Characterization of the Food Microbiota in Ready-to-Eat Mexican Foods †
Abstract
:1. Introduction
2. Materials and Methods
2.1. Ready-to-Eat Food Sampling
2.2. DNA Extraction
2.3. Semiconductor DNA Sequencing of V3-16S rDNA Libraries
2.4. Data Analysis
3. Results
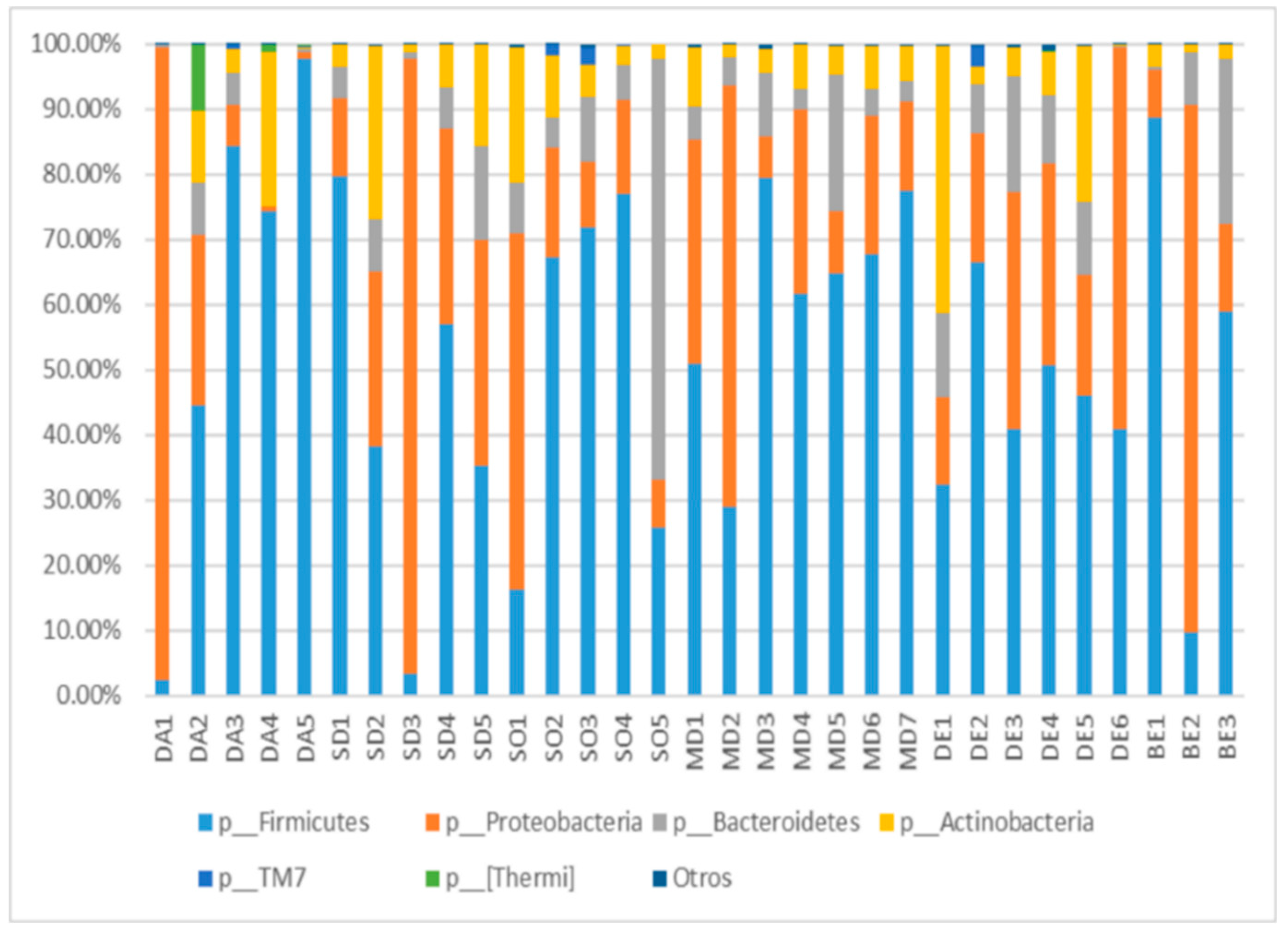
3.1. Abundance of Phyla in the Ready-to-Eat Food
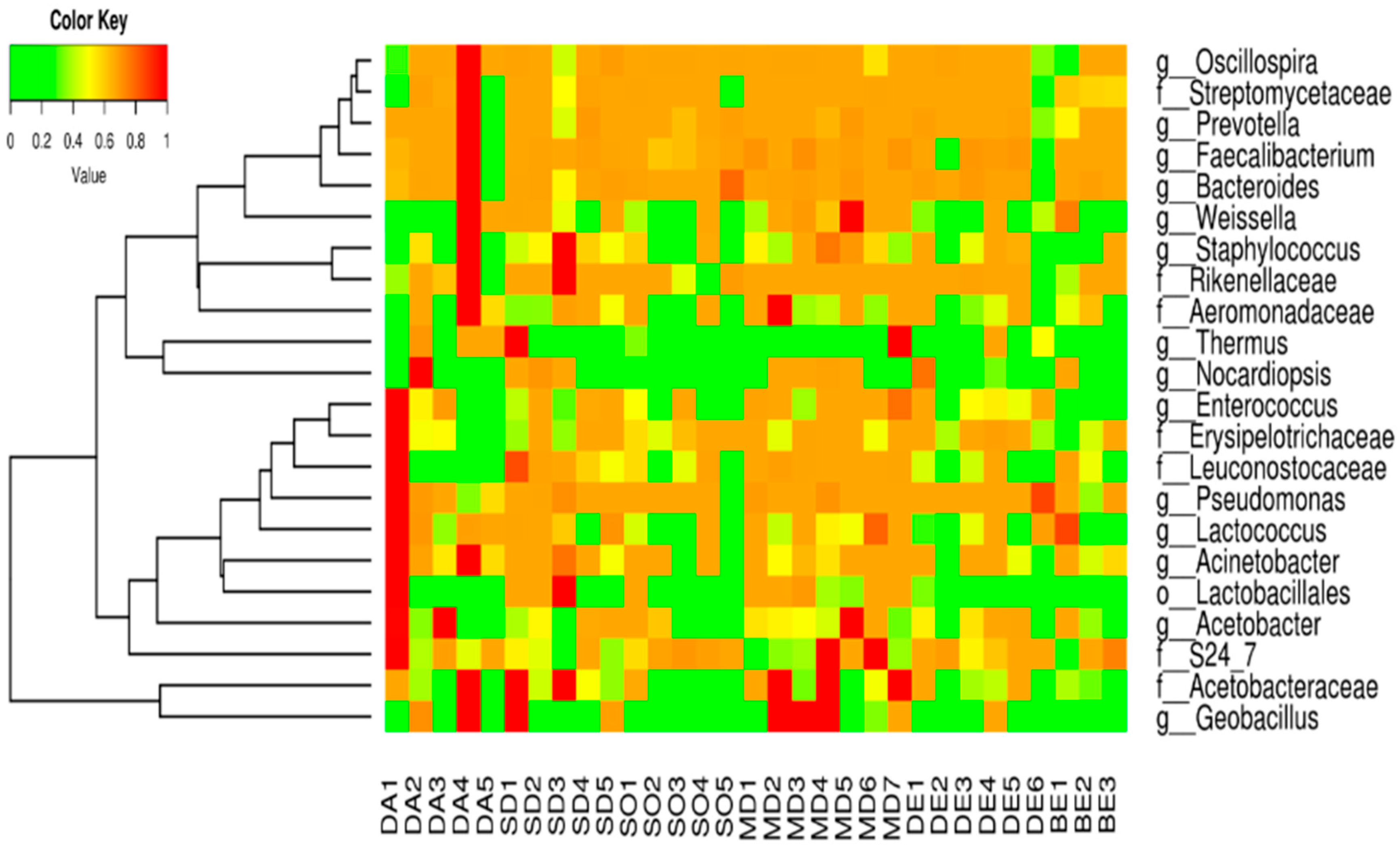
3.2. Abundance of Orders, Families, and Genera in the Ready-to-Eat Food
4. Discussion
5. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Hauptmann, A.L.; Paulová, P.; Hansen, L.H.; Sicheritz-Pontén, T.; Mulvad, G.; Nielsen, D.S. Microbiota in foods from Inuit traditional hunting. PLoS ONE 2020, 15, e0227819. [Google Scholar] [CrossRef] [PubMed]
- Suárez-Machín, C.; Garrido-Carralejo, N.A. Levadura Saccharomyces cerevisiae y la producción de alcohol. Revisión bibliográfica. ICIDCA. Sobre Los Derivados de La Caña de Azúcar 2016, 50, 20–28. [Google Scholar]
- Bayona, M.A. Microbiological evaluation of food acquired in streets of a northern area of Bogotá. Revista UDCA Actualidad y Divulgación Cientifica 2009, 12, 9–17. [Google Scholar]
- Murugesan, S.; Reyes-Mata, M.P.; Nirmalkar, K.; Chavez-Carbajal, A.; Juárez-Hernández, J.I.; Torres-Gómez, R.E.; García-Mena, J. Profiling of bacterial and fungal communities of Mexican cheeses by high throughput DNA sequencing. Food Res. Int. 2018, 113, 371–381. [Google Scholar] [CrossRef] [PubMed]
- Stavropoulou, E.; Bezirtzoglou, E. Predictive Modeling of Microbial Behavior in Food. Foods 2019, 8, 654. [Google Scholar] [CrossRef] [PubMed]
- Corona-Cervantes, K.; García-González, I.; Villalobos-Flores, L.E.; Hernández-Quiroz, F.; Piña-Escobedo, A.; Hoyo-Vadillo, C.; Rangel-Calvillo, M.N.; García-Mena, J. Human milk microbiota associated with early colonization of the neonatal gut in Mexican newborns. PeerJ 2020, 8, e9205. [Google Scholar] [CrossRef] [PubMed]
- Caporaso, J.G.; Kuczynski, J.; Stombaugh, J.; Bittinger, K.; Bushman, F.D.; Costello, E.K.; Fierer, N.; Peña, A.G.; Goodrich, J.K.; Gordon, J.I.; et al. QIIME allows analysis of high-throughput community sequencing data. Nat. Methods 2010, 7, 335–336. [Google Scholar] [CrossRef] [PubMed]
- McMurdie, P.J.; Holmes, S. phyloseq: An R package for reproducible interactive analysis and graphics of microbiome census data. PloS ONE 2013, 8, e61217. [Google Scholar] [CrossRef] [PubMed]
- Hernández-Quiroz, F.; Nirmalkar, K.; Villalobos-Flores, L.E.; Murugesan, S.; Cruz-Narváez, Y.; Rico-Arzate, E.; García-Mena, J. Influence of moderate beer consumption on human gut microbiota and its impact on fasting glucose and β-cell function. Alcohol 2019, 85, 77–94. [Google Scholar] [CrossRef] [PubMed]
| ID | Local Mexican Spanish Name | English Name | Description |
|---|---|---|---|
| DA1 | Leche-Búlgara | Kefir | Fermented dairy product. |
| DA2 | Leche-Liconsa | Milk | Synthetic milk. |
| DA3 | Leche-Saborizada | Flavored milk | Strawberry flavored milk. |
| DA4 | Yogurt-bebible | Drinkable yogurt | Yogurt with a liquid consistency. |
| DA5 | Yogurt-Griego | Greek style yogurt | Yogurt with a higher amount of fat than normal. |
| SD1 | Ensalada-de-pollo | Chicken salad | Vegetable salad that includes chicken for protein. |
| SD2 | Ensalada-de-jamón | Ham salad | Vegetable salad that includes ham as a source of protein. |
| SD3 | Ensalada-del-chef | Chef’s salad | Vegetable salad with chicken, cubed cheese, and boiled egg. |
| SD4 | Ensalada-de-frutas | Salad with fruits | Vegetable salad that includes fruits within its ingredients. |
| SD5 | Ensalada-de-broccoli | Broccoli salad | Steamed broccoli with fresh vegetables. |
| SO1 | Sopa-de-papa | Potato soup | Tomato broth with diced potato chunks. |
| SO2 | Arroz-a-la-Mexicana | Mexican rice | Rice with tomato, carrot, and pea as main ingredients. |
| SO3 | Crema-de-poblano | Poblano cream | Crepe prepared with poblano pepper. |
| SO4 | Sopa-de-verduras | Vegetable soup | Tomato broth with diced vegetables. |
| SO5 | Arroz-chino | Chinese-style rice | Rice prepared as a typical oriental recipe. |
| MD1 | Alambre | Beef kabob cooked on a grill. | Cubes of broiled beef, bell pepper, onion, bacon, and melted cheese. |
| MD2 | Taco-de-canasta con chicharrón | Soft taco with pork rind. | Steamed soft corn tortillas stuffed with fried pork rind. |
| MD3 | Huevo-con-chorizo | Egg with chorizo | Fried egg accompanied by Spanish sausage. |
| MD4 | Taco-de-guisado | Stew taco | Taco with Mexican style-rice with sausages with tomato. |
| MD5 | Torta-de-jamón | Mexican ham torta | Mexican bolillo (crusty roll) filled with mayonnaise, avocado, ham, and basket cheese. |
| MD6 | Enfrijoladas | Mexican enfrijoladas | Fried tortillas, dipped in a slurry of refried beans, and stuffed. |
| MD7 | Torta-de-pierna | Mexican pork leg torta | Mexican bolillo (crusty roll) filed with mayonnaise, beans, avocado, tomato, smoked pork leg. |
| DE1 | Churros-azucarados | Sugary churros | Fried-dough pastry in oil, with granulated sugar. |
| DE2 | Pay-de-Piña | Pineapple Pie | Pie filled with pineapple jam. |
| DE3 | Frituras de maíz con limón y sal | Corn chips with lemon and salt | Deep fried corn flour chips with lemon and salt. |
| DE4 | Dona glaseada | Glassed donut | Dough fried in oil and glazed with sucrose after cooking. |
| DE5 | Gorditas-de-la-Villa | Villas’s soft cookies | Cornmeal (cacahuazintle), sucrose, baking soda, eggs, vanilla essence, water, and vegetable shortening. |
| DE6 | Chocoflan | Chocolate bread-flan | Chocolate flavor bread with a layer of sweetened egg custard with caramel topping. |
| BE1 | Jugo de naranja | Orange juice | Juice obtained from orange fruit. |
| BE2 | Agua de melón | Melon water | Water accompanied by blended melon and sugar to sweeten. |
| BE3 | Jugo verde | Green juice | Juice obtained by blending spinach, pineapple, orange juice and celery. |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Flores-Rivas, C.; Hernández-Quiroz, F.; Villalobos-Flores, L.E.; Piña-Escobedo, A.; Chavez-Carbajal, A.; Nirmalkar, K.; García-Mena, J. Characterization of the Food Microbiota in Ready-to-Eat Mexican Foods. Proceedings 2020, 66, 32. https://doi.org/10.3390/proceedings2020066032
Flores-Rivas C, Hernández-Quiroz F, Villalobos-Flores LE, Piña-Escobedo A, Chavez-Carbajal A, Nirmalkar K, García-Mena J. Characterization of the Food Microbiota in Ready-to-Eat Mexican Foods. Proceedings. 2020; 66(1):32. https://doi.org/10.3390/proceedings2020066032
Chicago/Turabian StyleFlores-Rivas, Cintia, Fernando Hernández-Quiroz, Loan Edel Villalobos-Flores, Alberto Piña-Escobedo, Alejandra Chavez-Carbajal, Khemlal Nirmalkar, and Jaime García-Mena. 2020. "Characterization of the Food Microbiota in Ready-to-Eat Mexican Foods" Proceedings 66, no. 1: 32. https://doi.org/10.3390/proceedings2020066032
APA StyleFlores-Rivas, C., Hernández-Quiroz, F., Villalobos-Flores, L. E., Piña-Escobedo, A., Chavez-Carbajal, A., Nirmalkar, K., & García-Mena, J. (2020). Characterization of the Food Microbiota in Ready-to-Eat Mexican Foods. Proceedings, 66(1), 32. https://doi.org/10.3390/proceedings2020066032






