the arom Multienzyme Complex of Neurospora crassa. Biochem. J. 1978, 161. Available online: https://sciwheel.com/work/item/8971159/resources/8495092/pdf. Published (accessed on 9 October 2020).
- Basu, M.K.; Carmel, L.; Rogozin, I.B.; Koonin, E.V. Evolution of protein domain promiscuity in eukaryotes. Genome Res. 2008, 18, 449–461. [Google Scholar] [CrossRef] [PubMed]
- Leino, L.; Tall, T.; Helander, M.; Saloniemi, I.; Saikkonen, K.; Ruuskanen, S.; Puigbò, P. Classification of the glyphosate target enzyme (5-enolpyruvylshikimate-3-phosphate synthase) Equal contribution of the authors. bioRxiv 2020. [Google Scholar] [CrossRef]
- Rainio, M.J.; Ruuskanen, S.; Helander, M.; Saikkonen, K.; Saloniemi, I.; Puigbò, P. Adaptation of bacteria to glyphosate: A microevolutionary perspective of the enzyme 5-enolpyruvylshikimate-3-phosphate (EPSP) synthase. bioRxiv 2020. [Google Scholar] [CrossRef]
- Balbuena, M.S.; Tison, L.; Hahn, M.L.; Greggers, U.; Menzel, R.; Farina, W.M. Effects of sublethal doses of glyphosate on honeybee navigation. J. Exp. Biol. 2015, 218, 2799–2805. [Google Scholar] [CrossRef] [PubMed]
- Vandenberg, L.N.; Blumberg, B.; Antoniou, M.N.; Benbrook, C.M.; Carroll, L.; Colborn, T.; Everett, L.G.; Hansen, M.; Landrigan, P.J.; Lanphear, B.P.; et al. Is it time to reassess current safety standards for glyphosate-based herbicides? J. Epidemiol. Community Health 2017, 71, 613–618. [Google Scholar] [CrossRef] [PubMed]
- Burchfield, S.L.; Bailey, D.C.; Todt, C.E.; Denney, R.D.; Negga, R.; Fitsanakis, V.A. Acute exposure to a glyphosate-containing herbicide formulation inhibits Complex II and increases hydrogen peroxide in the model organism Caenorhabditis elegans. Environ. Toxicol. Pharmacol. 2019, 66, 36–42. [Google Scholar] [CrossRef] [PubMed]
- Nerozzi, C.; Recuero, S.; Galeati, G.; Bucci, D.; Spinaci, M.; Yeste, M. Effects of Roundup and its main component, glyphosate, upon mammalian sperm function and survival. Sci. Rep. 2020, 10, 11026. [Google Scholar] [CrossRef] [PubMed]
- Gomes, M.P.; Le Manac’h, S.G.; Hénault-Ethier, L.; Labrecque, M.; Lucotte, M.; Juneau, P. Glyphosate-dependent inhibition of photosynthesis in willow. Front. Plant Sci. 2017, 8, 207. [Google Scholar] [CrossRef] [PubMed]
- Sonnhammer, E.L.L.; Eddy, S.R.; Durbin, R. Pfam: A comprehensive database of protein domain families based on seed alignments. Proteins Struct. Funct. Genet. 1997, 28, 405–420. [Google Scholar] [CrossRef]
- Price, M.N.; Dehal, P.S.; Arkin, A.P. FastTree 2 Approximately maximum-likelihood trees for large alignments. PLoS ONE 2010, 5, e9490. [Google Scholar] [CrossRef] [PubMed]
- Edgar, R.C. MUSCLE: A multiple sequence alignment method with reduced time and space complexity. BMC Bioinform. 2004, 5, 113. [Google Scholar] [CrossRef] [PubMed]
- Castresana, J. Selection of Conserved Blocks from Multiple Alignments for Their Use in Phylogenetic Analysis. Mol. Biol. Evol. 2000, 17, 540–552. [Google Scholar] [CrossRef] [PubMed]
- Shannon, P.; Markiel, A.; Ozier, O.; Baliga, N.S.; Wang, J.T.; Ramage, D.; Amin, N.; Schwikowski, B.; Ideker, T. Cytoscape: A software Environment for integrated models of biomolecular interaction networks. Genome Res. 2003, 13, 2498–2504. [Google Scholar] [CrossRef] [PubMed]
- Csurös, M. Ancestral reconstruction by asymmetric Wagner parsimony over continuous characters and squared parsimony over distributions. In Lecture Notes in Computer Science (Including Subseries Lecture Notes in Artificial Intelligence and Lecture Notes in Bioinformatics); Vol. 5267 LNBI.; Springer: Berlin/Heidelberg, Germany, 2008; pp. 72–86. [Google Scholar] [CrossRef]
- Jaworski, E.G. Mode of action of N-phosphonomethylglycine. Inhibition of aromatic amino acid biosynthsis. J. Agric. Food Chem. 1972, 20, 1195–1198. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).